Mechanistic Contributions of lncRNAs to Cellular Signaling Pathways Crucial to the Lifecycle of Human Papillomaviruses
Abstract
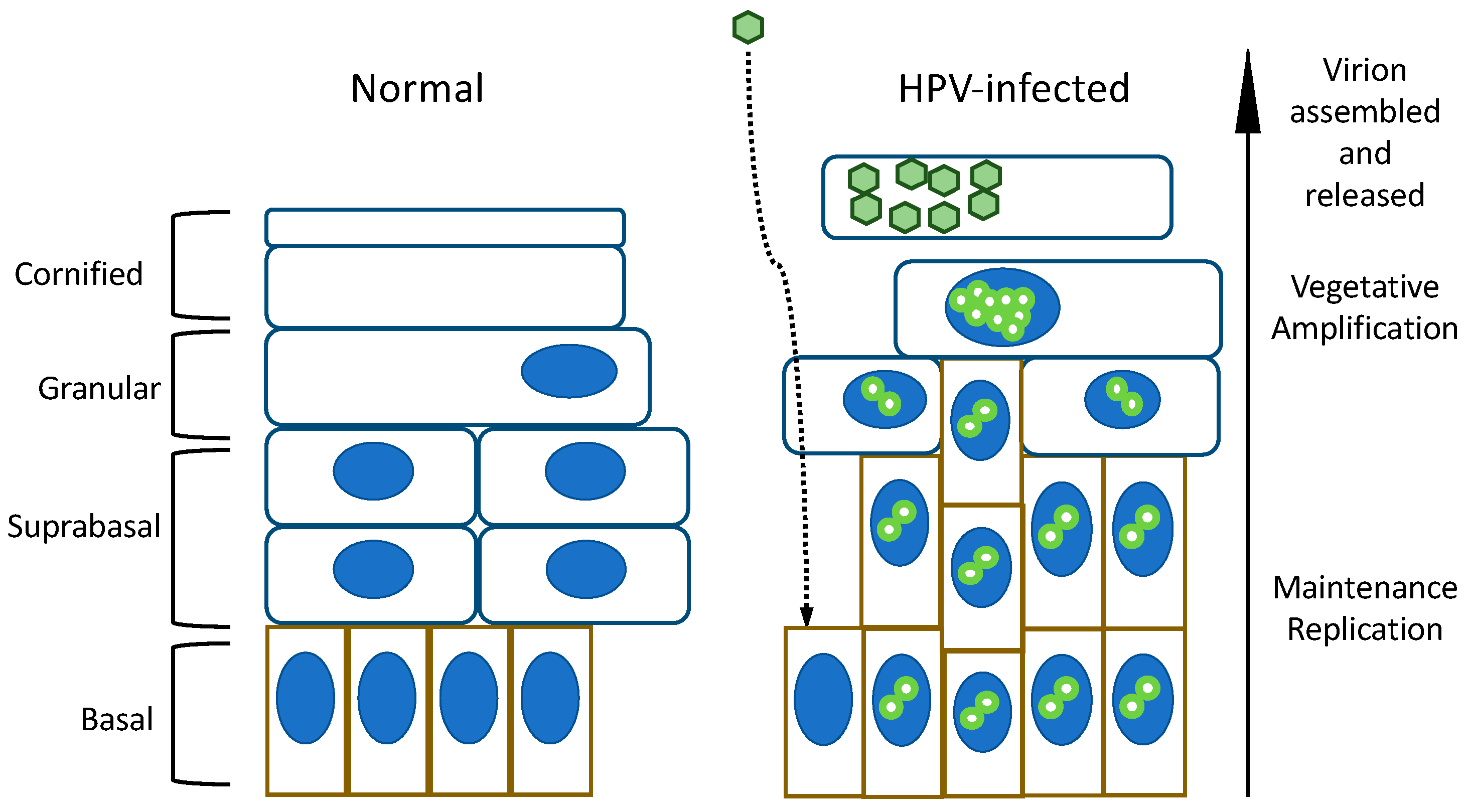
1. Introduction
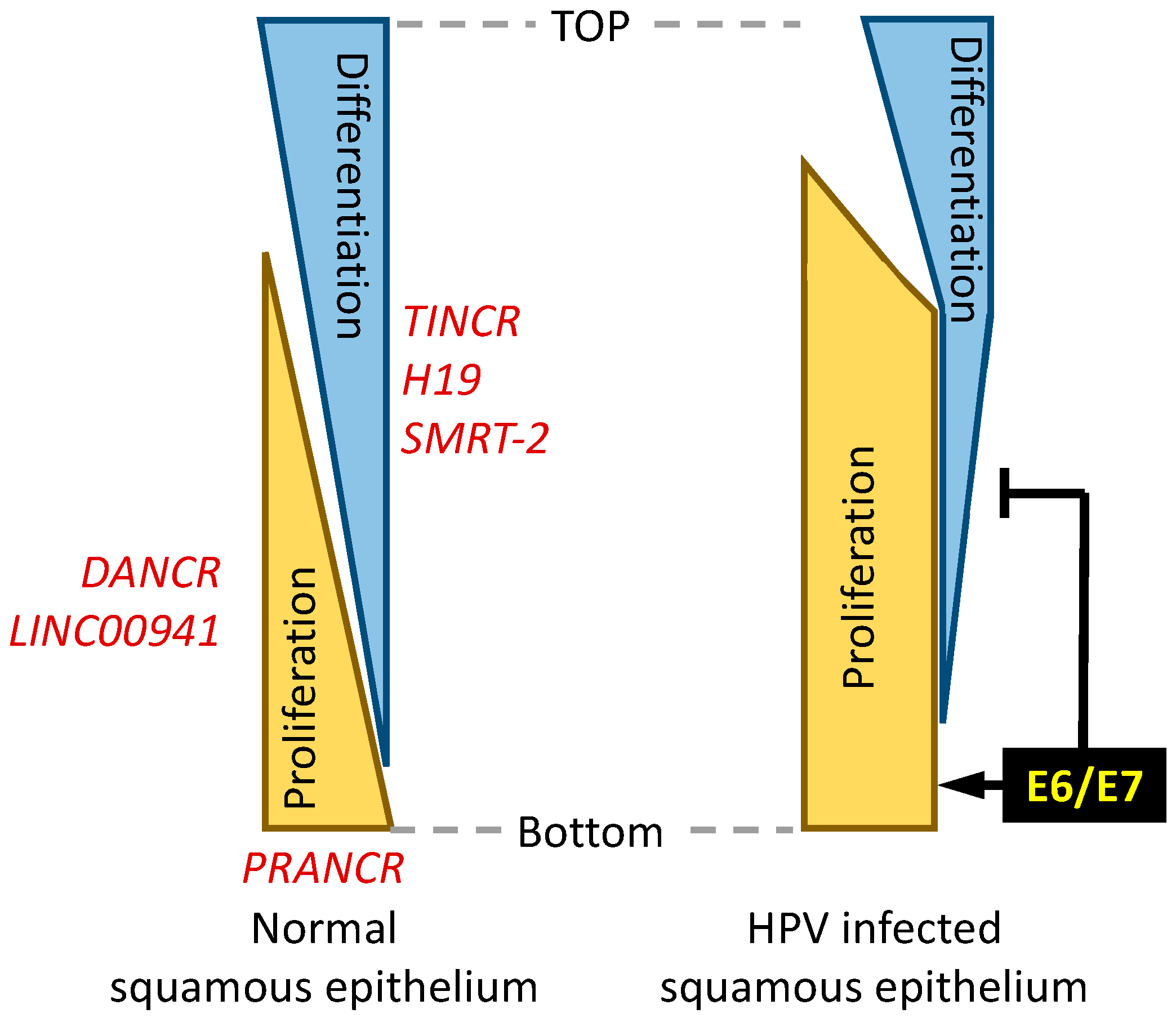
2. Regulation of Epithelial Cell Differentiation
2.1. TINCR and DANCR
2.2. Additional lncRNAs That Have Been Implicated in Modulating Differentiation
3. LncRNAs That Affect WNT/β-Catenin Signaling
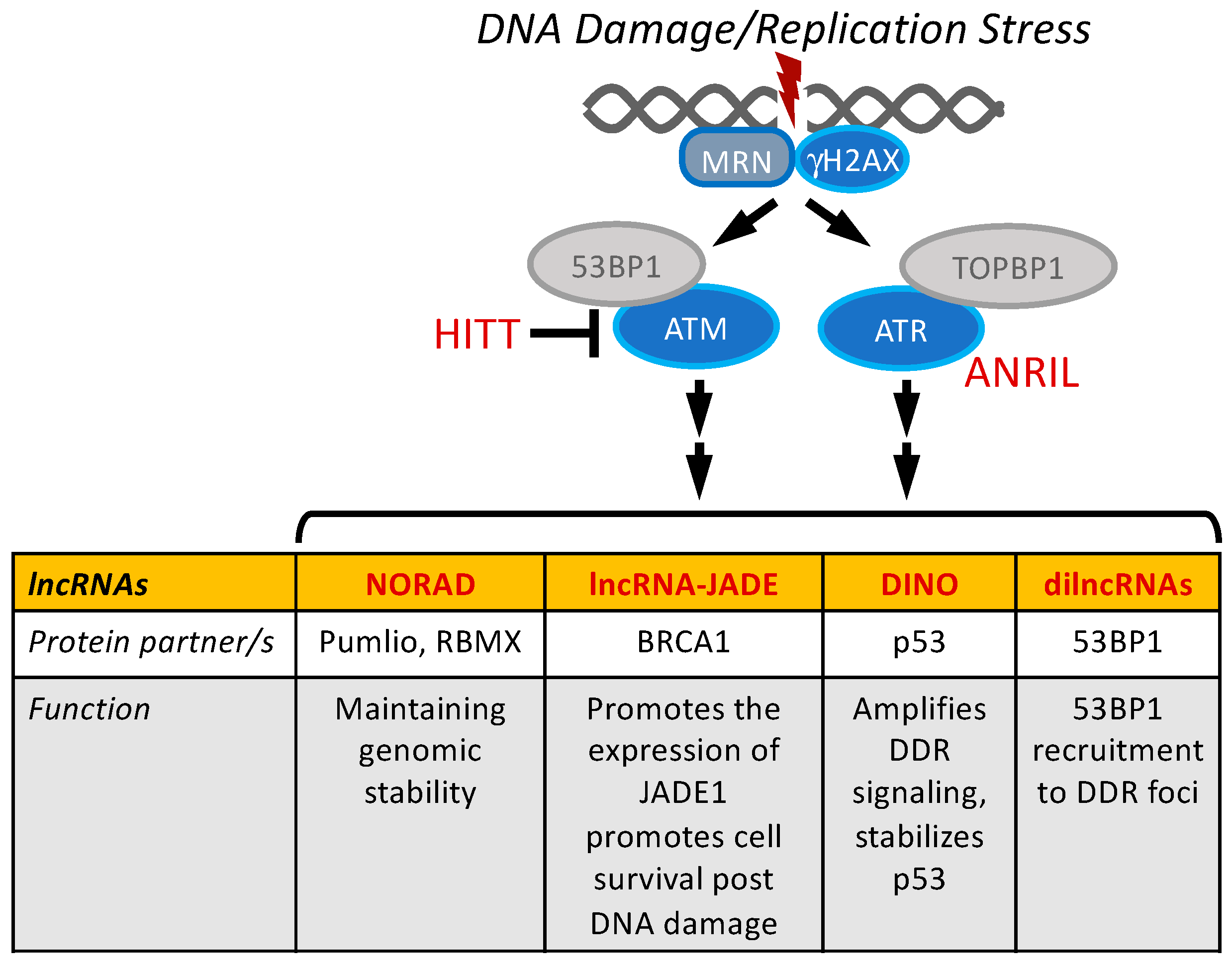
4. Modulation of DDR Signaling by lncRNAs
5. LncRNAs That Are Connected to p53 Activity
6. Conclusions
Funding
Conflicts of Interest
References
- Schiffman, M.; Castle, P.E.; Jeronimo, J.; Rodriguez, A.C.; Wacholder, S. Human papillomavirus and cervical cancer. Lancet 2007, 370, 890–907. [Google Scholar] [CrossRef]
- Doorbar, J.; Quint, W.; Banks, L.; Bravo, I.G.; Stoler, M.; Broker, T.R.; Stanley, M.A. The Biology and Life-Cycle of Human Papillomaviruses. Vaccine 2012, 30, F55–F70. [Google Scholar] [CrossRef] [PubMed]
- Coursey, T.L.; McBride, A.A. Hitchhiking of Viral Genomes on Cellular Chromosomes. Annu. Rev. Virol. 2019, 6, 275–296. [Google Scholar] [CrossRef] [PubMed]
- Moody, C.A.; Laimins, L.A. Human Papillomaviruses Activate the ATM DNA Damage Pathway for Viral Genome Amplification upon Differentiation. PLoS Pathog. 2009, 5, 1000605. [Google Scholar] [CrossRef]
- Gillespie, K.A.; Mehta, K.P.; Laimins, L.A.; Moody, C.A. Human Papillomaviruses Recruit Cellular DNA Repair and Homologous Recombination Factors to Viral Replication Centers. J. Virol. 2012, 86, 9520–9526. [Google Scholar] [CrossRef]
- Sakakibara, N.; Mitra, R.; McBride, A.A. The papillomavirus E1 helicase activates a cellular DNA damage response in viral replication foci. J. Virol. 2011, 85, 8981–8995. [Google Scholar] [CrossRef] [PubMed]
- White, E.A. Manipulation of Epithelial Differentiation by HPV Oncoproteins. Viruses 2019, 11, 369. [Google Scholar] [CrossRef]
- Della Fera, A.N.; Warburton, A.; Coursey, T.L.; Khurana, S.; McBride, A.A. Persistent Human Papillomavirus Infection. Viruses 2021, 13, 321. [Google Scholar] [CrossRef]
- McCance, D.J.; Kopant, R.; Fuchst, E.; Laimins, L.A. Human papillomavirus type 16 alters human epithelial cell differentiation in vitro (keratinocyte/transformation/cervical cancer). Proc. Natl. Acad. Sci. USA 1988, 85, 7169–7173. [Google Scholar] [CrossRef]
- Klymenko, T.; Gu, Q.; Herbert, I.; Stevenson, A.; Iliev, V.; Watkins, G.; Pollock, C.; Bhatia, R.; Cuschieri, K.; Herzyk, P.; et al. RNA-Seq Analysis of Differentiated Keratinocytes Reveals a Massive Response to Late Events during Human Papillomavirus 16 Infection, Including Loss of Epithelial Barrier Function. J. Virol. 2017, 91, 1001–1018. [Google Scholar] [CrossRef]
- Hatterschide, J.; Bohidar, A.E.; Grace, M.; Nulton, T.J.; Kim, H.W.; Windle, B.; Morgan, I.M.; Munger, K.; White, E.A. PTPN14 degradation by high-risk human papillomavirus E7 limits keratinocyte differentiation and contributes to HPV-mediated oncogenesis. Proc. Natl. Acad. Sci. USA 2019, 116, 7033–7042. [Google Scholar] [CrossRef]
- Hatterschide, J.; Castagnino, P.; Kim, H.W.; Sperry, S.M.; Montone, K.T.; Basu, D.; White, E.A. YAP1 activation by human papillomavirus E7 promotes basal cell identity in squamous epithelia. Elife 2022, 11, e75466. [Google Scholar] [CrossRef] [PubMed]
- Duensing, S.; Münger, K. The Human Papillomavirus Type 16 E6 and E7 Oncoproteins Independently Induce Numerical and Structural Chromosome Instability1. Cancer Res. 2002, 62, 7075–7082. [Google Scholar] [PubMed]
