Isolation, Characterization, and Genome Analysis of a Novel Bacteriophage, Escherichia Phage vB_EcoM-4HA13, Representing a New Phage Genus in the Novel Phage Family Chaseviridae
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Culturing Conditions
2.2. Phage Isolation and Propagation
2.3. Phage Host Range Determination
2.3.1. Turbidimetric Assay
2.3.2. Efficiency of Plating (EOP)
2.4. Virulence Index Determination
2.5. One-Step Growth Curve and Adsorption Assay
2.6. Transmission Emission Microscopy
2.7. DNA Extraction and Genome Sequencing
2.8. Genome Analyses
2.8.1. Genome Assembly and Annotation
2.8.2. In Silico Analyses
2.9. Proteome Analyses
2.9.1. CsCl Purification
2.9.2. Liquid Chromatography–Mass Spectrometry
3. Results and Discussion
3.1. Biological Characterizations
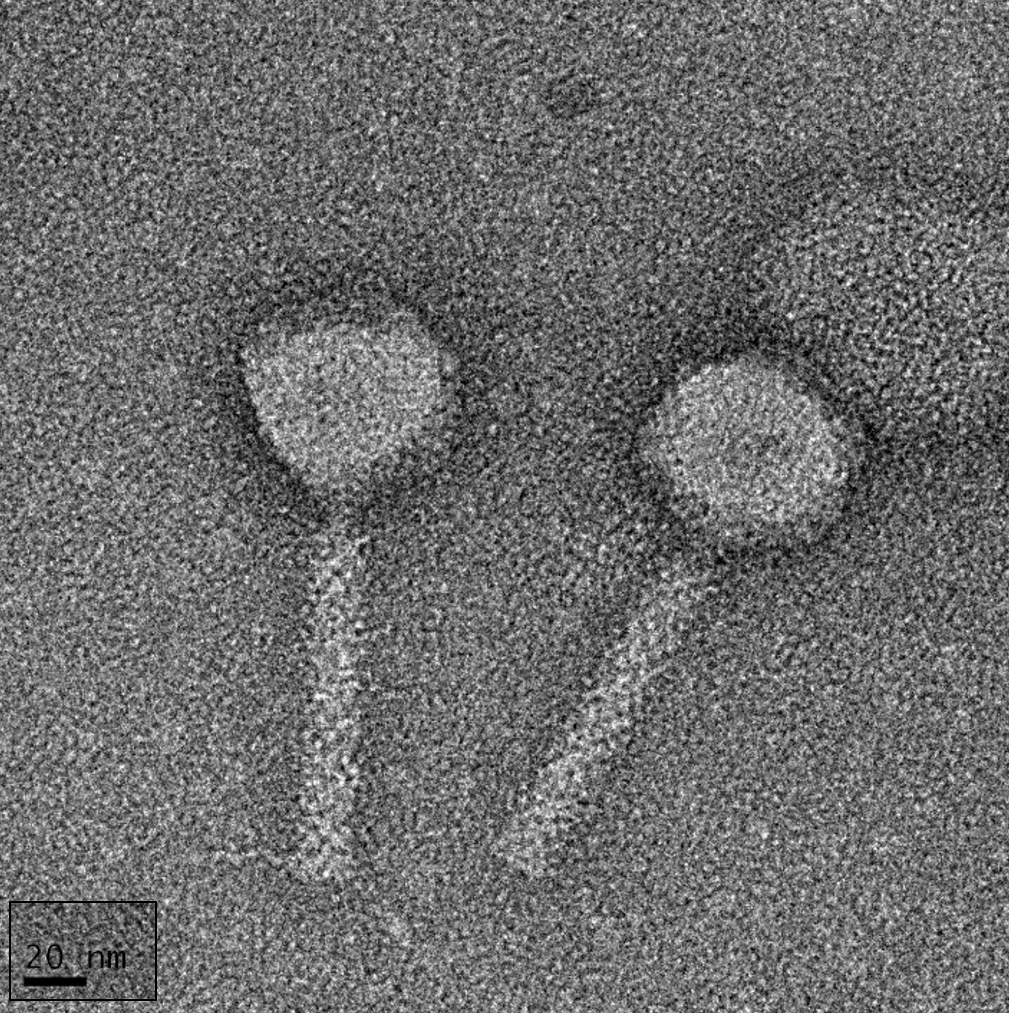
3.1.1. Morphology
3.1.2. Host Range
3.1.3. Virulence Index of 4HA13 against E. coli O111:NM
3.1.4. Kinetics
3.2. Genome Characterizations
3.2.1. General Features
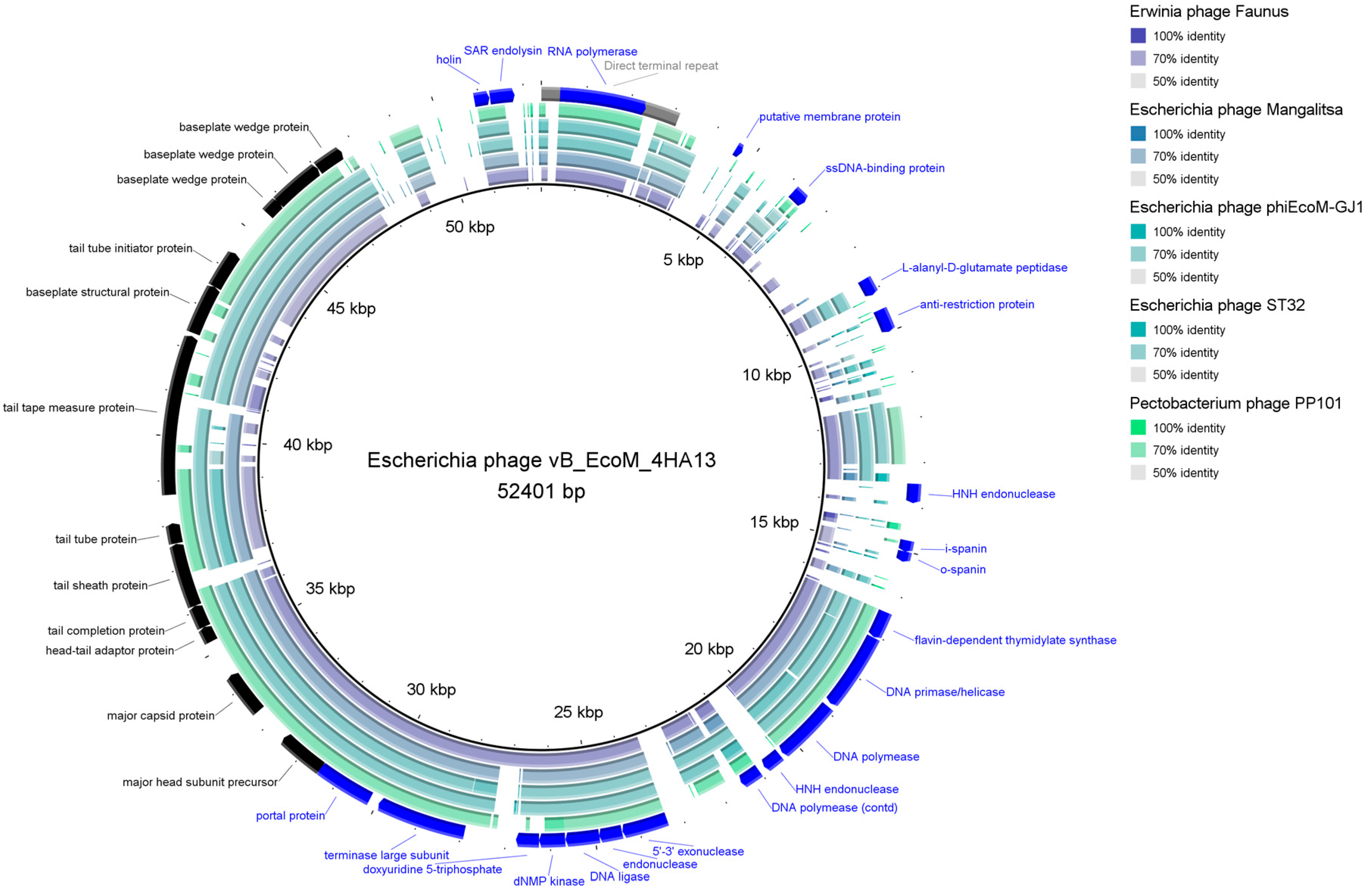
