Genomic Characterisation of Canis Familiaris Papillomavirus Type 24, a Novel Papillomavirus Associated with Extensive Pigmented Plaque Formation in a Pug Dog
Abstract
1. Introduction
2. Materials and Methods
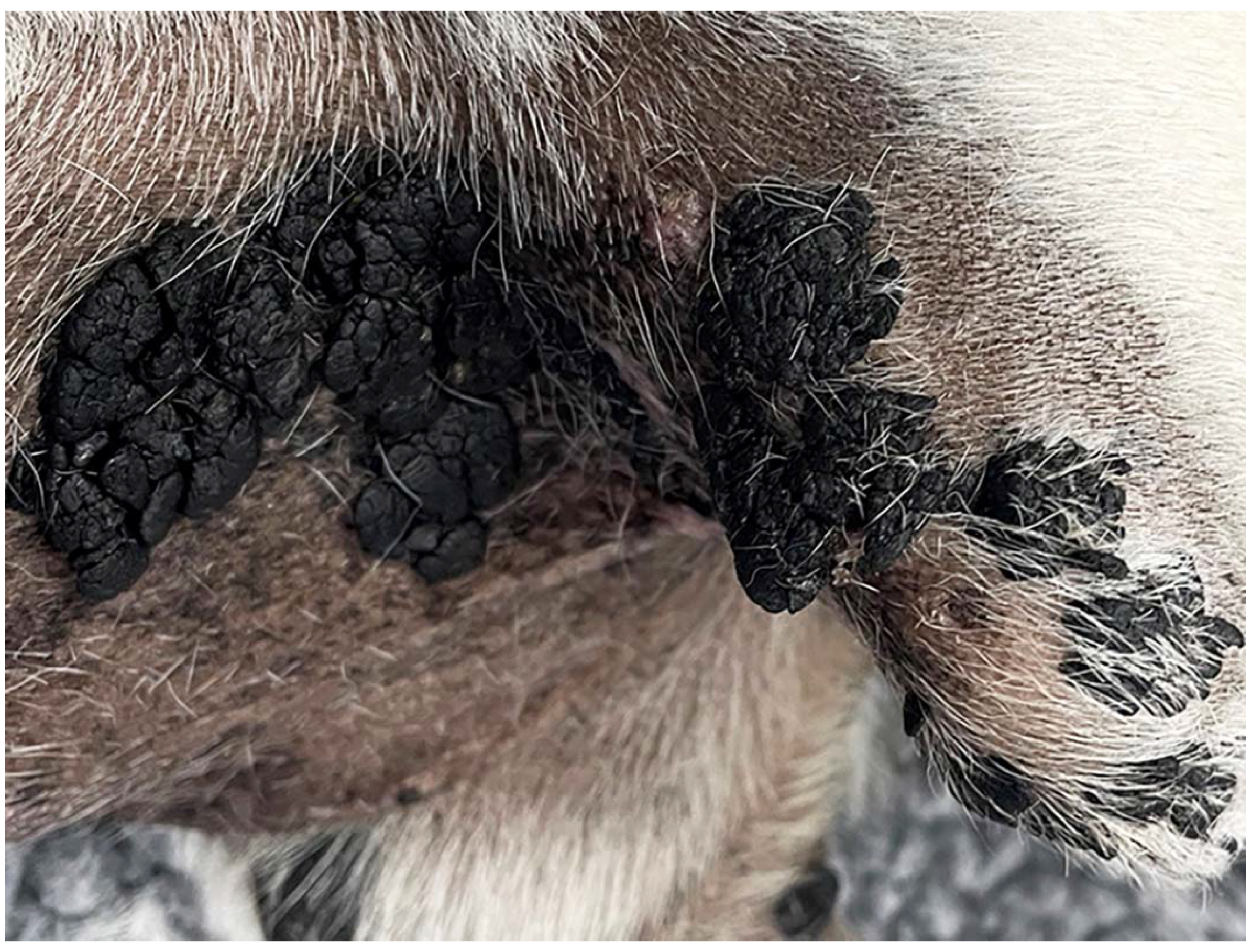
2.1. Case Summary and Sample Collection
2.2. Initial PCR and DNA Sequencing
2.3. DNA and Protein Sequence Analysis
2.4. Phylogenetic Analysis
2.5. Nucleotide Sequence Accession Number
3. Results
3.1. Initial PV DNA Amplification
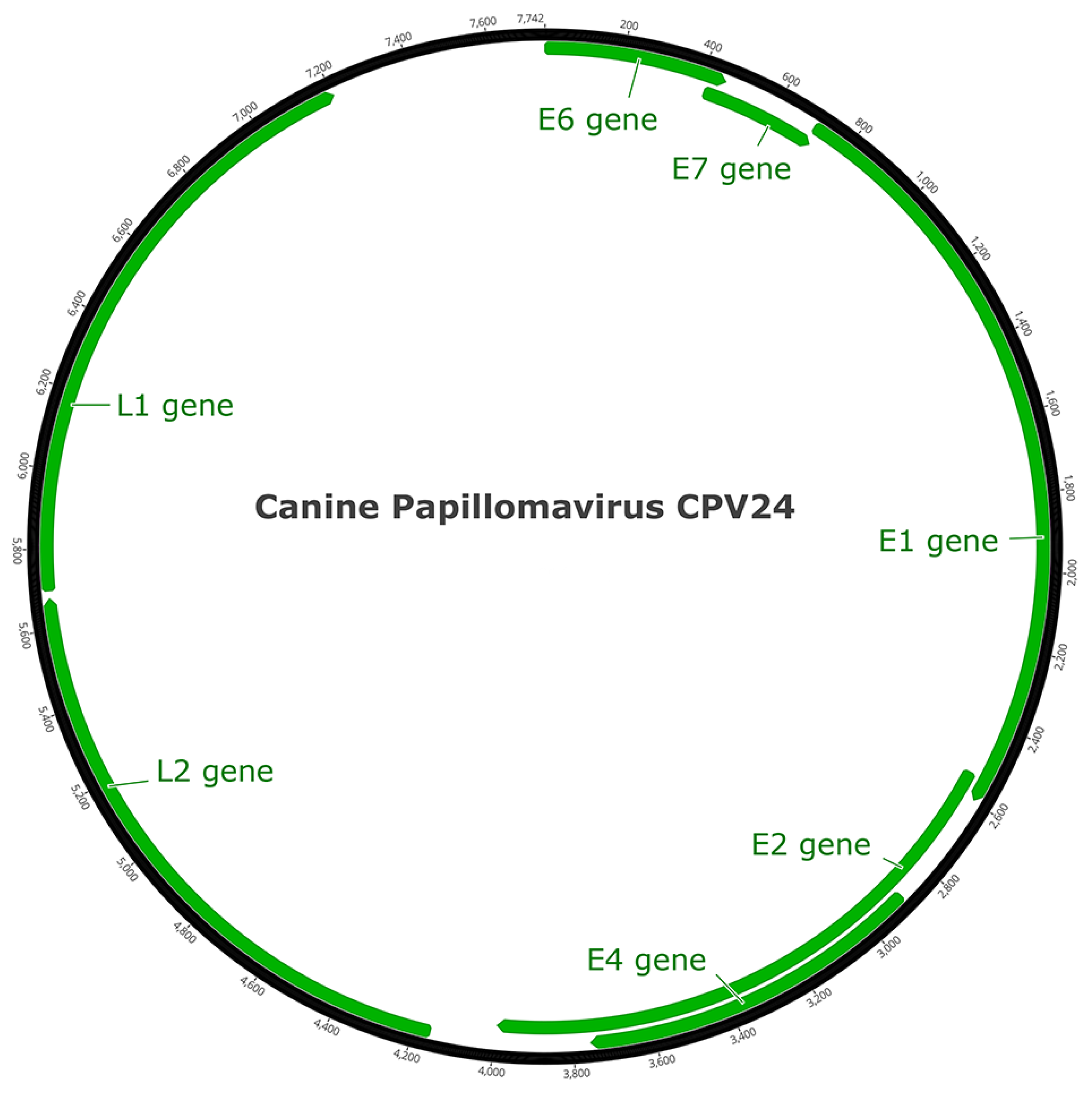
3.2. CPV24 Complete Gene Sequence
3.3. Open Reading Frame Organization of CPV24 Genes
3.4. Phylogenetic Analysis of CPV24
3.5. CPV24 Sequence Similarity to Other Papillomaviruses
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Munday, J.S.; Thomson, N.A.; Luff, J.A. Papillomaviruses in dogs and cats. Vet. J. 2017, 225, 23–31. [Google Scholar] [CrossRef] [PubMed]
- Bernard, H.U.; Burk, R.D.; Chen, Z.; van Doorslaer, K.; Hausen, H.; de Villiers, E.M. Classification of papillomaviruses (PVs) based on 189 PV types and proposal of taxonomic amendments. Virology 2010, 401, 70–79. [Google Scholar] [CrossRef] [PubMed]
- Knight, E.C.; Munday, J.S.; Stone, B.M.; Shipstone, M.A. Carbon dioxide laser treatment of extensive pigmented viral plaque lesions in a golden retriever dog. Vet. Dermatol. 2016, 27, 442-e117. [Google Scholar] [CrossRef] [PubMed]
- Munday, J.S.; Lam, A.T.H.; Sakai, M. Extensive progressive pigmented viral plaques in a Chihuahua dog. Vet. Dermatol. 2022, 33, 252–254. [Google Scholar] [CrossRef]
- Luff, J.; Rowland, P.; Mader, M.; Orr, C.; Yuan, H. Two canine papillomaviruses associated with metastatic squamous cell carcinoma in two related Basenji dogs. Vet. Pathol. 2016, 53, 1160–1163. [Google Scholar] [CrossRef]