- Hong, S.; Li, Y.; Kaminski, P.J.; Andrade, J.; Laimins, L.A. Pathogenesis of human papillomaviruses requires the atr/p62 autophagy-related pathway. mBio 2020, 11, e01628-20. [Google Scholar] [CrossRef] [PubMed]
- Rinn, J.L.; Chang, H.Y. Long Noncoding RNAs: Molecular Modalities to Organismal Functions. Annu. Rev. Biochem. 2020, 89, 283–308. [Google Scholar] [CrossRef]
- Necsulea, A.; Soumillon, M.; Warnefors, M.; Liechti, A.; Daish, T.; Zeller, U.; Baker, J.C.; Grützner, F.; Kaessmann, H. The evolution of lncRNA repertoires and expression patterns in tetrapods. Nature 2014, 505, 635–640. [Google Scholar] [CrossRef]
- Iyer, M.K.; Niknafs, Y.S.; Malik, R.; Singhal, U.; Sahu, A.; Hosono, Y.; Barrette, T.R.; Prensner, J.R.; Evans, J.R.; Zhao, S.; et al. The Landscape of Long Noncoding RNAs in the Human Transcriptome. Nat. Genet. 2015, 47, 199–208. [Google Scholar] [CrossRef]
- Yan, X.; Hu, Z.; Feng, Y.; Hu, X.; Yuan, J.; Zhao, S.D.; Zhang, Y.; Yang, L.; Shan, W.; He, Q.; et al. Comprehensive Genomic Characterization of Long Non-coding RNAs across Human Cancers. Cancer Cell 2015, 28, 529–540. [Google Scholar] [CrossRef]
- Ashouri, A.; Sayin, V.I.; Van den Eynden, J.; Singh, S.X.; Papagiannakopoulos, T.; Larsson, E. Pan-cancer transcriptomic analysis associates long non-coding RNAs with key mutational driver events. Nat. Commun. 2016, 7, 13197. [Google Scholar] [CrossRef]
- Schmitt, A.M.; Chang, H.Y. Long Noncoding RNAs in Cancer Pathways. Cancer Cell 2016, 29, 452–463. [Google Scholar] [CrossRef]
- Wang, C.; Li, D.; Zhang, L.; Jiang, S.; Liang, J.; Narita, Y.; Hou, I.; Zhong, Q.; Zheng, Z.; Xiao, H.; et al. RNA Sequencing Analyses of Gene Expression during Epstein-Barr Virus Infection of Primary B Lymphocytes. J. Virol. 2019, 93, 226–245. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Wang, P.; Li, Z.; Li, Z.; Han, D.; Wen, M.; Zhao, Q.; Zhang, L.; Ma, Y.; Liu, W.; et al. IRF3-binding lncRNA-ISIR strengthens interferon production in viral infection and autoinflammation. Cell Rep. 2021, 37, 109926. [Google Scholar] [CrossRef]
- Imam, H.; Bano, A.S.; Patel, P.; Holla, P.; Jameel, S. The lncRNA NRON modulates HIV-1 replication in a NFAT-dependent manner and is differentially regulated by early and late viral proteins. Sci. Rep. 2015, 5, 8639. [Google Scholar] [CrossRef] [PubMed]
- Yu, A.-M.; Choi, Y.H.; Tu, M.-J. RNA Drugs and RNA Targets for Small Molecules: Principles, Progress, and Challenges. Pharmacol. Rev. 2020, 72, 862–898. [Google Scholar] [CrossRef] [PubMed]
- Harden, M.E.; Prasad, N.; Griffiths, A.; Munger, K. Modulation of microRNA-mRNA Target Pairs by Human Papillomavirus 16 Oncoproteins. mBio 2017, 8, e02170-16. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.; Munger, K. The Role of Long Noncoding RNAs in Human Papillomavirus-associated Pathogenesis. Pathogens 2020, 9, 289. [Google Scholar] [CrossRef] [PubMed]
- Kretz, M.; Webster, D.E.; Flockhart, R.J.; Lee, C.S.; Zehnder, A.; Lopez-Pajares, V.; Qu, K.; Zheng, G.X.Y.; Chow, J.; Kim, G.E.; et al. Suppression of progenitor differentiation requires the long noncoding RNA ANCR. Genes Dev. 2012, 26, 338–343. [Google Scholar] [CrossRef]
- Klein, R.H.; Lin, Z.; Hopkin, A.S.; Gordon, W.; Tsoi, L.C.; Liang, Y.; Gudjonsson, J.E.; Andersen, B. GRHL3 binding and enhancers rearrange as epidermal keratinocytes transition between functional states. PLoS Genet. 2017, 13, e1006745. [Google Scholar] [CrossRef]
- Chen, S.Y.; Ishii, M.A.; Cheng, B.; Otten, A.B.C.; Sun, B.K. HOPX Is a ZNF750 Target that Promotes Late Epidermal Differentiation. J. Investig. Dermatol. 2019, 139, 2039–2042.e2032. [Google Scholar] [CrossRef]
