Identification and Characterization of Erwinia Phage IT22: A New Bacteriophage-Based Biocontrol against Erwinia amylovora
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Culture Conditions
2.2. Bacteriophage Isolation, Purification, and Titration
2.3. DNA Extraction, Whole Genome Sequencing (WGS), and Bioinformatic Analysis
2.4. PCR Assays
2.5. Host Range Determination
2.6. Microscopy
2.6.1. Transmission Electron Microscopy (TEM)
2.6.2. Fluorescence Microscopy (FM)
2.7. Turbidity Assay
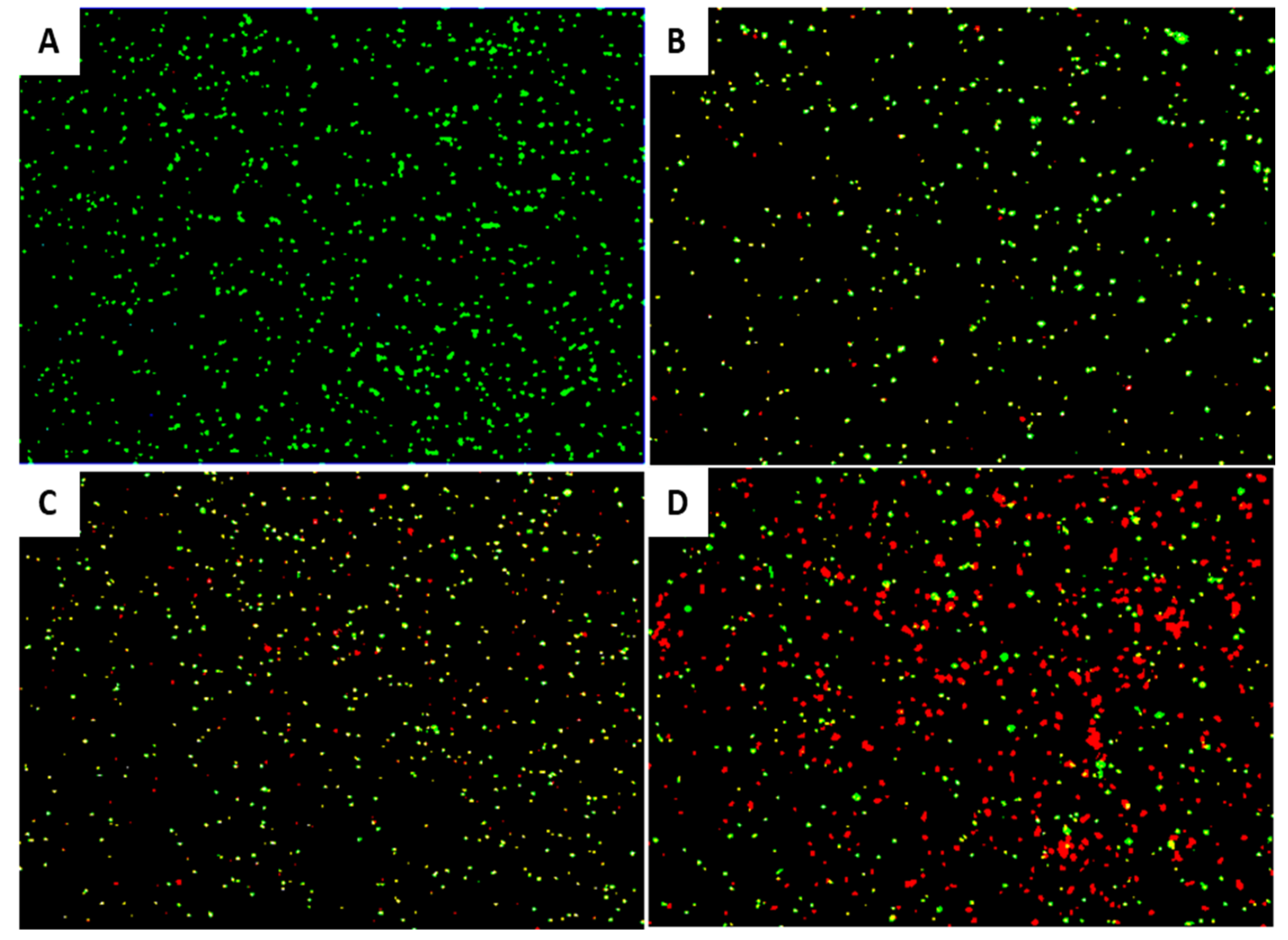
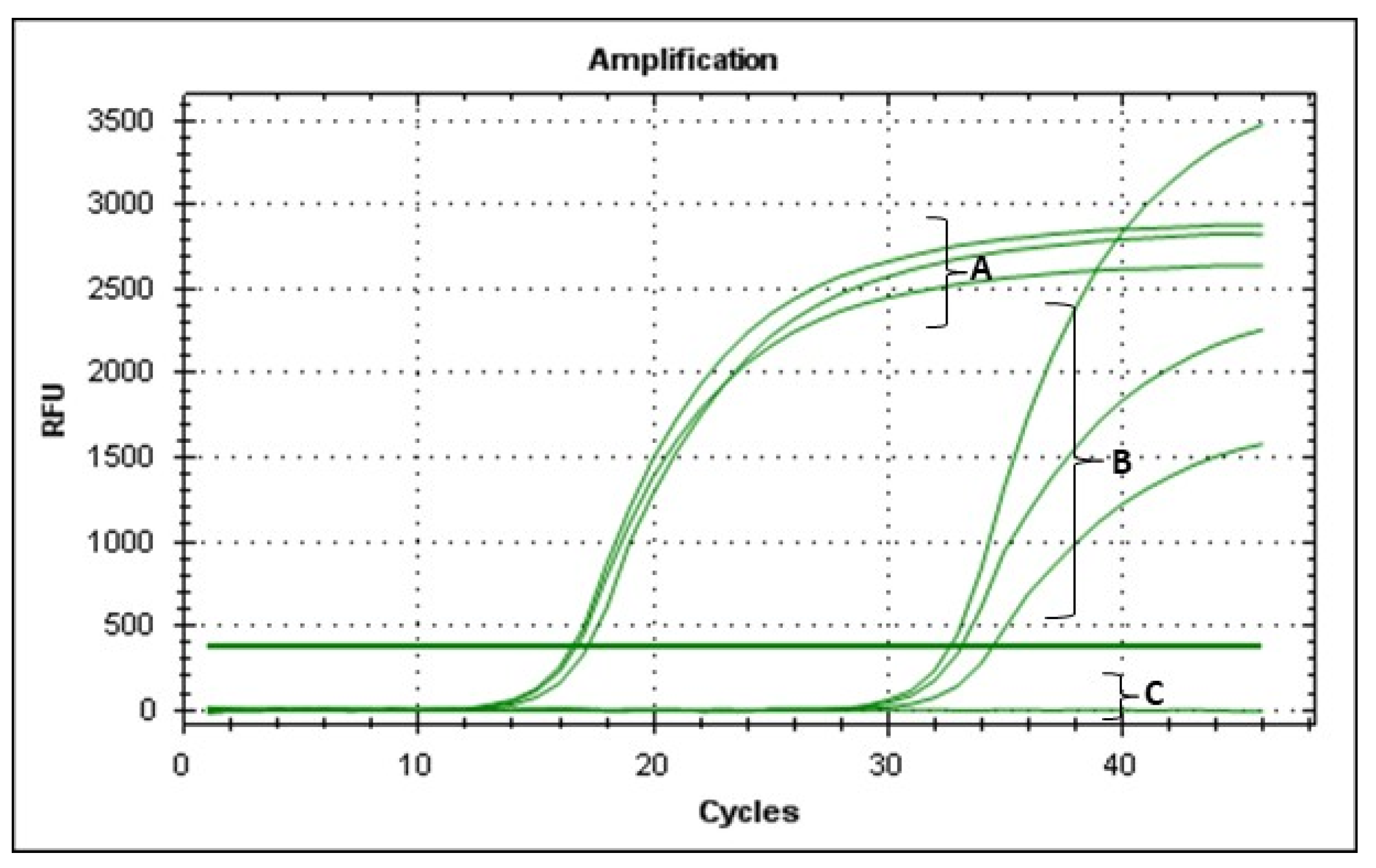
2.8. Viable-Quantitative PCR (v-qPCR)
2.9. Resistance to Environmental Stressors
2.10. Efficacy of EP-IT22 against E. amylovora on Pear Plants
3. Results and Discussion
3.1. Phage Isolation and Host Range Analysis
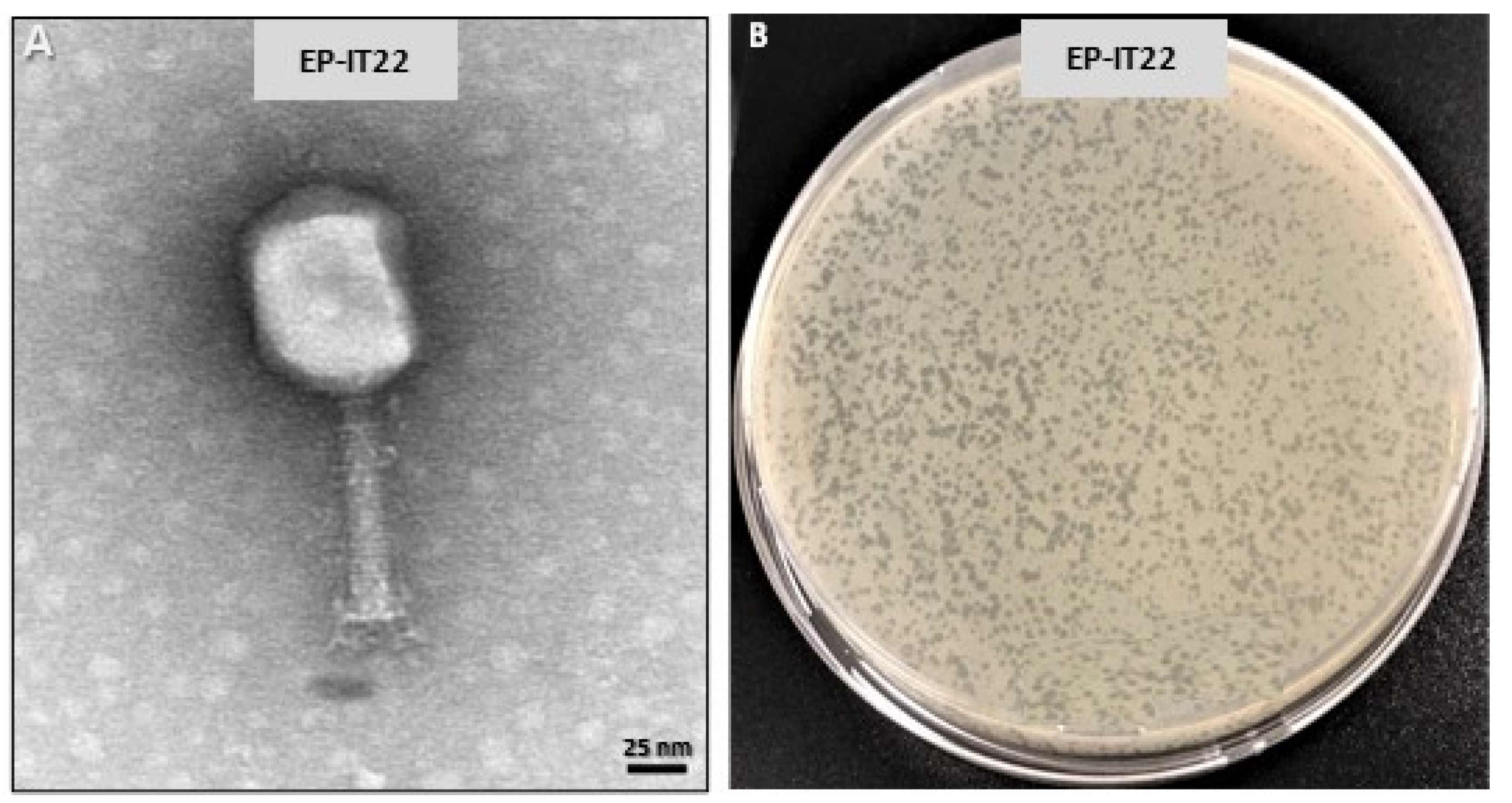
3.2. Biological Analysis of EP-IT22
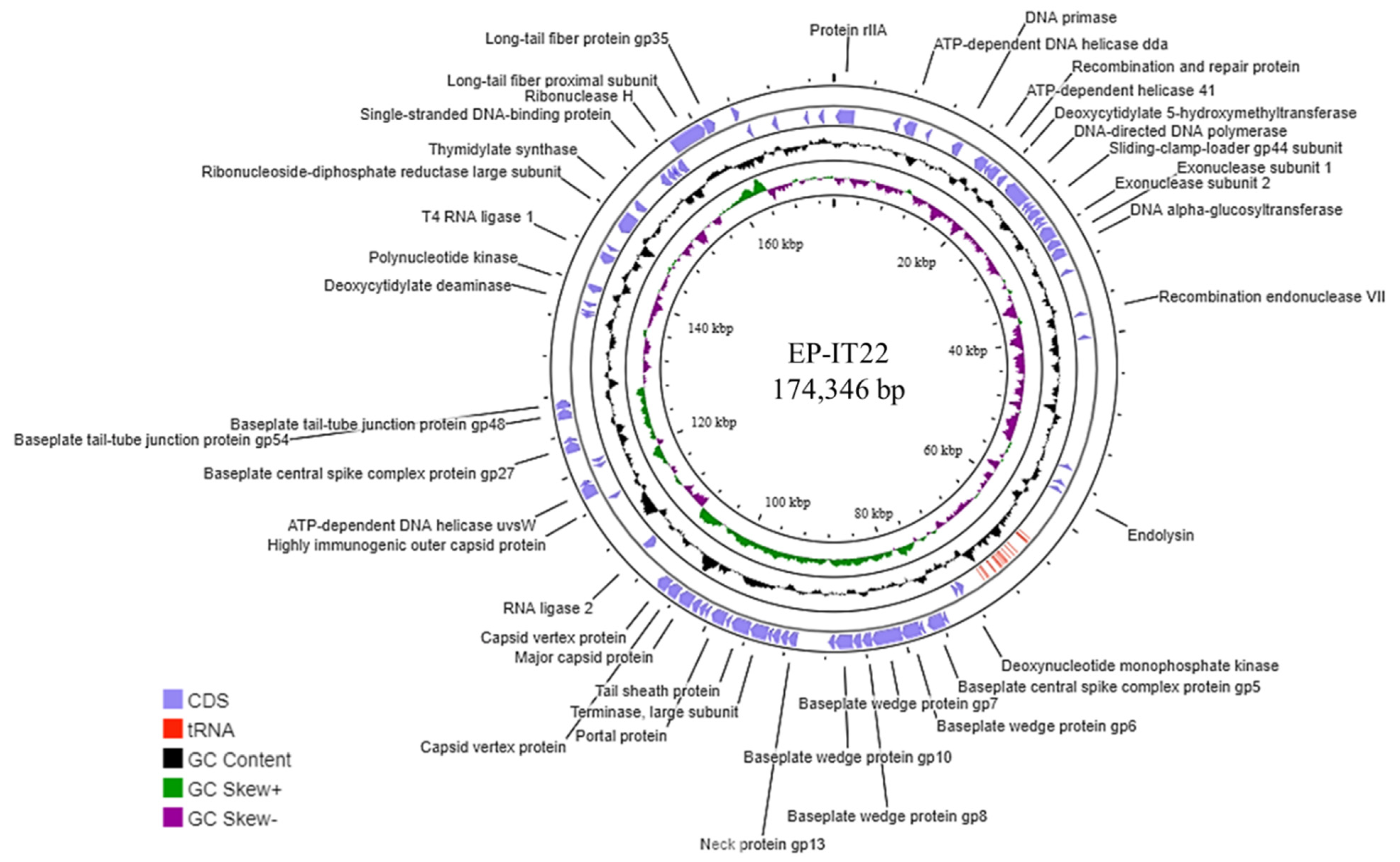
3.3. EP-IT22 Genomic Analysis
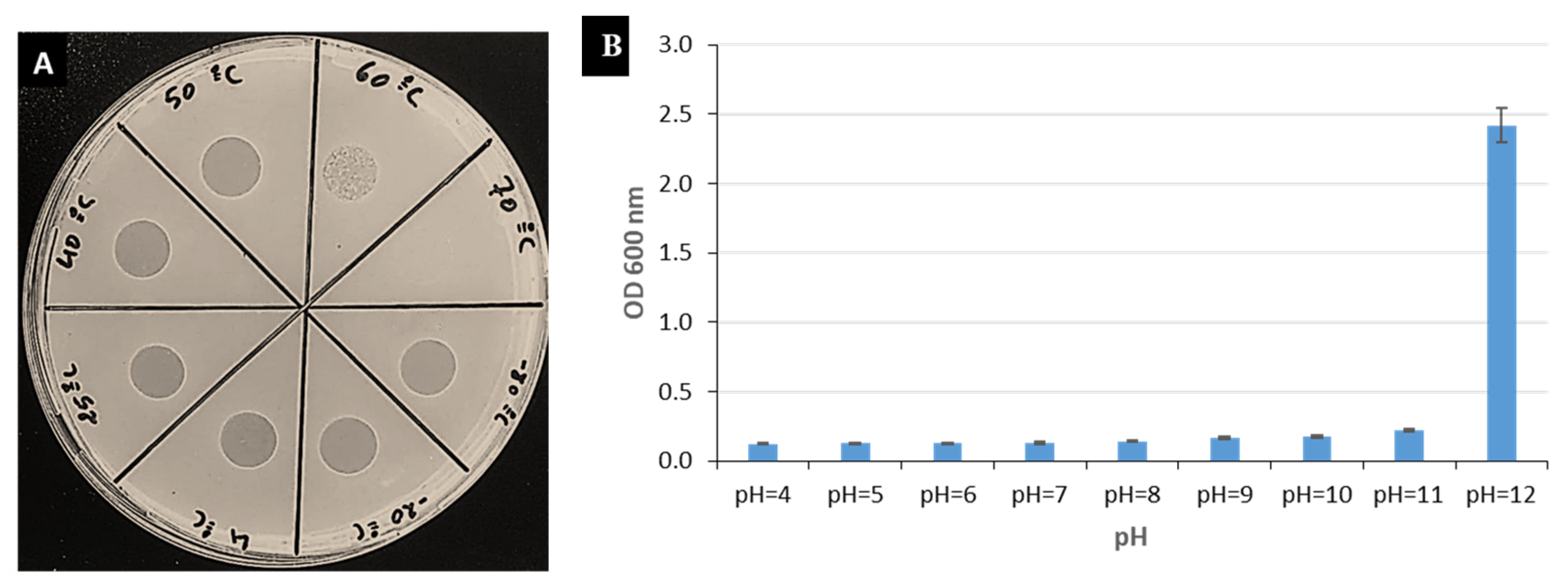
3.4. Resistance to Environmental Stresses
3.5. Growth Kinetics of EP-IT22
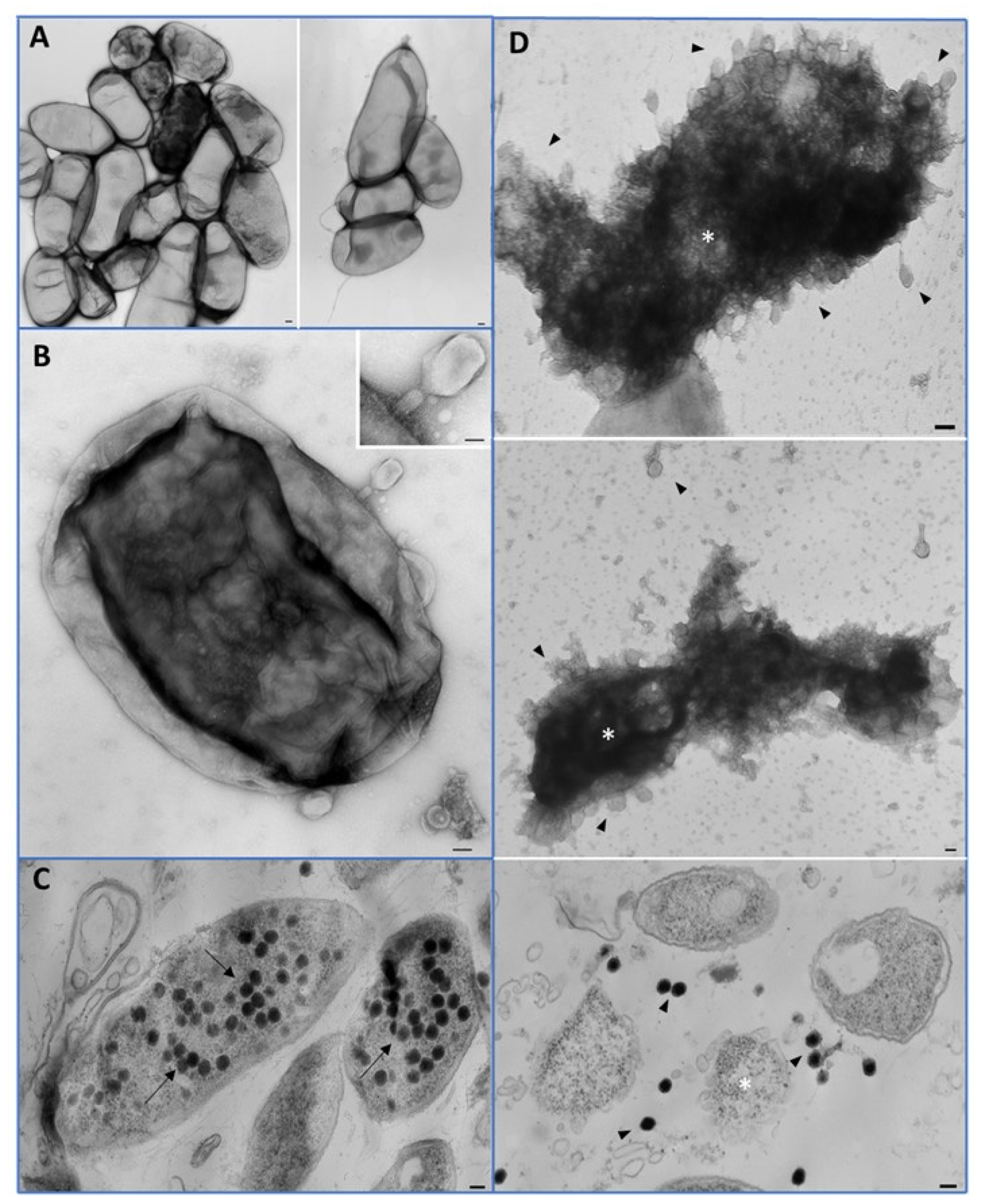
