Viral Small Terminase: A Divergent Structural Framework for a Conserved Biological Function
Abstract
1. Principles of Viral Genome Packaging and the Small Terminase Conundrum
2. Conservation of TerS in the Virosphere
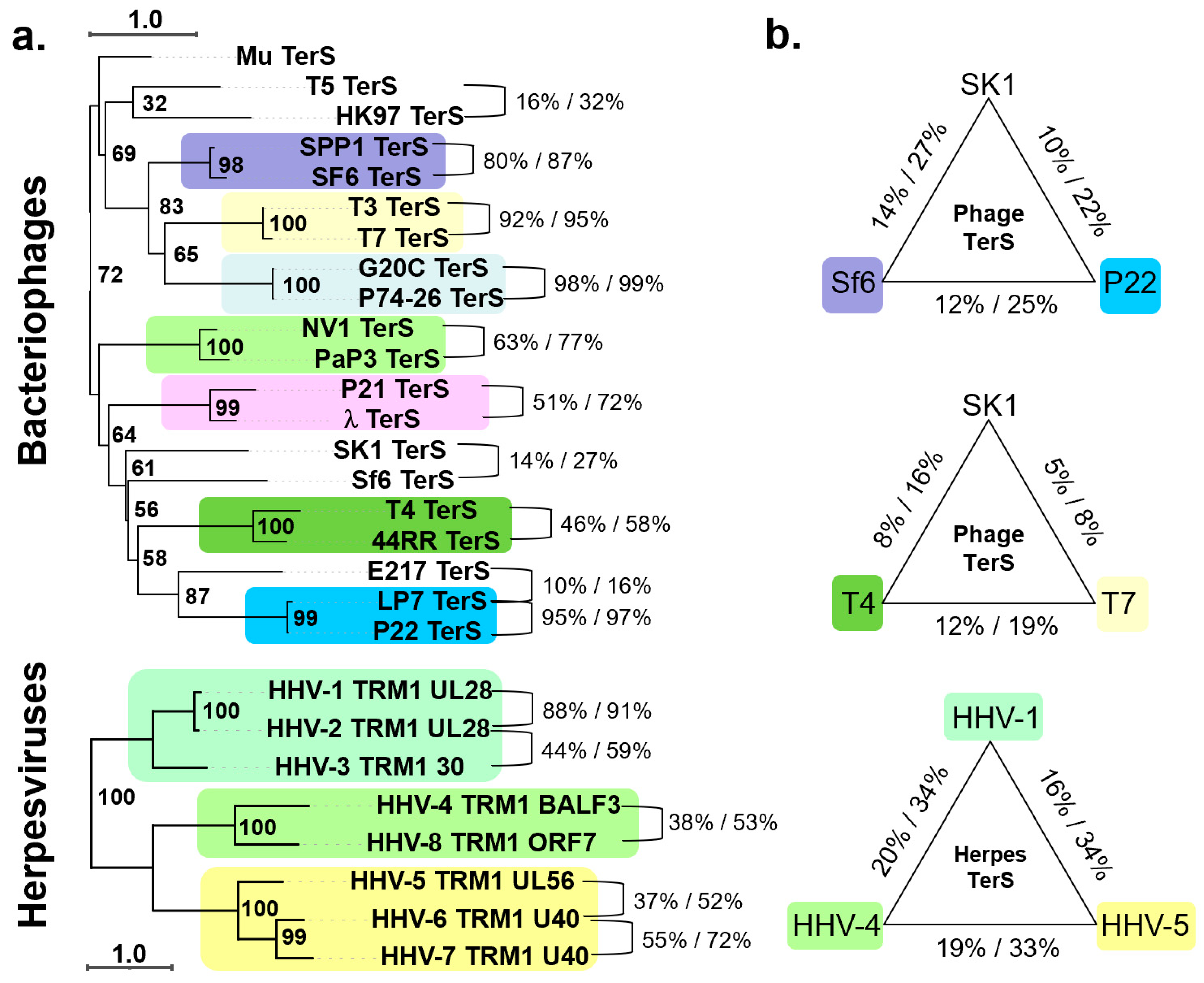
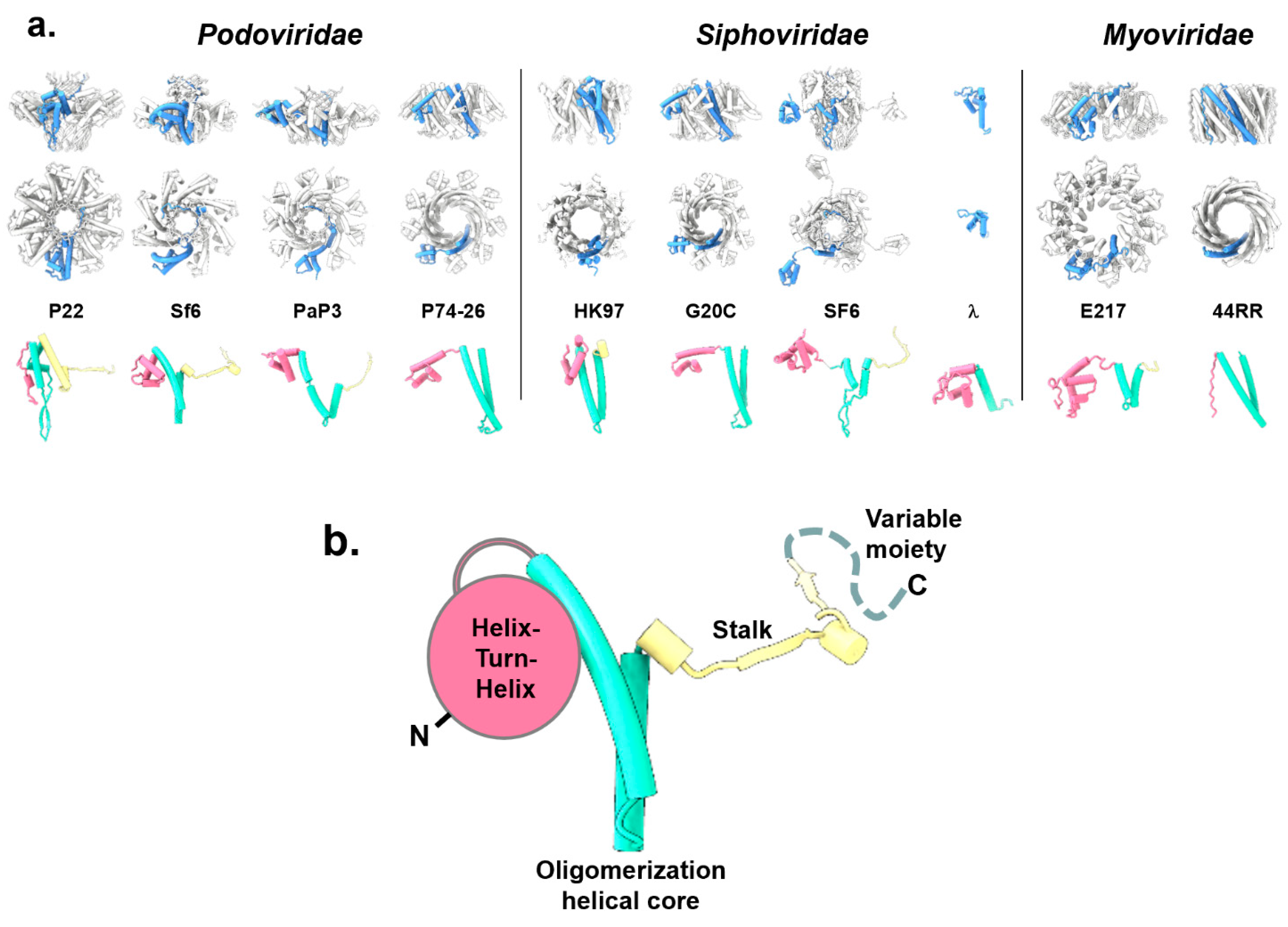
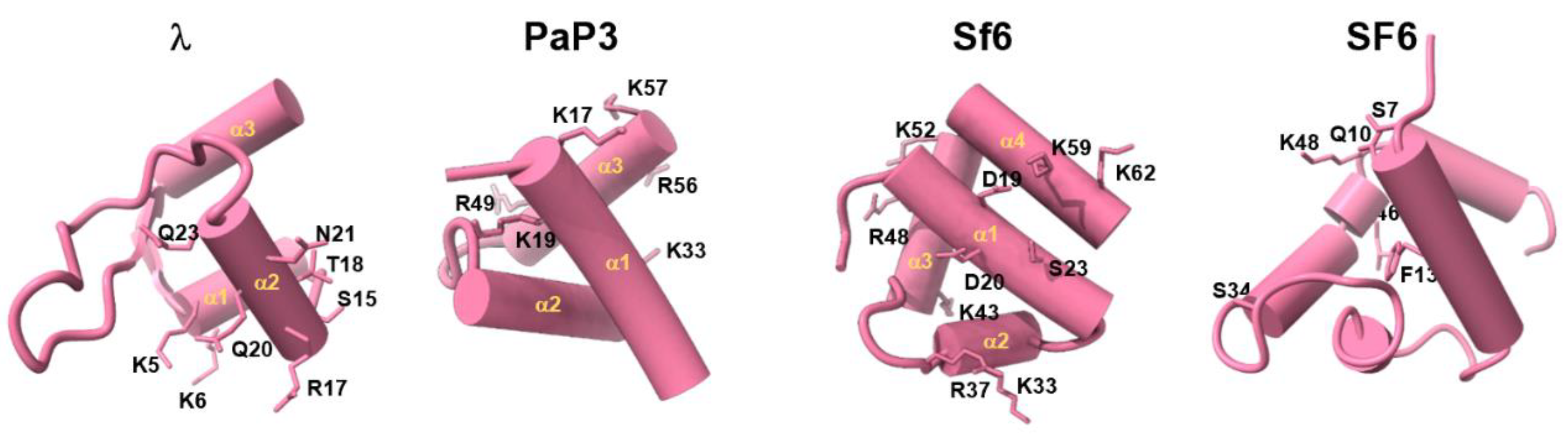