- Liang, H.; Zhang, C.; Guan, H.; Liu, J.; Cui, Y. LncRNA DANCR promotes cervical cancer progression by upregulating ROCK1 via sponging miR-335-5p. J. Cell. Physiol. 2019, 234, 7266–7278. [Google Scholar] [CrossRef]
- Kretz, M.; Siprashvili, Z.; Chu, C.; Webster, D.E.; Zehnder, A.; Qu, K.; Lee, C.S.; Flockhart, R.J.; Groff, A.F.; Chow, J.; et al. Control of somatic tissue differentiation by the long non-coding RNA TINCR. Nature 2013, 493, 231–235. [Google Scholar] [CrossRef] [PubMed]
- Eckhart, L.; Lachner, J.; Tschachler, E.; Rice, R.H. TINCR is not a non-coding RNA but encodes a protein component of cornified epidermal keratinocytes. Exp. Dermatol. 2020, 29, 376–379. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Pajares, V.; Qu, K.; Zhang, J.; Webster, D.E.; Barajas, B.C.; Siprashvili, Z.; Zarnegar, B.J.; Boxer, L.D.; Rios, E.J.; Tao, S.; et al. A LncRNA-MAF:MAFB Transcription Factor Network Regulates Epidermal Differentiation. Dev. Cell 2015, 32, 693–706. [Google Scholar] [CrossRef] [PubMed]
- Li, C.X.; Li, H.G.; Huang, L.T.; Kong, Y.W.; Chen, F.Y.; Liang, J.Y.; Yu, H.; Yao, Z.R. H19 lncRNA regulates keratinocyte differentiation by targeting miR-130b-3p. Cell Death Dis. 2017, 8, e3174. [Google Scholar] [CrossRef]
- Harmon, R.M.; Simpson, C.L.; Johnson, J.L.; Koetsier, J.L.; Dubash, A.D.; Najor, N.A.; Sarig, O.; Sprecher, E.; Green, K.J. Desmoglein-1/Erbin interaction suppresses ERK activation to support epidermal differentiation. J. Clin. Investig. 2013, 123, 1556–1570. [Google Scholar] [CrossRef]
- Getsios, S.; Simpson, C.L.; Kojima, S.-I.; Harmon, R.; Sheu, L.J.; Dusek, R.L.; Cornwell, M.; Green, K.J. Desmoglein 1–Dependent suppression of EGFR signaling promotes epidermal differentiation and morphogenesis. J. Cell Biol. 2009, 185, 1243–1258. [Google Scholar] [CrossRef]
- Cai, P.; Otten, A.B.C.; Cheng, B.; Ishii, M.A.; Zhang, W.; Huang, B.; Qu, K.; Sun, B.K. A genome-wide long noncoding RNA CRISPRi screen identifies PRANCR as a novel regulator of epidermal homeostasis. Genome Res. 2020, 30, 22–34. [Google Scholar] [CrossRef]
- Otten, A.B.C.; Amarbayar, O.; Cai, P.; Cheng, B.; Qu, K.; Sun, B.K. The noncoding RNA PRANCR regulates splicing of Fibronectin-1 to control keratinocyte proliferation and migration. bioRxiv 2021. [Google Scholar] [CrossRef]
- Lee, C.S.; Mah, A.; Aros, C.J.; Lopez-Pajares, V.; Bhaduri, A.; Webster, D.E.; Kretz, M.; Khavari, P.A. Cancer-Associated Long Noncoding RNA SMRT-2 Controls Epidermal Differentiation. J. Investig. Dermatol. 2018, 138, 1445–1449. [Google Scholar] [CrossRef]
- Ziegler, C.; Graf, J.; Faderl, S.; Schedlbauer, J.; Strieder, N.; Förstl, B.; Spang, R.; Bruckmann, A.; Merkl, R.; Hombach, S.; et al. The long non-coding RNA LINC00941 and SPRR5 are novel regulators of human epidermal homeostasis. EMBO Rep. 2019, 20, e46612. [Google Scholar] [CrossRef]
- Wu, N.; Jiang, M.; Liu, H.; Chu, Y.; Wang, D.; Cao, J.; Wang, Z.; Xie, X.; Han, Y.; Xu, B. LINC00941 promotes CRC metastasis through preventing SMAD4 protein degradation and activating the TGF-β/SMAD2/3 signaling pathway. Cell Death Differ. 2021, 28, 219–232. [Google Scholar] [CrossRef] [PubMed]
- Descargues, P.; Sil, A.K.; Sano, Y.; Korchynskyi, O.; Han, G.; Owens, P.; Wang, X.J.; Karin, M. IKKα is a critical coregulator of a Smad4-independent TGFβ-Smad2/3 signaling pathway that controls keratinocyte differentiation. Proc. Natl. Acad. Sci. USA 2008, 105, 2487–2492. [Google Scholar] [CrossRef] [PubMed]
- Tan, S.H.; Barker, N. Wnt Signaling in Adult Epithelial Stem Cells and Cancer. Prog. Mol. Biol. Transl. Sci. 2018, 153, 21–79. [Google Scholar] [CrossRef]