- Drummond, A.J.; Ashton, B.; Buxton, S.; Cheung, M.; Cooper, A.; Duran, C.; Field, M.; Heled, J.; Kearse, M.; Markowitz, S.; et al. Geneious; Version 5.3; Dotmatics: Boston, MA, USA, 2010. [Google Scholar]
- Katoh, K.; Misawa, K.; Kuma, K.; Miyata, T. MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002, 30, 3059–3066. [Google Scholar] [CrossRef]
- Guindon, S.; Dufayard, J.F.; Lefort, V.; Anisimova, M.; Hordijk, W.; Gascuel, O. New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst. Biol. 2010, 59, 307–321. [Google Scholar] [CrossRef]
- Lefort, V.; Longueville, J.E.; Gascuel, O. SMS: Smart Model Selection in PhyML. Mol. Biol. Evol. 2017, 34, 2422–2424. [Google Scholar] [CrossRef]
- Subramanian, B.; Gao, S.; Lercher, M.J.; Hu, S.; Chen, W.H. Evolview v3: A webserver for visualization, annotation, and management of phylogenetic trees. Nucleic Acids Res. 2019, 47, W270–W275. [Google Scholar] [CrossRef]
- Mantovani, F.; Banks, L. The human papillomavirus E6 protein and its contribution to malignant progression. Oncogene 2001, 20, 7874–7887. [Google Scholar] [CrossRef]
- Munger, K.; Baldwin, A.; Edwards, K.M.; Hayakawa, H.; Nguyen, C.L.; Owens, M.; Grace, M.; Huh, K. Mechanisms of human papillomavirus-induced oncogenesis. J. Virol. 2004, 78, 11451–11460. [Google Scholar] [CrossRef] [PubMed]
- Roman, A.; Munger, K. The papillomavirus E7 proteins. Virology 2013, 445, 138–168. [Google Scholar] [CrossRef] [PubMed]
- Ma, T.; Zou, N.; Lin, B.Y.; Chow, L.T.; Harper, J.W. Interaction between cyclin-dependent kinases and human papillomavirus replication-initiation protein E1 is required for efficient viral replication. Proc. Natl. Acad. Sci. USA 1999, 96, 382–387. [Google Scholar] [CrossRef] [PubMed]
- Van Doorslaer, K.; Rector, A.; Jenson, A.B.; Sundberg, J.P.; Van Ranst, M.; Ghim, S.J. Complete genomic characterization of a murine papillomavirus isolated from papillomatous lesions of a European harvest mouse (Micromys minutus). J. Gen. Virol. 2007, 88, 1484–1488. [Google Scholar] [CrossRef]