3. Diversification of the TerS Fold and Oligomeric State in Bacteriophages
4. TerS association with TerL and Modulation of TerL Catalytic Activities
5. TerS Function in cos versus pac Packagers
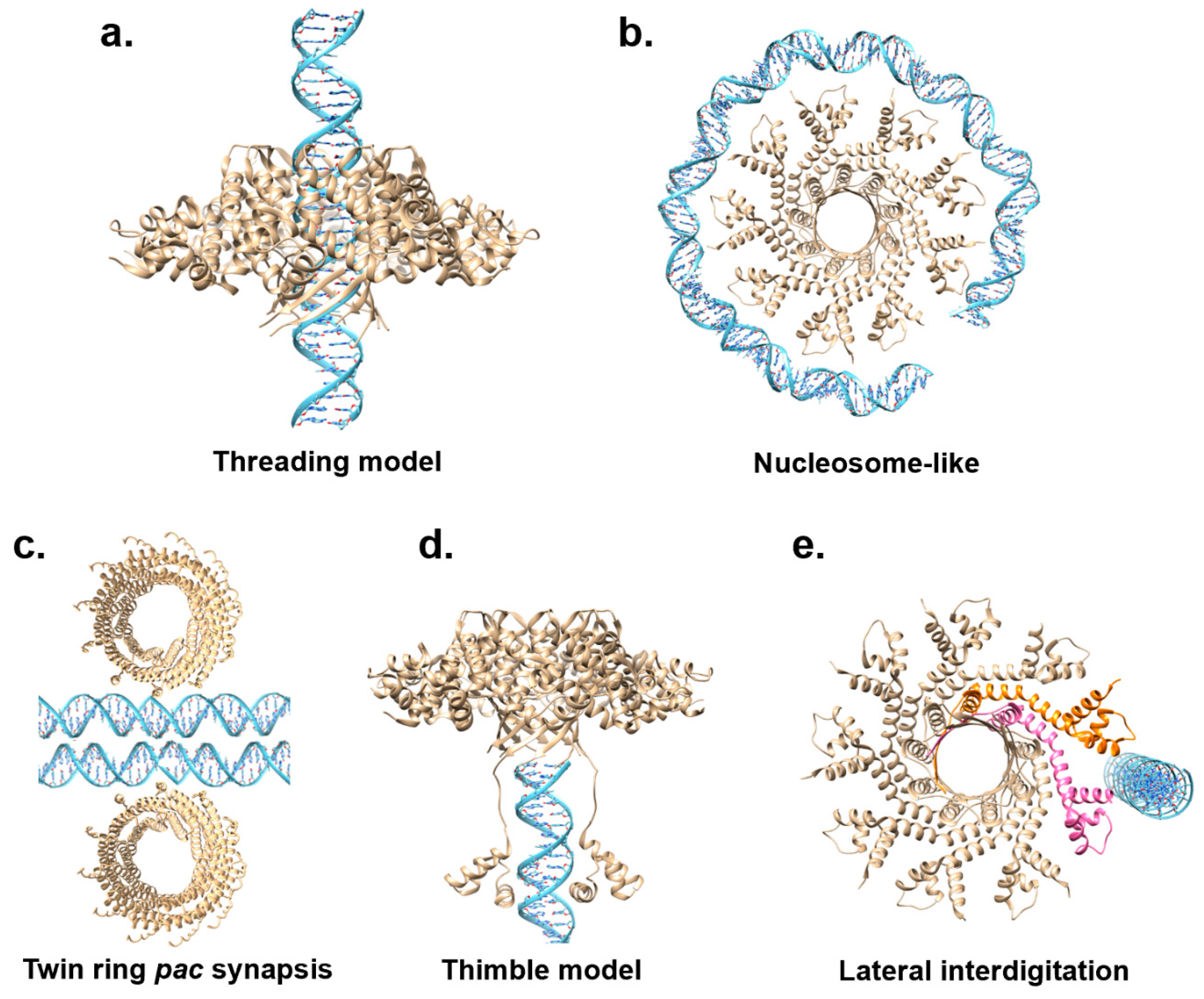
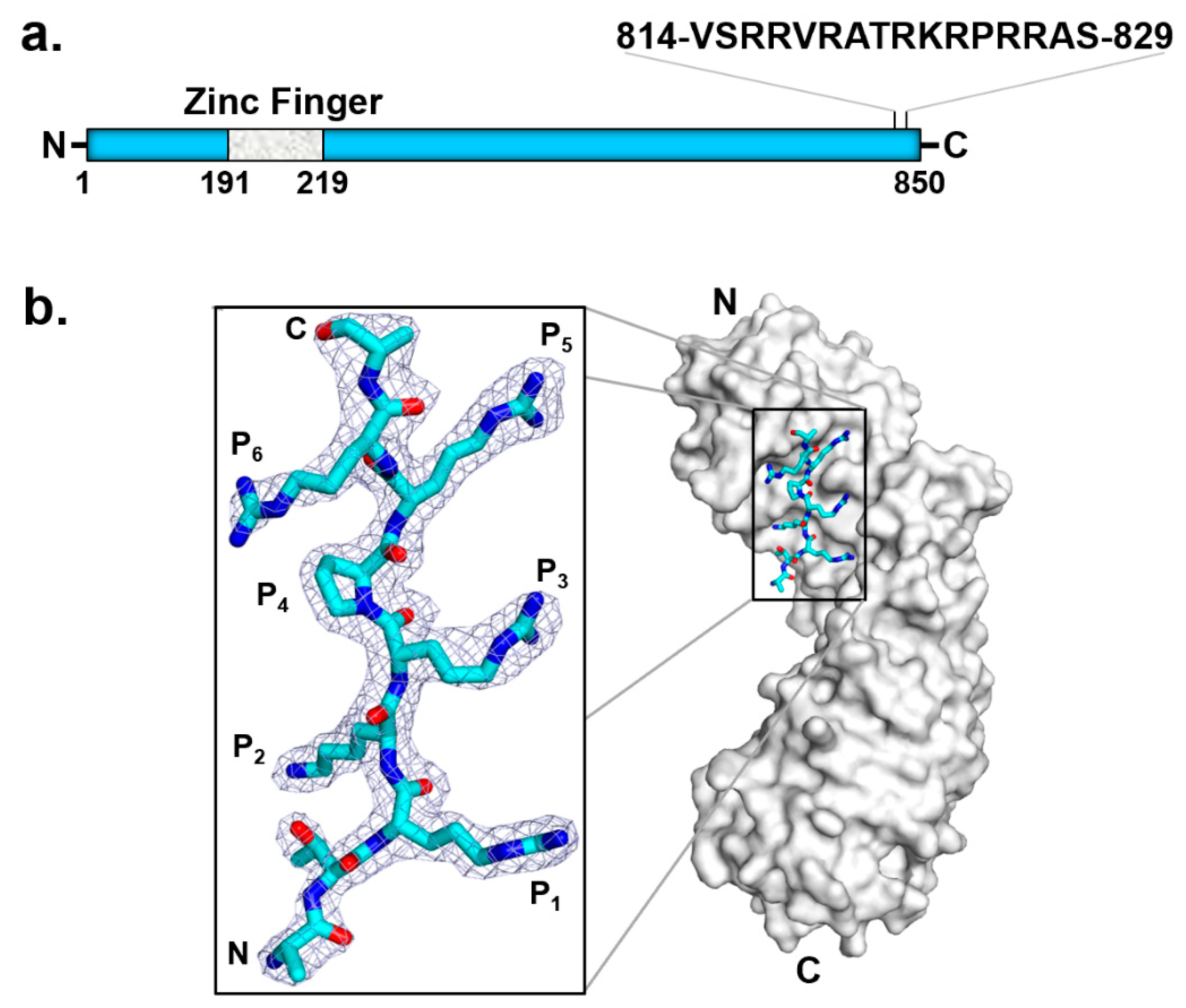
6. Mechanisms of DNA Recognition
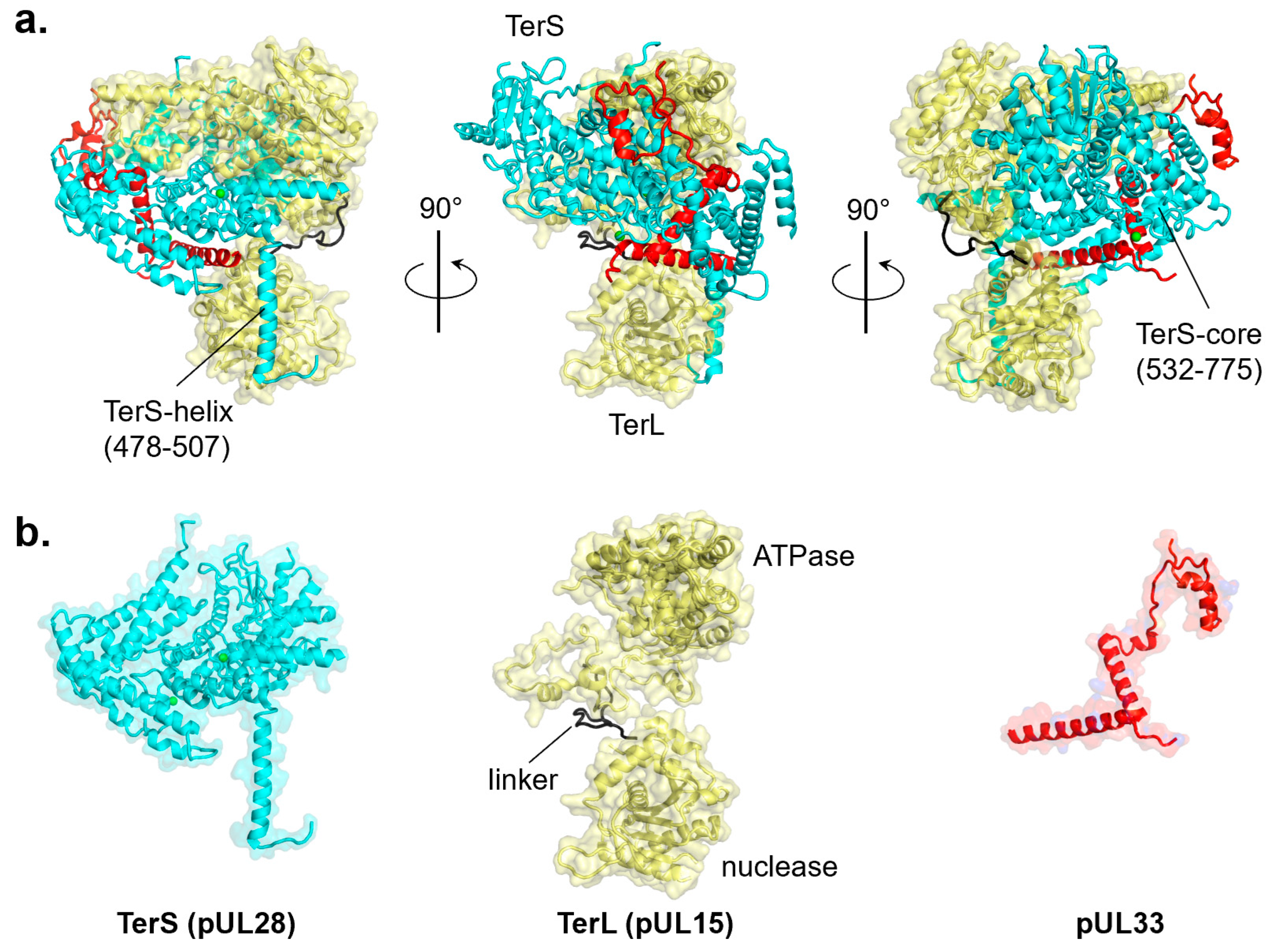
7. Functional Conservation of TerS in Herpesviruses
8. Conclusive Remarks: Challenges and Gaps to Be Filled
- (i)
- The gene encoding TerS has diverged more and faster than TerL or portal proteins, suggesting that this terminase subunit exerts a function that can be conserved with significant variations on the same structural framework. TerS is functionally conserved in herpesviruses but fundamentally reinvented from bacteriophages.