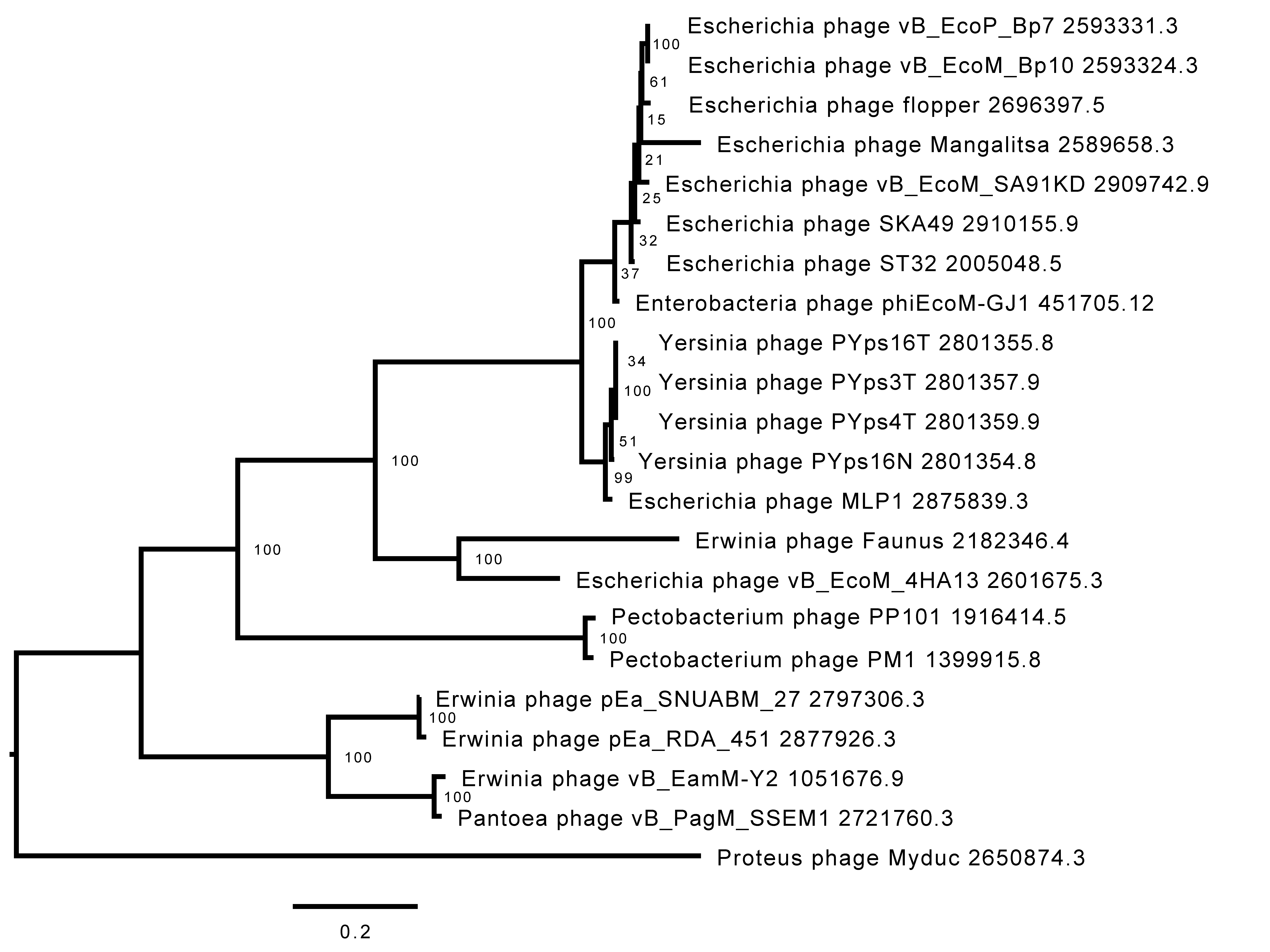
3.2.2. Comparative Analysis
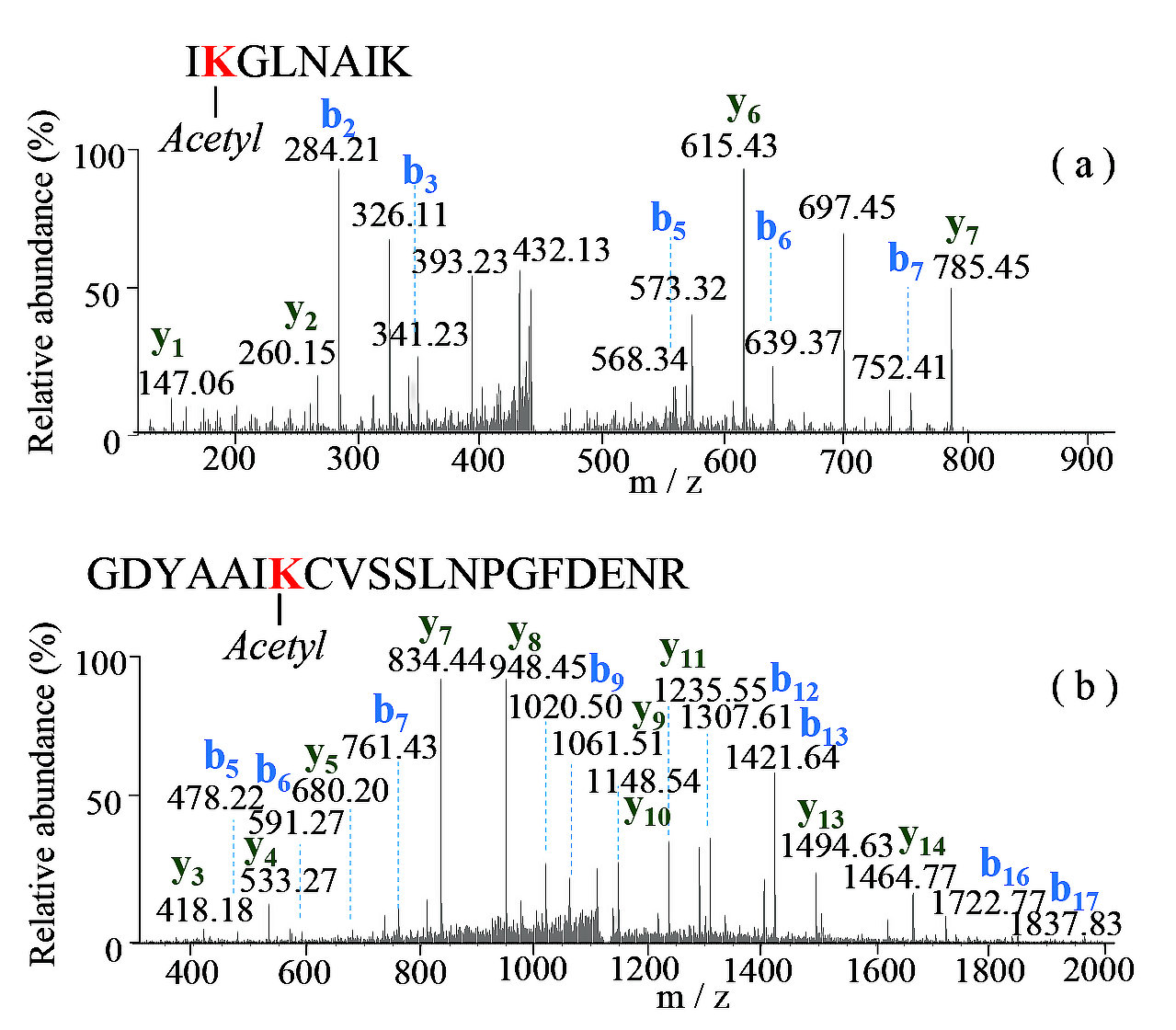
3.3. Proteome Characterizations
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Griffin, P.M.; Tauxe, R.V. The epidemiology of infections caused by Escherichia coli O157:H7, other enterohemorrhagic E. coli, and the associated hemolytic uremic syndrome. Epidemiol. Rev. 1991, 13, 60–98. [Google Scholar] [CrossRef] [PubMed]
- Jandhyala, D.M.; Thorpe, C.M.; Magun, B. Ricin and Shiga toxins: Effects on host cell signal transduction. Curr. Top. Microbiol. Immunol. 2012, 357, 41–65. [Google Scholar] [PubMed]
- Melton-Celsa, A.R. Shiga Toxin (Stx) Classification, Structure, and Function. Microbiol. Spectr. 2014, 2, EHEC-0024-2013. [Google Scholar] [CrossRef] [PubMed]
- Tesh, V.L. Activation of cell stress response pathways by Shiga toxins. Cell Microbiol. 2012, 14, 1–9. [Google Scholar] [CrossRef]
- National Outbreak Reporting System (NORS). Available online: https://wwwn.cdc.gov/norsdashboard/ (accessed on 12 August 2022).