- Doorbar, J. The E4 protein; structure, function and patterns of expression. Virology 2013, 445, 80–98. [Google Scholar] [CrossRef]
- Day, P.M.; Thompson, C.D.; Buck, C.B.; Pang, Y.Y.; Lowy, D.R.; Schiller, J.T. Neutralization of human papillomavirus with monoclonal antibodies reveals different mechanisms of inhibition. J. Virol. 2007, 81, 8784–8792. [Google Scholar] [CrossRef]
- Richards, R.M.; Lowy, D.R.; Schiller, J.T.; Day, P.M. Cleavage of the papillomavirus minor capsid protein, L2, at a furin consensus site is necessary for infection. Proc. Natl. Acad. Sci. USA 2006, 103, 1522–1527. [Google Scholar] [CrossRef]
- Titolo, S.; Brault, K.; Majewski, J.; White, P.W.; Archambault, J. Characterization of the minimal DNA binding domain of the human papillomavirus e1 helicase: Fluorescence anisotropy studies and characterization of a dimerization-defective mutant protein. J. Virol. 2003, 77, 5178–5191. [Google Scholar] [CrossRef][Green Version]
- Rector, A.; Lemey, P.; Tachezy, R.; Mostmans, S.; Ghim, S.J.; Van Doorslaer, K.; Roelke, M.; Bush, M.; Montali, R.J.; Joslin, J.; et al. Ancient papillomavirus-host co-speciation in Felidae. Genome Biol. 2007, 8, R57. [Google Scholar] [CrossRef]
- Antonsson, A.; Hansson, B.G. Healthy skin of many animal species harbors papillomaviruses which are closely related to their human counterparts. J. Virol. 2002, 76, 12537–12542. [Google Scholar] [CrossRef]
- Tobler, K.; Lange, C.; Carlotti, D.N.; Ackermann, M.; Favrot, C. Detection of a novel papillomavirus in pigmented plaques of four pugs. Vet. Dermatol. 2008, 19, 21–25. [Google Scholar] [CrossRef] [PubMed]