- (ii)
- TerS is essential in vivo but dispensable in vitro where an excess of TerL, viral DNA, and ATP are sufficient to promote energy-dependent genome packaging.
- (iii)
- Phage TerS are oligomeric in bacteriophages, but the stoichiometry of oligomerization varies mainly with nonamers, decamers, and octamers. This suggests that TerS can retain its function with a different stoichiometry of oligomerization. In herpesviruses, TerS is larger than TerL and adopts a helical solenoid-like structure.
- (iv)
- TerS interacts with TerL to promote genome packaging, but this association can be very transient or remarkably stable, suggesting that the heterotypic association of terminase subunits has diverged significantly, even in closely related phages.
- (v)
- TerS association with DNA is fundamentally different in cos versus pac packagers. Cos packagers encode TerSs that make strong and saturable interactions with dsDNA. In contrast, TerS from pac packagers makes weak, sequence-independent contacts with dsDNA.
- (vi)
- Both threading and nucleosome models are incomplete and unsupported by biochemical data. Neither model can explain the sequence-specific recognition of cos sites, a vital function of TerS. Whatever model is contemplated must consider how TerS can read DNA nucleobases. TerS contains Zn-binding domains in herpesvirus, possibly responsible for specific DNA recognition.
- (vii)
- HCMV TerS (pUL56) is the target of letermovir, an FDA-approved antiviral agent of great potential in treating HCMV opportunistic infection in transplant patients.
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Smith, D.E.; Tans, S.J.; Smith, S.B.; Grimes, S.; Anderson, D.L.; Bustamante, C. The bacteriophage straight phi29 portal motor can package DNA against a large internal force. Nature 2001, 413, 748–752. [Google Scholar] [CrossRef]
- Fuller, D.N.; Raymer, D.M.; Kottadiel, V.I.; Rao, V.B.; Smith, D.E. Single phage T4 DNA packaging motors exhibit large force generation, high velocity, and dynamic variability. Proc. Natl. Acad. Sci. USA 2007, 104, 16868–16873. [Google Scholar] [CrossRef] [PubMed]
- Sun, S.; Rao, V.; Rossmann, M.G. Genome packaging in viruses. Curr. Opin. Struct. Biol. 2010, 20, 114–120. [Google Scholar] [CrossRef] [PubMed]
- Rao, V.B.; Feiss, M. The Bacteriophage DNA Packaging Motor. Annu. Rev. Genet. 2008, 42, 647–681. [Google Scholar] [CrossRef] [PubMed]
- Casjens, S.R. The DNA-packaging nanomotor of tailed bacteriophages. Nat. Rev. Genet. 2011, 9, 647–657. [Google Scholar] [CrossRef] [PubMed]
- Guo, P.; Zhao, Z.; Haak, J.; Wang, S.; Wu, D.; Meng, B.; Weitao, T. Common mechanisms of DNA translocation motors in bacteria and viruses using one-way revolution mechanism without rotation. Biotechnol. Adv. 2014, 32, 853–872. [Google Scholar] [CrossRef] [PubMed]
- Smith, D.E. Single-molecule studies of viral DNA packaging. Curr. Opin. Virol. 2011, 1, 134–141. [Google Scholar] [CrossRef]
- Serwer, P.; Jiang, W. Dualities in the analysis of phage DNA packaging motors. Bacteriophage 2012, 2, 239–255. [Google Scholar] [CrossRef][Green Version]
- Olia, A.S.; Prevelige, P.E., Jr.; Johnson, J.E.; Cingolani, G. Three-dimensional structure of a viral genome-delivery portal vertex. Nat. Struct. Mol. Biol. 2011, 18, 597–603. [Google Scholar] [CrossRef] [PubMed]
- Lokareddy, R.K.; Sankhala, R.S.; Roy, A.; Afonine, P.V.; Motwani, T.; Teschke, C.M.; Parent, K.N.; Cingolani, G. Portal protein functions akin to a DNA-sensor that couples genome-packaging to icosahedral capsid maturation. Nat. Commun. 2017, 8, 14310. [Google Scholar] [CrossRef] [PubMed]
- Bayfield, O.W.; Klimuk, E.; Winkler, D.C.; Hesketh, E.L.; Chechik, M.; Cheng, N.; Dykeman, E.C.; Minakhin, L.; Ranson, N.A.; Severinov, K.; et al. Cryo-EM structure and in vitro DNA packaging of a thermophilic virus with supersized T=7 capsids. Proc. Natl. Acad. Sci. USA 2019, 116, 3556–3561. [Google Scholar] [CrossRef] [PubMed]
- Kala, S.; Cumby, N.; Sadowski, P.D.; Hyder, B.Z.; Kanelis, V.; Davidson, A.R.; Maxwell, K.L. HNH proteins are a widespread component of phage DNA packaging machines. Proc. Natl. Acad. Sci. USA 2014, 111, 6022–6027. [Google Scholar] [CrossRef]
- Xin, W.; Feiss, M. Function of IHF in λ DNA Packaging: I. Identification of the Strong Binding Site for Integration Host Factor and the Locus for Intrinsic Bending in cosB. J. Mol. Biol. 1993, 230, 492–504. [Google Scholar] [CrossRef] [PubMed]
- Andrews, B.T.; Catalano, C.E. The Enzymology of a Viral Genome Packaging Motor Is Influenced by the Assembly State of the Motor Subunits. Biochemistry 2012, 51, 9342–9353. [Google Scholar] [CrossRef]
- Poteete, A.R.; Botstein, D. Purification and properties of proteins essential to DNA encapsulation by phage P. Virology 1979, 95, 565–573. [Google Scholar] [CrossRef]
- McNulty, R.; Lokareddy, R.K.; Roy, A.; Yang, Y.; Lander, G.C.; Heck, A.J.; Johnson, J.E.; Cingolani, G. Architecture of the Complex Formed by Large and Small Terminase Subunits from Bacteriophage P. J. Mol. Biol. 2015, 427, 3285–3299. [Google Scholar] [CrossRef]
- Maluf, N.K.; Yang, Q.; Catalano, C.E. Self-association Properties of the Bacteriophage λ Terminase Holoenzyme: Implications for the DNA Packaging Motor. J. Mol. Biol. 2005, 347, 523–542. [Google Scholar] [CrossRef]
- Maluf, N.K.; Gaussier, H.; Bogner, E.; Feiss, M.; Catalano, C.E. Assembly of Bacteriophage Lambda Terminase into a Viral DNA Maturation and Packaging Machine. Biochemistry 2006, 45, 15259–15268. [Google Scholar] [CrossRef] [PubMed]