- Kintz, E.; Brainard, J.; Hooper, L.; Hunter, P. Transmission pathways for sporadic Shiga-toxin producing E. coli infections: A systematic review and meta-analysis. Int. J. Hyg. Environ. Health 2017, 220, 57–67. [Google Scholar] [CrossRef]
- Kim, J.S.; Lee, M.S.; Kim, J.H. Recent Updates on Outbreaks of Shiga Toxin-Producing Escherichia coli and Its Potential Reservoirs. Front. Cell Infect. Microbiol. 2020, 10, 273. [Google Scholar] [CrossRef]
- Public Health Notices—Archived Health Investigations. Available online: https://www.canada.ca/en/public-health/services/public-health-notices.html (accessed on 12 August 2022).
- Reports of Selected E. coli Outbreak Investigations. Available online: https://www.cdc.gov/ecoli/outbreaks.html (accessed on 12 August 2022).
- Mathusa, E.C.; Chen, Y.; Enache, E.; Hontz, L. Non-O157 Shiga toxin-producing Escherichia coli in foods. J. Food Prot. 2010, 73, 1721–1736. [Google Scholar] [CrossRef]
- Gould, L.H.; Mody, R.K.; Ong, K.L.; Clogher, P.; Cronquist, A.B.; Garman, K.N.; Lathrop, S.; Medus, C.; Spina, N.L.; Webb, T.H.; et al. Increased recognition of non-O157 Shiga toxin-producing Escherichia coli infections in the United States during 2000-2010: Epidemiologic features and comparison with E. coli O157 infections. Foodborne Pathog. Dis. 2013, 10, 453–460. [Google Scholar] [CrossRef]
- Glassman, H.; Ferrato, C.; Chui, L. Epidemiology of Non-O157 Shiga Toxin-Producing Escherichia coli in the Province of Alberta, Canada, from 2018 to 2021. Microorganisms 2022, 10, 814. [Google Scholar] [CrossRef]
- Ray, L.C.; Collins, J.P.; Griffin, P.M.; Shah, H.J.; Boyle, M.M.; Cieslak, P.R.; Dunn, J.; Lathrop, S.; McGuire, S.; Rissman, T.; et al. Decreased Incidence of Infections Caused by Pathogens Transmitted Commonly Through Food During the COVID-19 Pandemic—Foodborne Diseases Active Surveillance Network, 10 U.S. Sites, 2017–2020. MMWR Morb. Mortal. Wkly. Rep. 2021, 70, 1332–1336. [Google Scholar] [CrossRef]
- Pinto, G.; Almeida, C.; Azeredo, J. Bacteriophages to control Shiga toxin-producing E. coli—Safety and regulatory challenges. Crit. Rev. Biotechnol. 2020, 40, 1081–1097. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Qu, K.; Li, X.; Cao, Z.; Wang, X.; Li, Z.; Song, Y.; Xu, Y. Use of Bacteriophages to Control Escherichia coli O157:H7 in Domestic Ruminants, Meat Products, and Fruits and Vegetables. Foodborne Pathog. Dis. 2017, 14, 483–493. [Google Scholar] [CrossRef] [PubMed]
- Korf, I.H.E.; Meier-Kolthoff, J.P.; Adriaenssens, E.M.; Kropinski, A.M.; Nimtz, M.; Rohde, M.; van Raaij, M.J.; Wittmann, J. Still Something to Discover: Novel Insights into Escherichia coli Phage Diversity and Taxonomy. Viruses 2019, 11, 454. [Google Scholar] [CrossRef] [PubMed]
- Hinkley, T.C.; Garing, S.; Clute-Reinig, N.; Spencer, E.; Jain, P.; Alonzo, L.F.; Le Ny, A.M. Genome sequences of 38 bacteriophages infecting Escherichia coli, isolated from raw sewage. Microbiol. Resour. Announc. 2020, 9, e00909–e00920. [Google Scholar] [CrossRef] [PubMed]
- Maffei, E.; Shaidullina, A.; Burkolter, M.; Heyer, Y.; Estermann, F.; Druelle, V.; Sauer, P.; Willi, L.; Michaelis, S.; Hilbi, H.; et al. Systematic exploration of Escherichia coli phage–host interactions with the BASEL phage collection. PLoS Biol. 2021, 19, e3001424. [Google Scholar] [CrossRef]
- Dion, M.B.; Oechslin, F.; Moineau, S. Phage diversity, genomics and phylogeny. Nat. Rev. Microbiol. 2020, 18, 125–138. [Google Scholar] [CrossRef]