- Alves, C.; Weber, M.N.; Guimarães, L.L.B.; Cibulski, S.P.; da Silva, F.R.C.; Daudt, C.; Budaszewski, R.F.; Silva, M.S.; Mayer, F.Q.; Bianchi, R.M.; et al. Canine papillomavirus type 16 associated to squamous cell carcinoma in a dog: Virological and pathological findings. Braz. J. Microbiol. 2020, 51, 2087–2094. [Google Scholar] [CrossRef] [PubMed]
- Beziat, V.; Casanova, J.L.; Jouanguy, E. Human genetic and immunological dissection of papillomavirus-driven diseases: New insights into their pathogenesis. Curr. Opin. Virol. 2021, 51, 9–15. [Google Scholar] [CrossRef] [PubMed]
- Stokking, L.B.; Ehrhart, E.J.; Lichtensteiger, C.A.; Campbell, K.L. Pigmented epidermal plaques in three dogs. J. Am. Anim. Hosp. Assoc. 2004, 40, 411–417. [Google Scholar] [CrossRef] [PubMed]
- Luff, J.A.; Affolter, V.K.; Yeargan, B.; Moore, P.F. Detection of six novel papillomavirus sequences within canine pigmented plaques. J. Vet. Diagn. Investig. 2012, 24, 576–580. [Google Scholar] [CrossRef] [PubMed]
- Luff, J.; Mader, M.; Rowland, P.; Britton, M.; Fass, J.; Yuan, H. Viral genome integration of canine papillomavirus 16. Papillomavirus Res. 2019, 7, 88–96. [Google Scholar] [CrossRef] [PubMed]

| ORF | ORF Location | Length (nt) | Length (aa) | Molecular Mass (kDa) | pI |
|---|---|---|---|---|---|
| E1 | 705–2594 | 1890 | 629 | 71.27 | 5.48 |
| E2 | 2536–3993 | 1458 | 485 | 53.85 | 6.69 |
| E4 | 2891–3757 | 867 | 228 | 31.6 | 7.61 |
| E6 | 1–456 | 456 | 151 | 17.33 | 8.32 |
| E7 | 416–715 | 300 | 99 | 11.16 | 4.2 |
| L1 | 5697–7202 | 1506 | 501 | 56.18 | 8.15 |
| L2 | 4158–5678 | 1521 | 506 | 53.68 | 5.91 |
| Papillomavirus | Host Species | Classification | L1 Similarity (%) |
|---|---|---|---|
| Canine familiaris papillomavirus 4 (EF584537) | Domestic dog | Chipapillomavirus 2 | 78.2 |
| Canine familiaris papillomavirus 12 (JQ754321) | Domestic dog | Chipapillomavirus 1 | 69.7 |