- Heming, J.D.; Huffman, J.B.; Jones, L.M.; Homa, F.L. Isolation and Characterization of the Herpes Simplex Virus 1 Terminase Complex. J. Virol. 2014, 88, 225–236. [Google Scholar] [CrossRef] [PubMed]
- Sun, S.; Kondabagil, K.; Draper, B.; Alam, T.I.; Bowman, V.D.; Zhang, Z.; Hegde, S.; Fokine, A.; Rossmann, M.G.; Rao, V.B. The Structure of the Phage T4 DNA Packaging Motor Suggests a Mechanism Dependent on Electrostatic Forces. Cell 2008, 135, 1251–1262. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Zhang, X.; Gao, S.; Rao, P.A.; Padilla-Sanchez, V.; Chen, Z.; Sun, S.; Xiang, Y.; Subramaniam, S.; Rao, V.B.; et al. Cryo-EM structure of the bacteriophage T4 portal protein assembly at near-atomic resolution. Nat. Commun. 2015, 6, 7548. [Google Scholar] [CrossRef] [PubMed]
- Dauden, M.I.; Martín-Benito, J.; Sánchez-Ferrero, J.C.; Pulido-Cid, M.; Valpuesta, J.M.; Carrascosa, J.L. Large Terminase Conformational Change Induced by Connector Binding in Bacteriophage T. J. Biol. Chem. 2013, 288, 16998–17007. [Google Scholar] [CrossRef] [PubMed]
- Mao, H.; Saha, M.; Reyes-Aldrete, E.; Sherman, M.B.; Woodson, M.; Atz, R.; Grimes, S.; Jardine, P.J.; Morais, M.C. Structural and Molecular Basis for Coordination in a Viral DNA Packaging Motor. Cell Rep. 2016, 14, 2017–2029. [Google Scholar] [CrossRef] [PubMed]
- Strauss, H.; King, J. Steps in the stabilization of newly packaged DNA during phage P22 Morphogenesis. J. Mol. Biol. 1984, 172, 523–543. [Google Scholar] [CrossRef]
- Zhao, H.; Christensen, T.E.; Kamau, Y.N.; Tang, L. Structures of the phage Sf6 large terminase provide new insights into DNA translocation and cleavage. Proc. Natl. Acad. Sci. USA 2013, 110, 8075–8080. [Google Scholar] [CrossRef]
- Sun, S.; Kondabagil, K.; Gentz, P.M.; Rossmann, M.G.; Rao, V.B. The Structure of the ATPase that Powers DNA Packaging into Bacteriophage T4 Procapsids. Mol. Cell 2007, 25, 943–949. [Google Scholar] [CrossRef] [PubMed]
- Hilbert, B.J.; Hayes, J.A.; Stone, N.P.; Duffy, C.M.; Sankaran, B.; Kelch, B.A. Structure and mechanism of the ATPase that powers viral genome packaging. Proc. Natl. Acad. Sci. USA 2015, 112, E3792–E3799. [Google Scholar] [CrossRef] [PubMed]
- Lokareddy, R.K.; Hou, C.-F.D.; Doll, S.G.; Li, F.; Gillilan, R.E.; Forti, F.; Horner, D.S.; Briani, F.; Cingolani, G. Terminase Subunits from the Pseudomonas-Phage E217. J. Mol. Biol. 2022, 434, 167799. [Google Scholar] [CrossRef] [PubMed]
- Parent, K.N.; Schrad, J.R.; Cingolani, G. Breaking Symmetry in Viral Icosahedral Capsids as Seen through the Lenses of X-ray Crystallography and Cryo-Electron Microscopy. Viruses 2018, 10, 67. [Google Scholar] [CrossRef] [PubMed]
- Bhardwaj, A.; Olia, A.S.; Cingolani, G. Architecture of viral genome-delivery molecular machines. Curr. Opin. Struct. Biol. 2014, 25, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Woodson, M.; Pajak, J.; Mahler, B.P.; Zhao, W.; Zhang, W.; Arya, G.; White, M.A.; Jardine, P.J.; Morais, M.C. A viral genome packaging motor transitions between cyclic and helical symmetry to translocate dsDNA. Sci. Adv. 2021, 7, eabc1955. [Google Scholar] [CrossRef] [PubMed]
- Dai, L.; Singh, D.; Lu, S.; Kottadiel, V.I.; Vafabakhsh, R.; Mahalingam, M.; Chemla, Y.R.; Ha, T.; Rao, V.B. A viral genome packaging ring-ATPase is a flexibly coordinated pentamer. Nat. Commun. 2021, 12, 6548. [Google Scholar] [CrossRef] [PubMed]
- Pajak, J.; Dill, E.; Reyes-Aldrete, E.; White, M.A.; Kelch, B.A.; Jardine, P.J.; Arya, G.; Morais, M.C. Atomistic basis of force generation, translocation, and coordination in a viral genome packaging motor. Nucleic Acids Res. 2021, 49, 6474–6488. [Google Scholar] [CrossRef] [PubMed]
- Rao, V.; Black, L.W. Cloning, overexpression and purification of the terminase proteins gp16 and gp17 of bacteriophage T4: Construction of a defined in-vitro DNA packaging system using purified terminase proteins. J. Mol. Biol. 1988, 200, 475–488. [Google Scholar] [CrossRef]
- Hamada, K.; Fujisawa, H.; Minagawa, T. A defined in vitro system for packaging of bacteriophage T3 DNA. Virology 1986, 151, 119–123. [Google Scholar] [CrossRef]
- Rubinchik, S.; Parris, W.; Gold, M. The in Vitro Translocase Activity of λ Terminase and Its Subunits. Kinetic and biochemical analysis. J. Biol. Chem. 1995, 270, 20059–20066. [Google Scholar] [CrossRef]
- Schmieger, H. Pac sites are indispensable for in vivo packaging of DNA by phage P22. Mol. Gen. Genet. 1984, 195, 252–255. [Google Scholar] [CrossRef]
- Schmieger, H.; Koch, E. In vitro assay of packaging protein gp3 of Salmonella phage P22. Intervirology 1987, 28, 157–162. [Google Scholar] [CrossRef]
- Al-Zahrani, A.S.; Kondabagil, K.; Gao, S.; Kelly, N.; Ghosh-Kumar, M.; Rao, V.B. The Small Terminase, gp16, of Bacteriophage T4 Is a Regulator of the DNA Packaging Motor. J. Biol. Chem. 2009, 284, 24490–24500. [Google Scholar] [CrossRef]
- Kondabagil, K.R.; Zhang, Z.; Rao, V.B. The DNA Translocating ATPase of Bacteriophage T4 Packaging Motor. J. Mol. Biol. 2006, 363, 786–799. [Google Scholar] [CrossRef]
- Black, L.W.; Peng, G. Mechanistic Coupling of Bacteriophage T4 DNA Packaging to Components of the Replication-dependent Late Transcription Machinery. J. Biol. Chem. 2006, 281, 25635–25643. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, L.; Alonso, J.C.; Tavares, P. A Defined in Vitro System for DNA Packaging by the Bacteriophage SPP1: Insights into the Headful Packaging Mechanism. J. Mol. Biol. 2005, 353, 529–539. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Kottadiel, V.I.; Vafabakhsh, R.; Dai, L.; Chemla, Y.R.; Ha, T.; Rao, V.B. A Promiscuous DNA Packaging Machine from Bacteriophage T. PLOS Biol. 2011, 9, e1000592. [Google Scholar] [CrossRef] [PubMed]