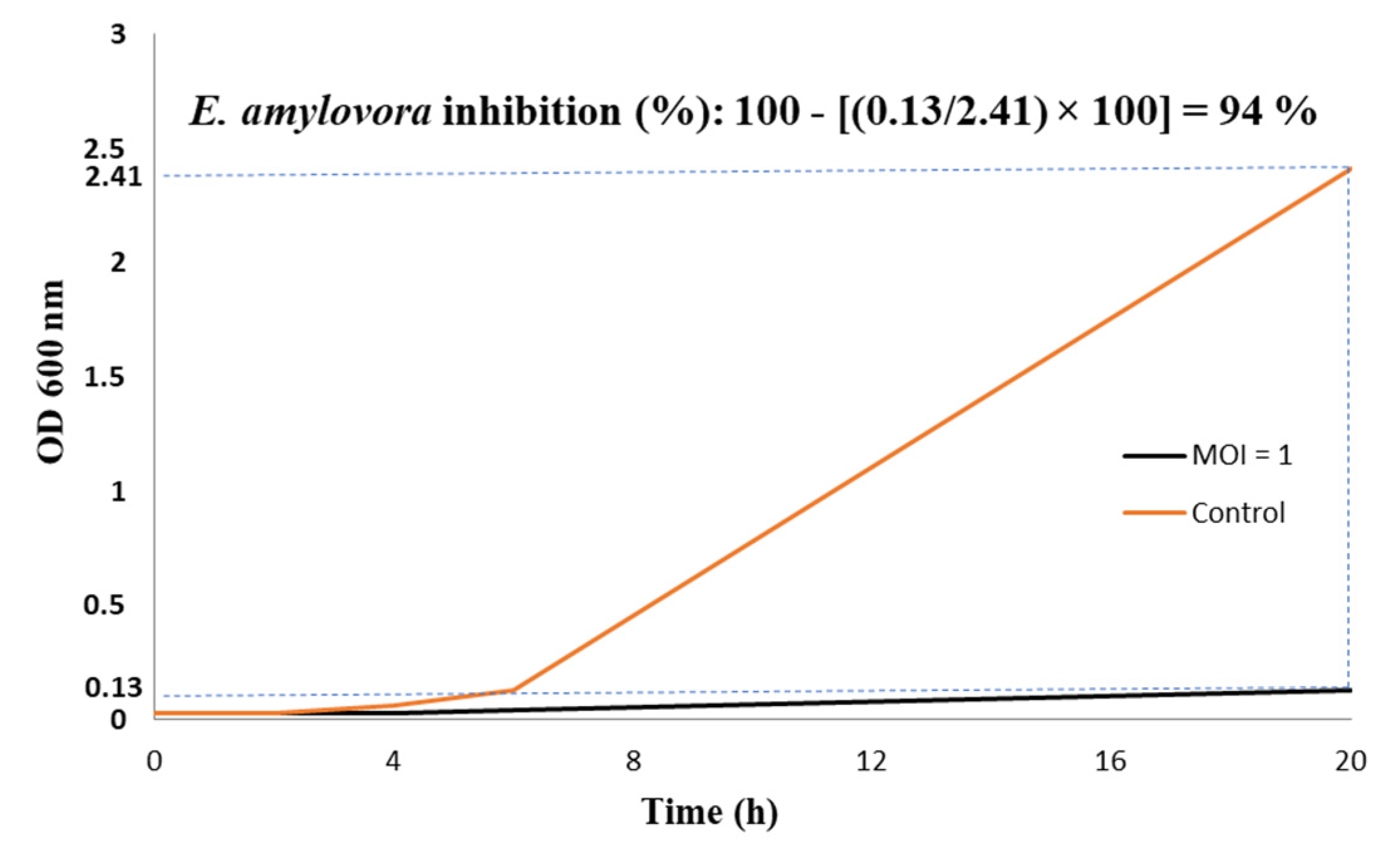
3.6. In Vitro EP-IT22 Bacteriolytic Activity against E. amylovora
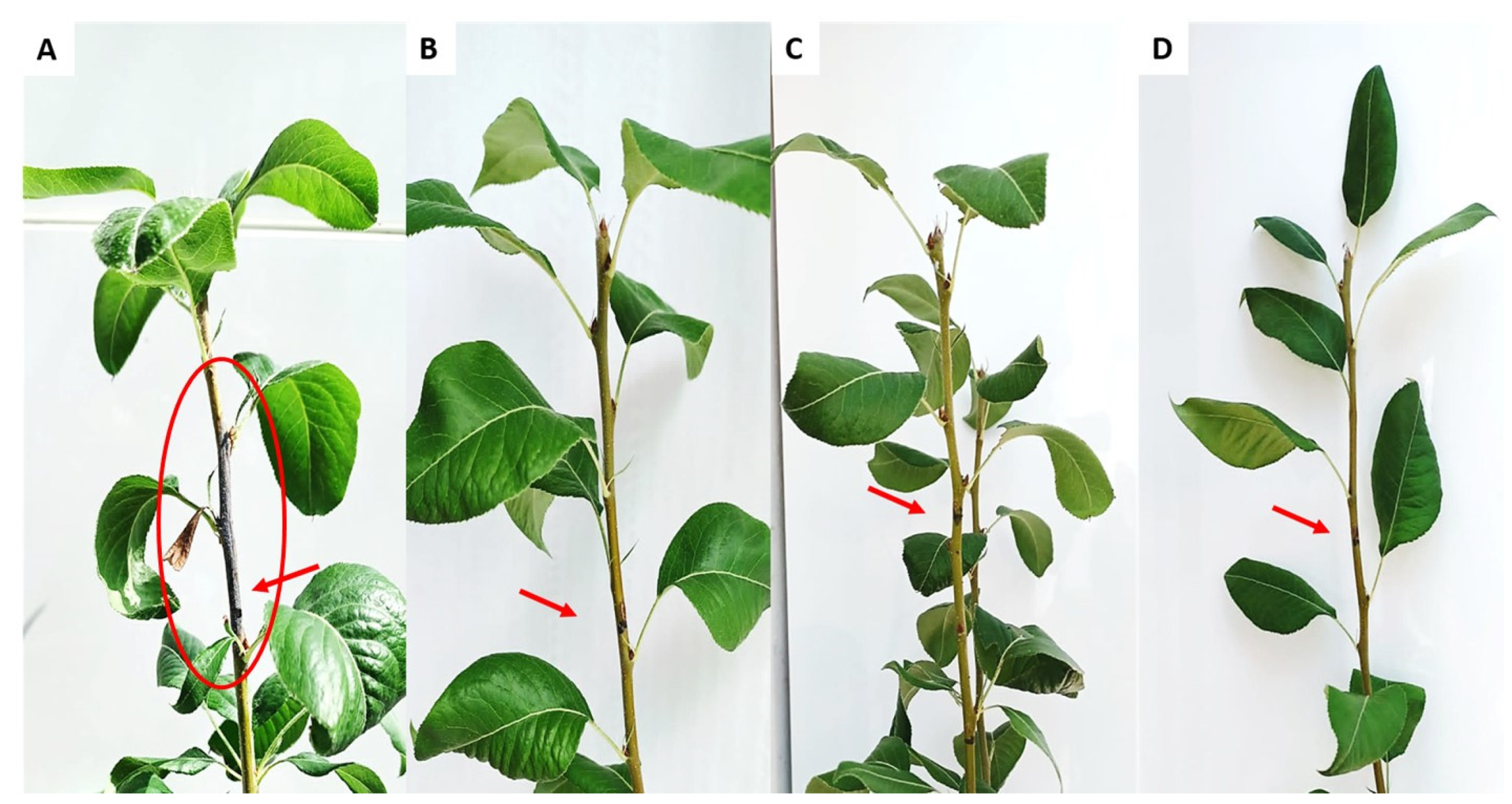
3.7. In Planta Biocontrol Assay
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Dagher, F.; Olishevska, S.; Philion, V.; Zheng, J.; Déziel, E. Development of a Novel Biological Control Agent Targeting the Phytopathogen Erwinia Amylovora. Heliyon 2020, 6, e05222. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Tian, Y.; Wang, L.; Geng, G.; Zhao, W.; Hu, B.; Zhao, Y. Fire Blight Disease, a Fast-Approaching Threat to Apple and Pear Production in China. J. Integr. Agric. 2019, 18, 815–820. [Google Scholar] [CrossRef]
- Mechan Llontop, M.E.; Hurley, K.; Tian, L.; Bernal Galeano, V.A.; Wildschutte, H.K.; Marine, S.C.; Yoder, K.S.; Vinatzer, B.A. Exploring Rain as Source of Biological Control Agents for Fire Blight on Apple. Front. Microbiol. 2020, 11, 199. [Google Scholar] [CrossRef] [PubMed]
- Piqué, N.; Miñana-Galbis, D.; Merino, S.; Tomás, J.M. Virulence Factors of Erwinia Amylovora: A Review. Int. J. Mol. Sci. 2015, 16, 12836–12854. [Google Scholar] [CrossRef]