- Rim, E.Y.; Clevers, H.; Nusse, R. The Wnt Pathway: From Signaling Mechanisms to Synthetic Modulators. Annu. Rev. Biochem. 2022, 91, 571–598. [Google Scholar] [CrossRef] [PubMed]
- Üren, A.; Fallen, S.; Yuan, H.; Usubütün, A.; Küçükali, T.; Schlegel, R.; Toretsky, J.A. Activation of the Canonical Wnt Pathway during Genital Keratinocyte Transformation: A Model for Cervical Cancer Progression. Cancer Res. 2005, 65, 6199–6206. [Google Scholar] [CrossRef] [PubMed]
- Webster, M.T.; Rozycka, M.; Sara, E.; Davis, E.; Smalley, M.; Young, N.; Dale, T.C.; Wooster, R. Sequence variants of the axin gene in breast, colon, and other cancers: An analysis of mutations that interfere with GSK3 binding. Genes Chromosomes Cancer 2000, 28, 443–453. [Google Scholar] [CrossRef]
- Su, T.H.; Chang, J.G.; Yeh, K.T.; Lin, T.H.; Lee, T.P.; Chen, J.C.; Lin, C.C. Mutation analysis of CTNNB1 (beta-catenin) and AXIN1, the components of Wnt pathway, in cervical carcinomas. Oncol. Rep. 2003, 10, 1195–1200. [Google Scholar]
- Bulut, G.; Fallen, S.; Beauchamp, E.M.; Drebing, L.E.; Sun, J.; Berry, D.L.; Kallakury, B.; Crum, C.P.; Toretsky, J.A.; Schlegel, R.; et al. Beta-Catenin Accelerates Human Papilloma Virus Type-16 Mediated Cervical Carcinogenesis in Transgenic Mice. PLoS ONE 2011, 6, e27243. [Google Scholar] [CrossRef]
- Lichtig, H.; Gilboa, D.A.; Jackman, A.; Gonen, P.; Levav-Cohen, Y.; Haupt, Y.; Sherman, L. HPV16 E6 augments Wnt signaling in an E6AP-dependent manner. Virology 2010, 396, 47–58. [Google Scholar] [CrossRef]
- Bello, J.O.M.; Nieva, L.O.; Paredes, A.C.; Gonzalez, A.M.F.; Zavaleta, L.R.; Lizano, M. Regulation of the Wnt/β-Catenin Signaling Pathway by Human Papillomavirus E6 and E7 Oncoproteins. Viruses 2015, 7, 4734–4755. [Google Scholar] [CrossRef]
- Tian, W.; Lei, N.; Guo, R.; Yuan, Z.; Chang, L. Long non-coding RNA DANCR promotes cervical cancer growth via activation of the Wnt/β-catenin signaling pathway. Cancer Cell Int. 2020, 20, 61. [Google Scholar] [CrossRef] [PubMed]
- Yuan, S.X.; Wang, J.; Yang, F.; Tao, Q.F.; Zhang, J.; Wang, L.L.; Yang, Y.; Liu, H.; Wang, Z.G.; Xu, Q.G.; et al. Long noncoding RNA DANCR increases stemness features of hepatocellular carcinoma by derepression of CTNNB1. Hepatology 2016, 63, 499–511. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Gao, Y. CCAT-1 promotes proliferation and inhibits apoptosis of cervical cancer cells via the Wnt signaling pathway. Oncotarget 2017, 8, 68059. [Google Scholar] [CrossRef][Green Version]
- Salmerón-Bárcenas, E.G.; Illades-Aguiar, B.; del Moral-Hernández, O.; Ortega-Soto, A.; Hernández-Sotelo, D. HOTAIR knockdown decreased the activity Wnt/β-catenin signaling pathway and increased the mRNA levels of its negative regulators in HeLa cells. Cell. Physiol. Biochem. 2019, 53, 948–960. [Google Scholar] [CrossRef] [PubMed]
- Tsai, M.C.; Manor, O.; Wan, Y.; Mosammaparast, N.; Wang, J.K.; Lan, F.; Shi, Y.; Segal, E.; Chang, H.Y. Long noncoding RNA as modular scaffold of histone modification complexes. Science 2010, 329, 689–693. [Google Scholar] [CrossRef]
- Lee, M.; Kim, H.J.; Kim, S.W.; Park, S.A.; Chun, K.H.; Cho, N.H.; Song, Y.S.; Kim, Y.T. The long non-coding RNA HOTAIR increases tumour growth and invasion in cervical cancer by targeting the Notch pathway. Oncotarget 2016, 7, 44558. [Google Scholar] [CrossRef]
- Liu, M.; Jia, J.; Wang, X.; Liu, Y.; Wang, C.; Fan, R. Long non-coding RNA HOTAIR promotes cervical cancer progression through regulating BCL2 via targeting miR-143-3p. Cancer Biol. Ther. 2018, 19, 391–399. [Google Scholar] [CrossRef]