- Jackson, E.N.; Laski, F.; Andres, C. Bacteriophage P22 mutants that alter the specificity of DNA packaging. J. Mol. Biol. 1982, 154, 551–563. [Google Scholar] [CrossRef]
- Shinder, G.; Gold, M. The Nul subunit of bacteriophage lambda terminase binds to specific sites in cos DNA. J. Virol. 1988, 62, 387–392. [Google Scholar] [CrossRef] [PubMed]
- Lin, H.; Simon, M.N.; Black, L.W. Purification and Characterization of the Small Subunit of Phage T4 Terminase, gp16, Required for DNA Packaging. J. Biol. Chem. 1997, 272, 3495–3501. [Google Scholar] [CrossRef]
- Casjens, S.; Sampson, L.; Randall, S.; Eppler, K.; Wu, H.; Petri, J.; Schmieger, H. Molecular genetic analysis of bacteriophage P22 gene 3 product, a protein involved in the initiation of headful DNA packaging. J. Mol. Biol. 1992, 227, 1086–1099. [Google Scholar] [CrossRef]
- Chai, S.; Kruft, V.; Alonso, J.C. Analysis of the Bacillus subtilis Bacteriophages SPP1 and SF6 Gene 1 Product: A Protein Involved in the Initiation of Headful Packaging. Virology 1994, 202, 930–939. [Google Scholar] [CrossRef]
- Efron, B.; Halloran, E.; Holmes, S. Bootstrap confidence levels for phylogenetic trees. Proc. Natl. Acad. Sci. USA 1996, 93, 13429. [Google Scholar] [CrossRef]
- Casjens, S.R.; Thuman-Commike, P.A. Evolution of mosaically related tailed bacteriophage genomes seen through the lens of phage P22 virion assembly. Virology 2011, 411, 393–415. [Google Scholar] [CrossRef]
- Heming, J.D.; Conway, J.F.; Homa, F.L. Herpesvirus Capsid Assembly and DNA Packaging. Adv. Anat. Embryol. Cell Biol. 2017, 223, 119–142. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Yang, Q.; Wang, M.; Jia, R.; Chen, S.; Zhu, D.; Liu, M.; Wu, Y.; Zhao, X.; Zhang, S.; et al. Terminase Large Subunit Provides a New Drug Target for Herpesvirus Treatment. Viruses 2019, 11, 219. [Google Scholar] [CrossRef] [PubMed]
- Sievers, F.; Wilm, A.; Dineen, D.; Gibson, T.J.; Karplus, K.; Li, W.; Lopez, R.; McWilliam, H.; Remmert, M.; Söding, J.; et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011, 7, 539. [Google Scholar] [CrossRef] [PubMed]
- Madeira, F.; Pearce, M.; Tivey, A.R.N.; Basutkar, P.; Lee, J.; Edbali, O.; Madhusoodanan, N.; Kolesnikov, A.; Lopez, R. Search and sequence analysis tools services from EMBL-EBI in 2022. Nucleic Acids Res. 2022, 50, W276–W279. [Google Scholar] [CrossRef]
- Guindon, S.; Dufayard, J.-F.; Lefort, V.; Anisimova, M.; Hordijk, W.; Gascuel, O. New Algorithms and Methods to Estimate Maximum-Likelihood Phylogenies: Assessing the Performance of PhyML 3. Syst. Biol. 2010, 59, 307–321. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef]
- Stothard, P. The Sequence Manipulation Suite: JavaScript Programs for Analyzing and Formatting Protein and DNA Sequences. Biotechniques 2000, 28, 1102–1104. [Google Scholar] [CrossRef]
- De Beer, T.; Fang, J.; Ortega, M.; Yang, Q.; Maes, L.; Duffy, C.; Berton, N.; Sippy, J.; Overduin, M.; Feiss, M.; et al. Insights into specific DNA recognition during the assembly of a viral genome packaging machine. Mol. Cell. 2002, 9, 981–991. [Google Scholar] [CrossRef]
- Sun, S.; Gao, S.; Kondabagil, K.; Xiang, Y.; Rossmann, M.G.; Rao, V.B. Structure and function of the small terminase component of the DNA packaging machine in T4-like bacteriophages. Proc. Natl. Acad. Sci. USA 2012, 109, 817–822. [Google Scholar] [CrossRef]
- Fung, H.K.H.; Grimes, S.; Huet, A.; Duda, R.L.; Chechik, M.; Gault, J.; Robinson, C.V.; Hendrix, R.W.; Jardine, P.J.; Conway, J.F.; et al. Structural basis of DNA packaging by a ring-type ATPase from an archetypal viral system. Nucleic Acids Res. 2022; in press. [Google Scholar] [CrossRef]
- Zhao, H.; Kamau, Y.N.; Christensen, T.E.; Tang, L. Structural and Functional Studies of the Phage Sf6 Terminase Small Subunit Reveal a DNA-Spooling Device Facilitated by Structural Plasticity. J. Mol. Biol. 2012, 423, 413–426. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Finch, C.J.; Sequeira, R.D.; Johnson, B.A.; Johnson, J.E.; Casjens, S.R.; Tang, L. Crystal structure of the DNA-recognition component of the bacterial virus Sf6 genome-packaging machine. Proc. Natl. Acad. Sci. USA 2010, 107, 1971–1976. [Google Scholar] [CrossRef] [PubMed]
- Roy, A.; Bhardwaj, A.; Datta, P.; Lander, G.C.; Cingolani, G. Small Terminase Couples Viral DNA Binding to Genome-Packaging ATPase Activity. Structure 2012, 20, 1403–1413. [Google Scholar] [CrossRef] [PubMed]
- Roy, A.; Bhardwaj, A.; Cingolani, G. Crystallization of the nonameric small terminase subunit of bacteriophage P22. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2011, 67, 104–110. [Google Scholar] [CrossRef] [PubMed]
- Němeček, D.; Gilcrease, E.B.; Kang, S.; Prevelige, P.E.; Casjens, S.; Thomas, G.J. Subunit Conformations and Assembly States of a DNA-translocating Motor: The Terminase of Bacteriophage P. J. Mol. Biol. 2007, 374, 817–836. [Google Scholar] [CrossRef] [PubMed]
- Němeček, D.; Lander, G.C.; Johnson, J.E.; Casjens, S.R.; Thomas, G.J. Assembly Architecture and DNA Binding of the Bacteriophage P22 Terminase Small Subunit. J. Mol. Biol. 2008, 383, 494–501. [Google Scholar] [CrossRef] [PubMed]