- Skoneczny, H.; Kubiak, K.; Spiralski, M.; Kotlarz, J.; Mikiciński, A.; Puławska, J. Fire Blight Disease Detection for Apple Trees: Hyperspectral Analysis of Healthy, Infected and Dry Leaves. Remote Sens. 2020, 12, 2101. [Google Scholar] [CrossRef]
- Norelli, J.L.; Jones, A.L.; Aldwinckle, H.S. Fire Blight Management in the Twenty-First Century: Using New Technologies That Enhance Host Resistance in Apple. Plant Dis. 2003, 87, 756–765. [Google Scholar] [CrossRef]
- Cvjetkovic, B.; Halupecki, E.; Å poljaric, J. The occurrence and control of fire blight in croatia. Acta Hortic. 1999, 489, 71–78. [Google Scholar] [CrossRef]
- Zurn, J.D.; Norelli, J.L.; Montanari, S.; Bell, R.; Bassil, N.V. Dissecting Genetic Resistance to Fire Blight in Three Pear Populations. Phytopathology 2020, 110, 1305–1311. [Google Scholar] [CrossRef]
- Escursell, M.M.; Roschi, A.; Smits, T.H.M.; Rezzonico, F. Characterization and Direct Molecular Discrimination of RpsL Mutations Leading to High Streptomycin Resistance in Erwinia Amylovora. J Plant Pathol 2021, 103, 99–108. [Google Scholar] [CrossRef]