| Canine familiaris papillomavirus 9 (JF800656) | Domestic dog | Chipapillomavirus 3 | 69.0 |
| Canine familiaris papillomavirus 16 (KP099966) | Domestic dog | Chipapillomavirus 2 | 68.0 |
| Canine familiaris papillomavirus 3 (DQ295066) | Domestic dog | Chipapillomavirus 1 | 67.5 |
| Sus scrofa papillomavirus 1 (EF395818) | Domestic pig | Dyodeltapapillomavirus | 60.3 |
| Felis catis papillomavirus 2 (EU796884) | Domestic cat | Dyothetapapillomavirus 1 | 59.6 |
| Canine familiaris papillomavirus 1 (D55633) | Domestic dog | Lambdapapillomavirus | 56.1 |
| Canine familiaris papillomavirus 2 (AY722648) | Domestic dog | Taupapillomavirus | 54.1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Munday, J.S.; Gedye, K.; Knox, M.A.; Ravens, P.; Lin, X. Genomic Characterisation of Canis Familiaris Papillomavirus Type 24, a Novel Papillomavirus Associated with Extensive Pigmented Plaque Formation in a Pug Dog. Viruses 2022, 14, 2357. https://doi.org/10.3390/v14112357
Munday JS, Gedye K, Knox MA, Ravens P, Lin X. Genomic Characterisation of Canis Familiaris Papillomavirus Type 24, a Novel Papillomavirus Associated with Extensive Pigmented Plaque Formation in a Pug Dog. Viruses. 2022; 14(11):2357. https://doi.org/10.3390/v14112357
Chicago/Turabian StyleMunday, John S., Kristene Gedye, Matthew A. Knox, Philippa Ravens, and Xiaoxiao Lin. 2022. "Genomic Characterisation of Canis Familiaris Papillomavirus Type 24, a Novel Papillomavirus Associated with Extensive Pigmented Plaque Formation in a Pug Dog" Viruses 14, no. 11: 2357. https://doi.org/10.3390/v14112357
APA StyleMunday, J. S., Gedye, K., Knox, M. A., Ravens, P., & Lin, X. (2022). Genomic Characterisation of Canis Familiaris Papillomavirus Type 24, a Novel Papillomavirus Associated with Extensive Pigmented Plaque Formation in a Pug Dog. Viruses, 14(11), 2357. https://doi.org/10.3390/v14112357