- Büttner, C.R.; Chechik, M.; Ortiz-Lombardía, M.; Smits, C.; Ebong, I.-O.; Chechik, V.; Jeschke, G.; Dykeman, E.; Benini, S.; Robinson, C.V.; et al. Structural basis for DNA recognition and loading into a viral packaging motor. Proc. Natl. Acad. Sci. USA 2012, 109, 811–816. [Google Scholar] [CrossRef] [PubMed]
- Niazi, M.; Florio, T.J.; Yang, R.; Lokareddy, R.K.; Swanson, N.A.; Gillilan, R.E.; Cingolani, G. Biophysical analysis of Pseudomonas-phage PaP3 small terminase suggests a mechanism for sequence-specific DNA-binding by lateral interdigitation. Nucleic Acids Res. 2020, 48, 11721–11736. [Google Scholar] [CrossRef] [PubMed]
- Loredo-Varela, J.; Chechik, M.; Levdikov, V.M.; Abd-El-Aziz, A.; Minakhin, L.; Severinov, K.; Smits, C.; Antson, A.A. The putative small terminase from the thermophilic dsDNA bacteriophage G20C is a nine-subunit oligomer. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2013, 69, 876–879. [Google Scholar] [CrossRef] [PubMed]
- Hayes, J.A.; Hilbert, B.J.; Gaubitz, C.; Stone, N.P.; Kelch, B.A. A thermophilic phage uses a small terminase protein with a fixed helix–turn–helix geometry. J. Biol. Chem. 2020, 295, 3783–3793. [Google Scholar] [CrossRef]
- Kühlbrandt, W. The Resolution Revolution. Science 2014, 343, 1443–1444. [Google Scholar] [CrossRef]
- Yang, Y.; Yang, P.; Wang, N.; Chen, Z.; Su, D.; Zhou, Z.H.; Rao, Z.; Wang, X. Architecture of the herpesvirus genome-packaging complex and implications for DNA translocation. Protein Cell 2020, 11, 339–351. [Google Scholar] [CrossRef] [PubMed]
- Roy, A.; Cingolani, G. Structure of P22 Headful Packaging Nuclease. J. Biol. Chem. 2012, 287, 28196–28205. [Google Scholar] [CrossRef] [PubMed]
- Benini, S.; Chechik, M.; Lombardía, M.O.; Polier, S.; Leech, A.; Shevtsov, M.B.; Alonso, J.C. The 1.58 Å resolution structure of the DNA-binding domain of bacteriophage SF6 small terminase provides new hints on DNA binding. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2013, 69, 376–381. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Berton, N.; Manning, A.M.C.; Catalano, C.E. Domain Structure of gpNu1, a Phage Lambda DNA Packaging Protein. Biochemistry 1999, 38, 14238–14247. [Google Scholar] [CrossRef] [PubMed]
- Frackman, S.; Siegele, D.A.; Feiss, M. The terminase of bacteriophage λ: Functional domains for cosB binding and multimer assembly. J. Mol. Biol. 1985, 183, 225–238. [Google Scholar] [CrossRef]
- Yang, T.-C.; Ortiz, D.; Nosaka, L.; Lander, G.C.; Catalano, C.E. Thermodynamic Interrogation of the Assembly of a Viral Genome Packaging Motor Complex. Biophys. J. 2015, 109, 1663–1675. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Beilstein, F.; Higgs, M.R.; Stow, N.D. Mutational Analysis of the Herpes Simplex Virus Type 1 DNA Packaging Protein UL. J. Virol. 2009, 83, 8938–8945. [Google Scholar] [CrossRef]
- Baumann, R.G.; Black, L.W. Isolation and Characterization of T4 Bacteriophage gp17 Terminase, a Large Subunit Multimer with Enhanced ATPase Activity. J. Biol. Chem. 2003, 278, 4618–4627. [Google Scholar] [CrossRef]
- Leffers, G.; Rao, V. Biochemical Characterization of an ATPase Activity Associated with the Large Packaging Subunit gp17 from Bacteriophage T. J. Biol. Chem. 2000, 275, 37127–37136. [Google Scholar] [CrossRef] [PubMed]
- Gual, A.; Camacho, A.G.; Alonso, J.C. Functional Analysis of the Terminase Large Subunit, G2P, of Bacillus subtilis Bacteriophage SPP1. J. Biol. Chem. 2000, 275, 35311–35319. [Google Scholar] [CrossRef]
- Hilbert, B.J.; Hayes, J.A.; Stone, N.P.; Xu, R.-G.; Kelch, B.A. The large terminase DNA packaging motor grips DNA with its ATPase domain for cleavage by the flexible nuclease domain. Nucleic Acids Res. 2017, 45, 3591–3605. [Google Scholar] [CrossRef]
- Ghosh-Kumar, M.; Alam, T.I.; Draper, B.; Stack, J.D.; Rao, V.B. Regulation by interdomain communication of a headful packaging nuclease from bacteriophage T4. Nucleic Acids Res. 2010, 39, 2742–2755. [Google Scholar] [CrossRef]
- Alam, T.I.; Draper, B.; Kondabagil, K.; Rentas, F.J.; Ghosh-Kumar, M.; Sun, S.; Rossmann, M.G.; Rao, V.B. The Headful Packaging Nuclease of Bacteriophage T4. Mol. Microbiol. 2008, 69, 1180–1190. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Catalano, C.E. ATP serves as a nucleotide switch coupling the genome maturation and packaging motor complexes of a virus assembly machine. Nucleic Acids Res. 2020, 48, 5006–5015. [Google Scholar] [CrossRef]
- Renault, L.; Kuhlmann, J.; Henkel, A.; Wittinghofer, A. Structural Basis for Guanine Nucleotide Exchange on Ran by the Regulator of Chromosome Condensation (RCC1). Cell 2001, 105, 245–255. [Google Scholar] [CrossRef]
- Casjens, S.R.; Gilcrease, E.B. Determining DNA Packaging Strategy by Analysis of the Termini of the Chromosomes in Tailed-Bacteriophage Virions. In Methods in Molecular Biology; Springer: Berlin/Heidelberg, Germany, 2009; Volume 502, pp. 91–111. [Google Scholar] [CrossRef]
- Catalano, C.E. (Ed.) Viral Genome Packaging Machines: Genetics, Structure and Mechanism; Kluwer Academic/Plenum Publishers: New York, NY, USA, 2005; pp. 1–4. [Google Scholar]
- Catalano, C.E.; Cue, D.; Feiss, M. Virus DNA packaging: The strategy used by phage? Mol. Microbiol. 1995, 16, 1075–1086. [Google Scholar] [CrossRef]
- Feiss, M.; Kobayashi, I.; Widner, W. Separate sites for binding and nicking of bacteriophage lambda DNA by terminase. Proc. Natl. Acad. Sci. USA 1983, 80, 955–959. [Google Scholar] [CrossRef] [PubMed]
- Casjens, S.; Weigele, P. Headful DNA Packaging by Bacteriophage P22 in Viral Genome Packaging Machines: Genetics, Structure and Mechanism. In The Bacteriophages; Catalano, C., Ed.; Landes Publishing: Georgetown, TX, USA; Oxford University Press: Oxford, UK, 2005; pp. 457–468. [Google Scholar]