- Evrenosoğlu, Y.; Mertoğlu, K.; Acarsoy Bilgin, N.; Misirli, A.; Altay, Y. An Analysis on Some Reciprocal Pear Hybridization Combinations in Terms of Transferring Resistance to Fire Blight. Erwerbs-Obstbau 2020, 62, 189–194. [Google Scholar] [CrossRef]
- Paraschivu, M.; Ciobanu, A.; Cotuna, O.; Paraschivu, M. Assessment of the Bacterium Erwinia Amylovora Attack on Several Pear Varieties (Pyrus communis L.) and the Influence on Fruits Sugar Content. Agricultural Sciences & Veterinary Medicine University, Bucharest. Scientific Papers. Series B. Horticulture 2020, 64, 163–168. [Google Scholar]
- Garcia, P.; Martinez, B.; Obeso, J.M.; Rodriguez, A. Bacteriophages and Their Application in Food Safety. Lett. Appl. Microbiol. 2008, 47, 479–485. [Google Scholar] [CrossRef] [PubMed]
- Sabri, M.; Benkirane, R.; Habbadi, K.; Sadik, S.; Ou-Zine, M.; Diouri, M.; Achbani, E.H. Phages as a Potential Biocontrol of Phytobacteria. Arch. Phytopathol. Plant Prot. 2021, 54, 1277–1291. [Google Scholar] [CrossRef]
- Korniienko, N.; Kharina, A.; Budzanivska, I.; Burketová, L.; Kalachova, T. Phages of Phytopathogenic Bacteria: High Potential, but Challenging Application. Plant Prot. Sci. 2022, 58, 81–91. [Google Scholar] [CrossRef]