- Sharma, S.; Mandal, P.; Sadhukhan, T.; Roy Chowdhury, R.; Ranjan Mondal, N.; Chakravarty, B.; Chatterjee, T.; Roy, S.; Sengupta, S. Bridging Links between Long Noncoding RNA HOTAIR and HPV Oncoprotein E7 in Cervical Cancer Pathogenesis. Sci. Rep. 2015, 5, 11724. [Google Scholar] [CrossRef]
- Niu, Z.; Wang, F.; Lv, S.; Lv, Y.; Liu, M.; Fu, L.; Yao, Y.; Wang, L.; Lin, W.; Yuan, F. HNRNPU-AS1 Regulates Cell Proliferation and Apoptosis via the MicroRNA 205-5p/AXIN2 Axis and Wnt/β-Catenin Signaling Pathway in Cervical Cancer. Mol. Cell. Biol. 2021, 41, e0011521. [Google Scholar] [CrossRef]
- Song, H.; Liu, Y.; Liang, H.; Jin, X.; Liu, L. SPINT1-AS1 Drives Cervical Cancer Progression via Repressing miR-214 Biogenesis. Front. Cell Dev. Biol. 2021, 9, 691140. [Google Scholar] [CrossRef]
- Hong, S.; Cheng, S.; Iovane, A.; Laimins, L.A. STAT-5 Regulates Transcription of the Topoisomerase IIβ-Binding Protein 1 (TopBP1) Gene To Activate the ATR Pathway and Promote Human Papillomavirus Replication. mBio 2015, 6, e02006–e02015. [Google Scholar] [CrossRef] [PubMed]
- Chappell, W.H.; Gautam, D.; Ok, S.T.; Johnson, B.A.; Anacker, D.C.; Moody, C.A. Homologous Recombination Repair Factors Rad51 and BRCA1 Are Necessary for Productive Replication of Human Papillomavirus 31. J. Virol. 2016, 90, 2639–2652. [Google Scholar] [CrossRef] [PubMed]
- McKinney, C.C.; Hussmann, K.L.; McBride, A.A. The Role of the DNA Damage Response throughout the Papillomavirus Life Cycle. Viruses 2015, 7, 2450–2469. [Google Scholar] [CrossRef] [PubMed]
- Spriggs, C.C.; Laimins, L.A. Human Papillomavirus and the DNA Damage Response: Exploiting Host Repair Pathways for Viral Replication. Viruses 2017, 9, 232. [Google Scholar] [CrossRef]
- Wan, G.; Hu, X.; Liu, Y.; Han, C.; Sood, A.K.; Calin, G.A.; Zhang, X.; Lu, X. A novel non-coding RNA lncRNA-JADE connects DNA damage signalling to histone H4 acetylation. EMBO J. 2013, 32, 2833–2847. [Google Scholar] [CrossRef]
- Yang, M.; Sun, Y.; Xiao, C.; Ji, K.; Zhang, M.; He, N.; Wang, J.; Wang, Q.; Sun, Z.; Wang, Y.; et al. Integrated Analysis of the Altered lncRNAs and mRNAs Expression in 293T Cells after Ionizing Radiation Exposure. Int. J. Mol. Sci. 2019, 20, 2968. [Google Scholar] [CrossRef]
- Doyon, Y.; Cayrou, C.; Ullah, M.; Landry, A.J.; Côté, V.; Selleck, W.; Lane, W.S.; Tan, S.; Yang, X.J.; Côté, J. ING tumor suppressor proteins are critical regulators of chromatin acetylation required for genome expression and perpetuation. Mol. Cell 2006, 21, 51–64. [Google Scholar] [CrossRef]
- Saksouk, N.; Avvakumov, N.; Champagne, K.S.; Hung, T.; Doyon, Y.; Cayrou, C.; Paquet, E.; Ullah, M.; Landry, A.J.; Côté, V.; et al. HBO1 HAT complexes target chromatin throughout gene coding regions via multiple PHD finger interactions with histone H3 tail. Mol. Cell 2009, 33, 257–265. [Google Scholar] [CrossRef]
- Miotto, B.; Struhl, K. HBO1 histone acetylase activity is essential for DNA replication licensing and inhibited by Geminin. Mol. Cell 2010, 37, 57–66. [Google Scholar] [CrossRef]
- Lalonde, M.E.; Avvakumov, N.; Glass, K.C.; Joncas, F.H.; Saksouk, N.; Holliday, M.; Paquet, E.; Yan, K.; Tong, Q.; Klein, B.J.; et al. Exchange of associated factors directs a switch in HBO1 acetyltransferase histone tail specificity. Genes Dev. 2013, 27, 2009–2024. [Google Scholar] [CrossRef]
- Borgal, L.; Habbig, S.; Hatzold, J.; Liebau, M.C.; Dafinger, C.; Sacarea, I.; Hammerschmidt, M.; Benzing, T.; Schermer, B. The ciliary protein nephrocystin-4 translocates the canonical Wnt regulator Jade-1 to the nucleus to negatively regulate β-catenin signaling. J. Biol. Chem. 2012, 287, 25370–25380. [Google Scholar] [CrossRef] [PubMed]