- Anany, H.; Mahadevan, P.; Turner, D.; Adriaenssens, E.M.; Kropinski, A.M. ICTV Virus Taxonomy Profile: Chaseviridae 2022. J. Gen. Virol. 2022, 103, 001715. [Google Scholar]
- Martinez-Soto, C.E.; Cucic, S.; Lin, J.T.; Kirst, S.; Mahmoud, E.S.; Khursigara, C.M.; Anany, H. PHIDA: A High Throughput Turbidimetric Data Analytic Tool to Compare Host Range Profiles of Bacteriophages Isolated Using Different Enrichment Methods. Viruses 2021, 13, 2120. [Google Scholar] [CrossRef]
- Storms, Z.; Teel, M.; Mercurio, K.; Sauvageau, D. The Virulence Index: A Metric for Quantitative Analysis of Phage Virulence. Phage 2020, 1, 27–36. [Google Scholar] [CrossRef]
- Kropinski, A.M. Practical Advice on the One-Step Growth Curve. Methods Mol. Biol. 2018, 1681, 41–47. [Google Scholar]
- Kropinski, A.M. Measurement of the rate of attachment of bacteriophage to cells. Methods Mol. Biol. 2009, 501, 151–155. [Google Scholar] [PubMed]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef] [PubMed]
- Darling, A.E.; Mau, B.; Perna, N.T. progressiveMauve: Multiple genome alignment with gene gain, loss and rearrangement. PLoS ONE 2010, 5, e11147. [Google Scholar] [CrossRef] [PubMed]
- McNair, K.; Aziz, R.K.; Pusch, G.D.; Overbeek, R.; Dutilh, B.E.; Edwards, R. Phage Genome Annotation Using the RAST Pipeline. Methods Mol. Biol. 2018, 1681, 231–238. [Google Scholar]
- Ramsey, J.; Rasche, H.; Maughmer, C.; Criscione, A.; Mijalis, E.; Liu, M.; Hu, J.C.; Young, R.; Gill, J.J. Galaxy and Apollo as a biologist-friendly interface for high-quality cooperative phage genome annotation. PLoS Comput. Biol. 2020, 16, e1008214. [Google Scholar] [CrossRef]
- Moller, S.; Croning, M.D.; Apweiler, R. Evaluation of methods for the prediction of membrane spanning regions. Bioinformatics 2001, 17, 646–653. [Google Scholar] [CrossRef]
- Kall, L.; Krogh, A.; Sonnhammer, E.L. Advantages of combined transmembrane topology and signal peptide prediction--the Phobius web server. Nucleic Acids Res. 2007, 35, W429–W432. [Google Scholar] [CrossRef]
- Alikhan, N.F.; Petty, N.K.; Ben Zakour, N.L.; Beatson, S.A. BLAST Ring Image Generator (BRIG): Simple prokaryote genome comparisons. BMC Genom. 2011, 12, 402. [Google Scholar] [CrossRef]
- Garneau, J.R.; Depardieu, F.; Fortier, L.C.; Bikard, D.; Monot, M. PhageTerm: A tool for fast and accurate determination of phage termini and packaging mechanism using next-generation sequencing data. Sci. Rep. 2017, 7, 8292. [Google Scholar] [CrossRef]
- Bortolaia, V.; Kaas, R.S.; Ruppe, E.; Roberts, M.C.; Schwarz, S.; Cattoir, V.; Philippon, A.; Allesoe, R.L.; Rebelo, A.R.; Florensa, A.F.; et al. ResFinder 4.0 for predictions of phenotypes from genotypes. J. Antimicrob. Chemother. 2020, 75, 3491–3500. [Google Scholar] [CrossRef]
- Joensen, K.G.; Scheutz, F.; Lund, O.; Hasman, H.; Kaas, R.S.; Nielsen, E.M.; Aarestrup, F.M. Real-time whole-genome sequencing for routine typing, surveillance, and outbreak detection of verotoxigenic Escherichia coli. J. Clin. Microbiol. 2014, 52, 1501–1510. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Li, X.; Xie, Y.; Bi, D.; Sun, J.; Li, J.; Tai, C.; Deng, Z.; Ou, H.Y. ICEberg 2.0: An updated database of bacterial integrative and conjugative elements. Nucleic Acids Res. 2019, 47, D660–D665. [Google Scholar] [CrossRef] [PubMed]
- Moraru, C.; Varsani, A.; Kropinski, A.M. VIRIDIC-A Novel Tool to Calculate the Intergenomic Similarities of Prokaryote-Infecting Viruses. Viruses 2020, 12, 1268. [Google Scholar] [CrossRef] [PubMed]
- NCBI GenBank. 1982. Available online: https://www.ncbi.nlm.nih.gov/nucleotide/ (accessed on 13 August 2022).