- Goodridge, L.D.; Abedon, S.T. Bacteriophage Biocontrol: The Technology Matures. Microbiol. Aust. 2008, 29, 48–49. [Google Scholar] [CrossRef]
- Połaska, M.; Sokołowska, B. Bacteriophages—A New Hope or a Huge Problem in the Food Industry. AIMS Microbiol. 2019, 5, 324. [Google Scholar] [CrossRef]
- Carstens, A.B.; Djurhuus, A.M.; Kot, W.; Hansen, L.H. A Novel Six-Phage Cocktail Reduces Pectobacterium Atrosepticum Soft Rot Infection in Potato Tubers under Simulated Storage Conditions. FEMS Microbiol. Lett. 2019, 366, fnz101. [Google Scholar] [CrossRef]
- Kropinski, A.M.; Mazzocco, A.; Waddell, T.E.; Lingohr, E.; Johnson, R.P. Enumeration of Bacteriophages by Double Agar Overlay Plaque Assay. In Bacteriophages; Springer: Berlin/Heidelberg, Germany, 2009; pp. 69–76. [Google Scholar]
- Stocks, S.M. Mechanism and Use of the Commercially Available Viability Stain, BacLight. Cytom. Part A 2004, 61A, 189–195. [Google Scholar] [CrossRef]