- Vohhodina, J.; Goehring, L.J.; Liu, B.; Kong, Q.; Botchkarev, V.V.; Huynh, M.; Liu, Z.; Abderazzaq, F.O.; Clark, A.P.; Ficarro, S.B.; et al. BRCA1 binds TERRA RNA and suppresses R-Loop-based telomeric DNA damage. Nat. Commun. 2021, 12, 3542. [Google Scholar] [CrossRef]
- D’Alessandro, G.; Whelan, D.R.; Howard, S.M.; Vitelli, V.; Renaudin, X.; Adamowicz, M.; Iannelli, F.; Jones-Weinert, C.W.; Lee, M.Y.; Matti, V.; et al. BRCA2 controls DNA:RNA hybrid level at DSBs by mediating RNase H2 recruitment. Nat. Commun. 2018, 9, 5376. [Google Scholar] [CrossRef] [PubMed]
- Feldstein, O.; Nizri, T.; Doniger, T.; Jacob, J.; Rechavi, G.; Ginsberg, D. The long non-coding RNA ERIC is regulated by E2F and modulates the cellular response to DNA damage. Mol. Cancer 2013, 12, 131. [Google Scholar] [CrossRef]
- Lee, S.; Kopp, F.; Chang, T.C.; Sataluri, A.; Chen, B.; Sivakumar, S.; Yu, H.; Xie, Y.; Mendell, J.T. Noncoding RNA NORAD Regulates Genomic Stability by Sequestering PUMILIO Proteins. Cell 2016, 164, 69–80. [Google Scholar] [CrossRef]
- Tichon, A.; Gil, N.; Lubelsky, Y.; Havkin Solomon, T.; Lemze, D.; Itzkovitz, S.; Stern-Ginossar, N.; Ulitsky, I. A conserved abundant cytoplasmic long noncoding RNA modulates repression by Pumilio proteins in human cells. Nat. Commun. 2016, 7, 12209. [Google Scholar] [CrossRef]
- Munschauer, M.; Nguyen, C.T.; Sirokman, K.; Hartigan, C.R.; Hogstrom, L.; Engreitz, J.M.; Ulirsch, J.C.; Fulco, C.P.; Subramanian, V.; Chen, J.; et al. The NORAD lncRNA assembles a topoisomerase complex critical for genome stability. Nature 2018, 561, 132–136. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Liu, H.; Shi, L.; Yu, X.; Gu, Y.; Sun, X. LINP1 facilitates DNA damage repair through non-homologous end joining (NHEJ) pathway and subsequently decreases the sensitivity of cervical cancer cells to ionizing radiation. Cell Cycle 2018, 17, 439–447. [Google Scholar] [CrossRef]
- Taiana, E.; Favasuli, V.; Ronchetti, D.; Todoerti, K.; Pelizzoni, F.; Manzoni, M.; Barbieri, M.; Fabris, S.; Silvestris, I.; Gallo Cantafio, M.E.; et al. Long non-coding RNA NEAT1 targeting impairs the DNA repair machinery and triggers anti-tumor activity in multiple myeloma. Leukemia 2020, 34, 234–244. [Google Scholar] [CrossRef]
- Farooq, U.; Notani, D. Transcriptional regulation of INK4/ARF locus by cis and trans mechanisms. Front. Cell Dev. Biol. 2022, 10, 948351. [Google Scholar] [CrossRef]
- Liu, L.; Chen, Y.; Huang, Y.; Cao, K.; Liu, T.; Shen, H.; Cui, J.; Li, B.; Cai, J.; Gao, F.; et al. Long non-coding RNA ANRIL promotes homologous recombination-mediated DNA repair by maintaining ATR protein stability to enhance cancer resistance. Mol. Cancer 2021, 20, 94. [Google Scholar] [CrossRef] [PubMed]
- Zhao, K.; Wang, X.; Xue, X.; Li, L.; Hu, Y. A long noncoding RNA sensitizes genotoxic treatment by attenuating ATM activation and homologous recombination repair in cancers. PLoS Biol. 2020, 18, e3000666. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.; Munger, K. Expression of the Long Noncoding RNA DINO in Human Papillomavirus-Positive Cervical Cancer Cells Reactivates the Dormant TP53 Tumor Suppressor through ATM/CHK2 Signaling. mBio 2020, 11, e01190-20. [Google Scholar] [CrossRef]
- Schmitt, A.M.; Garcia, J.T.; Hung, T.; Flynn, R.A.; Shen, Y.; Qu, K.; Payumo, A.Y.; Peres-Da-Silva, A.; Broz, D.K.; Baum, R.; et al. An inducible long noncoding RNA amplifies DNA damage signaling. Nat. Genet. 2016, 48, 1370–1376. [Google Scholar] [CrossRef] [PubMed]