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Davis, J.J.; Wattam, A.R.; Aziz, R.K.; Brettin, T.; Butler, R.; Butler, R.M.; Chlenski, P.; Conrad, N.; Dickerman, A.; Dietrich, E.M.; et al. The PATRIC Bioinformatics Resource Center: Expanding data and analysis capabilities. Nucleic Acids Res. 2020, 48, D606–D612. [Google Scholar] [CrossRef]
- Perkins, D.N.; Pappin, D.J.; Creasy, D.M.; Cottrell, J.S. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 1999, 20, 3551–3567. [Google Scholar] [CrossRef]
- Yildirim, Z.; Sakin, T.; Akcelik, M.; Akcelik, N. Identification and characterization of lytic bacteriophages specific to foodborne pathogenic Escherichia coli O157:H7. Food Sci. Technol. Int. 2021, 27, 56–72. [Google Scholar] [CrossRef] [PubMed]
- Necel, A.; Bloch, S.; Nejman-Falenczyk, B.; Grabski, M.; Topka, G.; Dydecka, A.; Kosznik-Kwasnicka, K.; Grabowski, L.; Jurczak-Kurek, A.; Wolkowicz, T.; et al. Characterization of a bacteriophage, vB_Eco4M-7, that effectively infects many Escherichia coli O157 strains. Sci. Rep. 2020, 10, 3743. [Google Scholar] [CrossRef]
- Pham-Khanh, N.H.; Sunahara, H.; Yamadeya, H.; Sakai, M.; Nakayama, T.; Yamamoto, H.; Truong Thi Bich, V.; Miyanaga, K.; Kamei, K. Isolation, Characterisation and Complete Genome Sequence of a Tequatrovirus Phage, Escherichia phage KIT03, Which Simultaneously Infects Escherichia coli O157:H7 and Salmonella enterica. Curr. Microbiol. 2019, 76, 1130–1137. [Google Scholar] [CrossRef]
- Yamaki, S.; Yamazaki, K.; Kawai, Y. Broad host range bacteriophage, EscoHU1, infecting Escherichia coli O157:H7 and Salmonella enterica: Characterization, comparative genomics, and applications in food safety. Int. J. Food Microbiol. 2022, 372, 109680. [Google Scholar] [CrossRef]
- Steensma, H.Y. Adsorption of the defective phage PBS Z1 to Bacillus subtilis 168 Wt. J. Gen. Virol. 1981, 52 Pt 1, 93–101. [Google Scholar] [CrossRef] [PubMed]
- Kropinski, A.M.; Warren, R.A. Isolation and properties of a Pseudomonas acidovorans bacteriophage. J. Gen. Virol. 1970, 6, 85–93. [Google Scholar] [CrossRef] [PubMed]
- Kelln, R.A.; Warren, R.A. Isolation and properties of a bacteriophage lytic for a wide range of pseudomonads. Can. J. Microbiol. 1971, 17, 677–682. [Google Scholar] [CrossRef] [PubMed]
- Storms, Z.J.; Sauvageau, D. Modeling tailed bacteriophage adsorption: Insight into mechanisms. Virology 2015, 485, 355–362. [Google Scholar] [CrossRef] [PubMed]
- Storms, Z.J.; Sauvageau, D. Evidence that the heterogeneity of a T4 population is the result of heritable traits. PLoS ONE 2014, 9, e116235. [Google Scholar] [CrossRef]
- Turner, D.; Adriaenssens, E.M.; Tolstoy, I.; Kropinski, A.M. Phage Annotation Guide: Guidelines for Assembly and High-Quality Annotation. Phage 2021, 2, 170–182. [Google Scholar] [CrossRef]
- Frottin, F.; Martinez, A.; Peynot, P.; Mitra, S.; Holz, R.C.; Giglione, C.; Meinnel, T. The proteomics of N-terminal methionine cleavage. Mol. Cell Proteom. 2006, 5, 2336–2349. [Google Scholar] [CrossRef]
- Helbig, A.O.; Gauci, S.; Raijmakers, R.; van Breukelen, B.; Slijper, M.; Mohammed, S.; Heck, A.J. Profiling of N-acetylated protein termini provides in-depth insights into the N-terminal nature of the proteome. Mol. Cell Proteom. 2010, 9, 928–939. [Google Scholar] [CrossRef]