- Elbeaino, T.; Incerti, O.; Dakroub, H.; Valentini, F.; Huang, Q. Development of an FTP-LAMP Assay Based on TaqMan Real-Time PCR and LAMP for the Specific Detection of Xylella Fastidiosa De Donno and Mulberry Strains in Both Plants and Insect Vectors. J. Microbiol. Methods 2020, 175, 105992. [Google Scholar] [CrossRef]
- Laforest, M.; Bisaillon, K.; Ciotola, M.; Cadieux, M.; Hébert, P.-O.; Toussaint, V.; Svircev, A.M. Rapid Identification of Erwinia Amylovora and Pseudomonas Syringae Species and Characterization of E. Amylovora Streptomycin Resistance Using Quantitative PCR Assays. Can. J. Microbiol. 2019, 65, 496–509. [Google Scholar] [CrossRef]
- Ackermann, H.-W. Phage Classification and Characterization. In Bacteriophages; Springer: Berlin/Heidelberg, Germany, 2009; pp. 127–140. [Google Scholar]
- Abedon, S.T.; Yin, J. Bacteriophage Plaques: Theory and Analysis. In Bacteriophages; Springer: Berlin/Heidelberg, Germany, 2009; pp. 161–174. [Google Scholar]
- Lucks, J.B.; Nelson, D.R.; Kudla, G.R.; Plotkin, J.B. Genome Landscapes and Bacteriophage Codon Usage. PLoS Comput. Biol. 2008, 4, e1000001. [Google Scholar] [CrossRef] [PubMed]
| Species | Strains | Hosts | Origins | Isolation | Lytic Activity of IT22 |