- Michelini, F.; Pitchiaya, S.; Vitelli, V.; Sharma, S.; Gioia, U.; Pessina, F.; Cabrini, M.; Wang, Y.; Capozzo, I.; Iannelli, F.; et al. Damage-induced lncRNAs control the DNA damage response through interaction with DDRNAs at individual double-strand breaks. Nat. Cell Biol. 2017, 19, 1400–1411. [Google Scholar] [CrossRef]
- Xu, J.; Liu, H.; Yang, Y.; Wang, X.; Liu, P.; Li, Y.; Meyers, C.; Banerjee, N.S.; Wang, H.K.; Cam, M.; et al. Genome-wide profiling of cervical rna-binding proteins identifies human papillomavirus regulation of rnaseh2a expression by viral e7 and e2f1. mBio 2019, 10, e02687-18. [Google Scholar] [CrossRef]
- Jang, M.K.; Shen, K.; McBride, A.A. Papillomavirus Genomes Associate with BRD4 to Replicate at Fragile Sites in the Host Genome. PLoS Pathog. 2014, 10, 1004117. [Google Scholar] [CrossRef]
- Khurana, S.; Markowitz, T.E.; Kabat, J.; McBride, A.A. Spatial and Functional Organization of Human Papillomavirus Replication Foci in the Productive Stage of Infection. mBio 2021, 12, e02684-21. [Google Scholar] [CrossRef]
- Kandoth, C.; McLellan, M.D.; Vandin, F.; Ye, K.; Niu, B.; Lu, C.; Xie, M.; Zhang, Q.; McMichael, J.F.; Wyczalkowski, M.A.; et al. Mutational landscape and significance across 12 major cancer types. Nature 2013, 502, 333–339. [Google Scholar] [CrossRef]
- Scheffner, M.; Munger, K.; Byrne, J.C.; Howley, P.M. The state of the p53 and retinoblastoma genes in human cervical carcinoma cell lines. Proc. Nati. Acad. Sci. USA 1991, 88, 5523–5527. [Google Scholar] [CrossRef]
- Scheffner, M.; Werness, B.A.; Huibregtse, J.M.; Levine, A.J.; Howley, P.M. The E6 oncoprotein encoded by human papillomavirus types 16 and 18 promotes the degradation of p53. Cell 1990, 63, 1129–1136. [Google Scholar] [CrossRef]
- Fakhry, C.; Westra, W.H.; Li, S.; Cmelak, A.; Ridge, J.A.; Pinto, H.; Forastiere, A.; Gillison, M.L. Improved Survival of Patients With Human Papillomavirus–Positive Head and Neck Squamous Cell Carcinoma in a Prospective Clinical Trial. JNCI J. Natl. Cancer Inst. 2008, 100, 261–269. [Google Scholar] [CrossRef] [PubMed]
- Fontan, C.T.; James, C.D.; Prabhakar, A.T.; Bristol, M.L.; Otoa, R.; Wang, X.; Karimi, E.; Rajagopalan, P.; Basu, D.; Morgan, I.M. A Critical Role for p53 during the HPV16 Life Cycle. Microbiol. Spectr. 2022, 10, e0068122. [Google Scholar] [CrossRef] [PubMed]
- Yugawa, T.; Handa, K.; Narisawa-Saito, M.; Ohno, S.-I.; Fujita, M.; Kiyono, T. Regulation of Notch1 Gene Expression by p53 in Epithelial Cells. Mol. Cell. Biol. 2007, 27, 3732. [Google Scholar] [CrossRef] [PubMed]
- Kranjec, C.; Holleywood, C.; Libert, D.; Griffin, H.; Mahmood, R.; Isaacson, E.; Doorbar, J. Modulation of basal cell fate during productive and transforming HPV-16 infection is mediated by progressive E6-driven depletion of Notch. J. Pathol. 2017, 242, 448–462. [Google Scholar] [CrossRef]
- Sánchez, Y.; Segura, V.; Marín-Béjar, O.; Athie, A.; Marchese, F.P.; González, J.; Bujanda, L.; Guo, S.; Matheu, A.; Huarte, M. Genome-wide analysis of the human p53 transcriptional network unveils a lncRNA tumour suppressor signature. Nat. Commun. 2014, 5, 5812. [Google Scholar] [CrossRef]
- Adriaens, C.; Standaert, L.; Barra, J.; Latil, M.; Verfaillie, A.; Kalev, P.; Boeckx, B.; Wijnhoven, P.W.G.; Radaelli, E.; Vermi, W.; et al. p53 induces formation of NEAT1 lncRNA-containing paraspeckles that modulate replication stress response and chemosensitivity. Nat. Med. 2016, 22, 861–868. [Google Scholar] [CrossRef] [PubMed]
- Mello, S.S.; Sinow, C.; Raj, N.; Mazur, P.K.; Bieging-Rolett, K.; Broz, D.K.; Imam, J.F.C.; Vogel, H.; Wood, L.D.; Sage, J.; et al. Neat1 is a p53-inducible lincRNA essential for transformation suppression. Genes Dev. 2017, 31, 1095–1108. [Google Scholar] [CrossRef]