- Zhang, J.; Sprung, R.; Pei, J.; Tan, X.; Kim, S.; Zhu, H.; Liu, C.F.; Grishin, N.V.; Zhao, Y. Lysine acetylation is a highly abundant and evolutionarily conserved modification in Escherichia coli. Mol. Cell Proteom. 2009, 8, 215–225. [Google Scholar] [CrossRef]
- Christensen, D.G.; Xie, X.; Basisty, N.; Byrnes, J.; McSweeney, S.; Schilling, B.; Wolfe, A.J. Post-translational Protein Acetylation: An Elegant Mechanism for Bacteria to Dynamically Regulate Metabolic Functions. Front. Microbiol. 2019, 10, 1604. [Google Scholar] [CrossRef]
- Moye, Z.D.; Woolston, J.; Sulakvelidze, A. Bacteriophage Applications for Food Production and Processing. Viruses 2018, 10, 205. [Google Scholar] [CrossRef] [PubMed]
- Fernandez, L.; Gutierrez, D.; Garcia, P.; Rodriguez, A. The Perfect Bacteriophage for Therapeutic Applications-A Quick Guide. Antibiotics 2019, 8, 126. [Google Scholar] [CrossRef] [PubMed]
- Hassan, A.Y.; Lin, J.T.; Ricker, N.; Anany, H. The Age of Phage: Friend or Foe in the New Dawn of Therapeutic and Biocontrol Applications? Pharmaceuticals 2021, 14, 199. [Google Scholar] [CrossRef]
- Picozzi, C.; Garcia, P.; Vives, M. Editorial: Bacteriophages to Fight Food-Borne Pathogens/Phages Struggling for Food Safety. Front. Microbiol. 2021, 12, 741387. [Google Scholar] [CrossRef]
- Vikram, A.; Woolston, J.; Sulakvelidze, A. Phage Biocontrol Applications in Food Production and Processing. Curr. Issues Mol. Biol. 2021, 40, 267–302. [Google Scholar] [CrossRef]
- Anany, H.; Brovko, L.Y.; Arutyunov, D.; Poshtiban, N.; Singh, A.; Singh, U.; Brook, M.; Szymanski, C.; Evoy, S.; Griffiths, M.W. Immobilization of Intact Phage and Phage-Derived Proteins for Detection and Biocontrol Purposes. Methods Mol. Biol. 2019, 1898, 89–105. [Google Scholar]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by His Majesty the King in Right of Canada as represented by the Minister of Agriculture and Agri-Food Canada the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lin, J.T.; Kirst, S.; Cucić, S.; Klem, A.; She, Y.-M.; Kropinski, A.M.; Anany, H. Isolation, Characterization, and Genome Analysis of a Novel Bacteriophage, Escherichia Phage vB_EcoM-4HA13, Representing a New Phage Genus in the Novel Phage Family Chaseviridae. Viruses 2022, 14, 2356. https://doi.org/10.3390/v14112356
Lin JT, Kirst S, Cucić S, Klem A, She Y-M, Kropinski AM, Anany H. Isolation, Characterization, and Genome Analysis of a Novel Bacteriophage, Escherichia Phage vB_EcoM-4HA13, Representing a New Phage Genus in the Novel Phage Family Chaseviridae. Viruses. 2022; 14(11):2356. https://doi.org/10.3390/v14112356
Chicago/Turabian StyleLin, Janet T., Sarah Kirst, Stevan Cucić, Alexandra Klem, Yi-Min She, Andrew M. Kropinski, and Hany Anany. 2022. "Isolation, Characterization, and Genome Analysis of a Novel Bacteriophage, Escherichia Phage vB_EcoM-4HA13, Representing a New Phage Genus in the Novel Phage Family Chaseviridae" Viruses 14, no. 11: 2356. https://doi.org/10.3390/v14112356
APA StyleLin, J. T., Kirst, S., Cucić, S., Klem, A., She, Y.-M., Kropinski, A. M., & Anany, H. (2022). Isolation, Characterization, and Genome Analysis of a Novel Bacteriophage, Escherichia Phage vB_EcoM-4HA13, Representing a New Phage Genus in the Novel Phage Family Chaseviridae. Viruses, 14(11), 2356. https://doi.org/10.3390/v14112356