|---|---|---|---|---|---|
| Erwinia amylovora | PGL Z1 * | Pyrus communis | Apulia/Italy | 2013 | ++ |
| Erwinia amylovora | PGL 102 * | Pyrus communis | Apulia/Italy | 2013 | ++ |
| Erwinia amylovora | OMP-BO 25.1 | Pyrus communis | Emilia-Romagna/Italy | 2000 | + |
| Erwinia amylovora | OMP-BO 8630.1 | Pyrus communis | Emilia-Romagna/Italy | 2010 | ++ |
| Erwinia amylovora | OMP-BO 1077.7 | Pyrus communis | Emilia-Romagna/Italy | 1994 | ++ |
| Erwinia amylovora | OMP-BO 347.1 | Crataegus monogyna | Emilia-Romagna/Italy | 2006 | ++ |
| Erwinia amylovora | OMP-BO 329.1 | Malus domestica | Emilia-Romagna/Italy | 2002 | ++ |
| Xanthomonas campestris pv. campestris | CFBP 1710 | Brassica oleracea var. botrytis | France | 1975 | − |
| Pseudomonas syringae pv. syringae | CFBP 311 | Pyrus communis | Indre et Loire-France | 1962 | − |
| Dickeya chrysanthemi | CFBP 1346 | Chrysanthemum maximum | Italy | 1969 | − |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sabri, M.; El Handi, K.; Valentini, F.; De Stradis, A.; Achbani, E.H.; Benkirane, R.; Resch, G.; Elbeaino, T. Identification and Characterization of Erwinia Phage IT22: A New Bacteriophage-Based Biocontrol against Erwinia amylovora. Viruses 2022, 14, 2455. https://doi.org/10.3390/v14112455
Sabri M, El Handi K, Valentini F, De Stradis A, Achbani EH, Benkirane R, Resch G, Elbeaino T. Identification and Characterization of Erwinia Phage IT22: A New Bacteriophage-Based Biocontrol against Erwinia amylovora. Viruses. 2022; 14(11):2455. https://doi.org/10.3390/v14112455
Chicago/Turabian StyleSabri, Miloud, Kaoutar El Handi, Franco Valentini, Angelo De Stradis, El Hassan Achbani, Rachid Benkirane, Grégory Resch, and Toufic Elbeaino. 2022. "Identification and Characterization of Erwinia Phage IT22: A New Bacteriophage-Based Biocontrol against Erwinia amylovora" Viruses 14, no. 11: 2455. https://doi.org/10.3390/v14112455
APA StyleSabri, M., El Handi, K., Valentini, F., De Stradis, A., Achbani, E. H., Benkirane, R., Resch, G., & Elbeaino, T. (2022). Identification and Characterization of Erwinia Phage IT22: A New Bacteriophage-Based Biocontrol against Erwinia amylovora. Viruses, 14(11), 2455. https://doi.org/10.3390/v14112455