- Li, X.L.; Subramanian, M.; Jones, M.F.; Chaudhary, R.; Singh, D.K.; Zong, X.; Gryder, B.; Sindri, S.; Mo, M.; Schetter, A.; et al. Long noncoding RNA PURPL suppresses basal p53 levels and promotes tumorigenicity in colorectal cancer. Cell Rep. 2017, 20, 2408–2423. [Google Scholar] [CrossRef]
- Mao, C.; Wang, X.; Liu, Y.; Wang, M.; Yan, B.; Jiang, Y.; Shi, Y.; Shen, Y.; Liu, X.; Lai, W.; et al. A G3BP1-Interacting lncRNA Promotes Ferroptosis and Apoptosis in Cancer via Nuclear Sequestration of p53. Cancer Res. 2018, 78, 3484–3496. [Google Scholar] [CrossRef]
- Ali, M.S.S.H.; Cheng, X.; Moran, M.; Haemmig, S.; Naldrett, M.J.; Alvarez, S.; Feinberg, M.W.; Sun, X. LncRNA Meg3 protects endothelial function by regulating the DNA damage response. Nucleic Acids Res. 2019, 47, 1505–1522. [Google Scholar] [CrossRef]
- Meza-Sosa, K.F.; Miao, R.; Navarro, F.; Zhang, Z.; Zhang, Y.; Hu, J.J.; Hartford, C.C.R.; Li, X.L.; Pedraza-Alva, G.; Pérez-Martínez, L.; et al. SPARCLE, a p53-induced lncRNA, controls apoptosis after genotoxic stress by promoting PARP-1 cleavage. Mol. Cell 2022, 82, 785–802.e710. [Google Scholar] [CrossRef] [PubMed]
- Olivero, C.E.; Martínez-Terroba, E.; Zimmer, J.; Liao, C.; Tesfaye, E.; Hooshdaran, N.; Schofield, J.A.; Bendor, J.; Fang, D.; Simon, M.D.; et al. p53 activates the long noncoding RNA Pvt1b to inhibit Myc and suppress tumorigenesis. Mol. Cell 2020, 77, 761–774. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Hao, Q.; Wang, S.; Cao, M.; Huang, Y.; Weng, X.; Wang, J.; Zhang, Z.; He, X.; Lu, H.; et al. Inactivation of the tumor suppressor p53 by long noncoding RNA RMRP. Proc. Natl. Acad. Sci. USA 2021, 118, e2026813118. [Google Scholar] [CrossRef]
- Marney, C.B.; Anderson, E.S.; Adnan, M.; Peng, K.L.; Hu, Y.; Weinhold, N.; Schmitt, A.M. p53-intact cancers escape tumor suppression through loss of long noncoding RNA Dino. Cell Rep. 2021, 35, 109329. [Google Scholar] [CrossRef] [PubMed]
- Martín-Caballero, J.; Flores, J.M.; García-Palencia, P.; Serrano, M. Tumor susceptibility of p21(Waf1/Cip1)-deficient mice. Cancer Res. 2001, 61, 6234–6238. [Google Scholar] [PubMed]
- Marney, C.B.; Anderson, E.S.; Baum, R.; Schmitt, A.M. A Unique Spectrum of Spontaneous Tumors in Dino Knockout Mice Identifies Tissue-Specific Requirements for Tumor Suppression. Cells 2022, 11, 1818. [Google Scholar] [CrossRef]
- Lane, D.P. Cancer. p53, guardian of the genome. Nature 1992, 358, 15–16. [Google Scholar] [CrossRef]
- Soussi, T. The history of p53: A perfect example of the drawbacks of scientific paradigms. EMBO Rep. 2010, 11, 822–826. [Google Scholar] [CrossRef]
- Marcel, V.; Van Long, F.N.; Diaz, J.J. 40 Years of Research Put p53 in Translation. Cancers 2018, 10, 152. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Arman, W.; Munger, K. Mechanistic Contributions of lncRNAs to Cellular Signaling Pathways Crucial to the Lifecycle of Human Papillomaviruses. Viruses 2022, 14, 2439. https://doi.org/10.3390/v14112439
Arman W, Munger K. Mechanistic Contributions of lncRNAs to Cellular Signaling Pathways Crucial to the Lifecycle of Human Papillomaviruses. Viruses. 2022; 14(11):2439. https://doi.org/10.3390/v14112439
Chicago/Turabian StyleArman, Warda, and Karl Munger. 2022. "Mechanistic Contributions of lncRNAs to Cellular Signaling Pathways Crucial to the Lifecycle of Human Papillomaviruses" Viruses 14, no. 11: 2439. https://doi.org/10.3390/v14112439
APA StyleArman, W., & Munger, K. (2022). Mechanistic Contributions of lncRNAs to Cellular Signaling Pathways Crucial to the Lifecycle of Human Papillomaviruses. Viruses, 14(11), 2439. https://doi.org/10.3390/v14112439