- Wu, H.; Sampson, L.; Parr, R.; Casjens, S. The DNA site utilized by bacteriophage P22 for initiation of DNA packaging. Mol. Microbiol. 2002, 45, 1631–1646. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, L.; Tavares, P.; Alonso, J.C. Headful DNA packaging: Bacteriophage SPP1 as a model system. Virus Res. 2013, 173, 247–259. [Google Scholar] [CrossRef]
- Chai, S.; Lurz, R.; Alonso, J.C. The Small Subunit of the Terminase Enzyme ofBacillus subtilisBacteriophage SPP1 forms a Specialized Nucleoprotein Complex with the Packaging Initiation Region. J. Mol. Biol. 1995, 252, 386–398. [Google Scholar] [CrossRef] [PubMed]
- Tavares, P.; Santos, M.; Lurz, R.; Morelli, G.; De Lencastre, H.; Trautner, T.A. Identification of a gene in Bacillus subtilis bacteriophage SPP1 determining the amount of packaged DNA. J. Mol. Biol. 1992, 225, 81–92. [Google Scholar] [CrossRef]
- Greive, S.J.; Fung, H.K.; Chechik, M.; Jenkins, H.T.; Weitzel, S.E.; Aguiar, P.M.; Brentnall, A.S.; Glousieau, M.; Gladyshev, G.V.; Potts, J.R.; et al. DNA recognition for virus assembly through multiple sequence-independent interactions with a helix-turn-helix motif. Nucleic Acids Res. 2015, 44, 776–789. [Google Scholar] [CrossRef] [PubMed]
- Gao, S.; Zhang, L.; Rao, V.B. Exclusion of small terminase mediated DNA threading models for genome packaging in bacteriophage T. Nucleic Acids Res. 2016, 44, 4425–4439. [Google Scholar] [CrossRef] [PubMed]
- Nolan, J.M.; Petrov, V.; Bertrand, C.; Krisch, H.M.; Karam, J.D. Genetic diversity among five T4-like bacteriophages. Virol. J. 2006, 3, 30. [Google Scholar] [CrossRef] [PubMed]
- Gao, S.; Rao, V.B. Specificity of Interactions among the DNA-packaging Machine Components of T4-related Bacteriophages. J. Biol. Chem. 2011, 286, 3944–3956. [Google Scholar] [CrossRef]
- Dedeo, C.L.; Cingolani, G.; Teschke, C.M. Portal Protein: The Orchestrator of Capsid Assembly for the dsDNA Tailed Bacteriophages and Herpesviruses. Annu. Rev. Virol. 2019, 6, 141–160. [Google Scholar] [CrossRef]
- Hou, C.-F.D.; Swanson, N.A.; Li, F.; Yang, R.; Lokareddy, R.K.; Cingolani, G. Cryo-EM Structure of a Kinetically Trapped Dodecameric Portal Protein from the Pseudomonas-phage PaP3. J. Mol. Biol. 2022, 434, 167537. [Google Scholar] [CrossRef]
- Dixit, A.B.; Ray, K.; Black, L.W. A viral small terminase subunit (TerS) twin ring pac synapsis DNA packaging model is supported by fluorescent fusion proteins. Virology 2019, 536, 39–48. [Google Scholar] [CrossRef] [PubMed]
- Nadal, M.; Mas, P.J.; Blanco, A.G.; Arnan, C.; Solà, M.; Hart, D.J.; Coll, M. Structure and inhibition of herpesvirus DNA packaging terminase nuclease domain. Proc. Natl. Acad. Sci. USA 2010, 107, 16078–16083. [Google Scholar] [CrossRef] [PubMed]
- Sigamani, S.S.; Zhao, H.; Kamau, Y.N.; Baines, J.D.; Tang, L. The Structure of the Herpes Simplex Virus DNA-Packaging Terminase pUL15 Nuclease Domain Suggests an Evolutionary Lineage among Eukaryotic and Prokaryotic Viruses. J. Virol. 2013, 87, 7140–7148. [Google Scholar] [CrossRef]
- Smits, C.; Chechik, M.; Kovalevskiy, O.; Shevtsov, M.B.; Foster, A.W.; Alonso, J.C.; Antson, A. Structural basis for the nuclease activity of a bacteriophage large terminase. EMBO Rep. 2009, 10, 592–598. [Google Scholar] [CrossRef]
- Sankhala, R.S.; Lokareddy, R.K.; Cingolani, G. Divergent Evolution of Nuclear Localization Signal Sequences in Herpesvirus Terminase Subunits. J. Biol. Chem. 2016, 291, 11420–11433. [Google Scholar] [CrossRef] [PubMed]
- Bogner, E.; Radsak, K.; Stinski, M.F. The gene product of human cytomegalovirus open reading frame UL56 binds the pac motif and has specific nuclease activity. J. Virol. 1998, 72, 2259–2264. [Google Scholar] [CrossRef]
- Giesen, K.; Radsak, K.; Bogner, E. Targeting of the gene product encoded by ORF UL56 of human cytomegalovirus into viral replication centers. FEBS Lett. 2000, 471, 215–218. [Google Scholar] [CrossRef]
- Giesen, K.; Radsak, K.; Bogner, E. The potential terminase subunit of human cytomegalovirus, pUL56, is translocated into the nucleus by its own nuclear localization signal and interacts with importin α. J. Gen. Virol. 2000, 81, 2231–2244. [Google Scholar] [CrossRef] [PubMed]
- Berger, J.M. SnapShot: Nucleic Acid Helicases and Translocases. Cell 2008, 134, 888–888.e1. [Google Scholar] [CrossRef]
- Savva, C.G.; Holzenburg, A.; Bogner, E. Insights into the structure of human cytomegalovirus large terminase subunit pUL56. FEBS Lett. 2004, 563, 135–140. [Google Scholar] [CrossRef]
- Kobe, B.; Kajava, A.V. When protein folding is simplified to protein coiling: The continuum of solenoid protein structures. Trends Biochem. Sci. 2000, 25, 509–515. [Google Scholar] [CrossRef]
- Cook, A.; Bono, F.; Jinek, M.; Conti, E. Structural Biology of Nucleocytoplasmic Transport. Annu. Rev. Biochem. 2007, 76, 647–671. [Google Scholar] [CrossRef] [PubMed]
- Bogner, E. Human cytomegalovirus terminase as a target for antiviral chemotherapy. Rev. Med. Virol. 2002, 12, 115–127. [Google Scholar] [CrossRef] [PubMed]
- Champier, G.; Couvreux, A.; Hantz, S.; Rametti, A.; Mazeron, M.-C.; Bouaziz, S.; Denis, F.; Alain, S. Putative functional domains of human cytomegalovirus pUL56 involved in dimerization and benzimidazole D-ribonucleoside activity. Antivir. Ther. 2008, 13, 643–654. [Google Scholar] [CrossRef] [PubMed]
- Lischka, P.; Hewlett, G.; Wunberg, T.; Baumeister, J.; Paulsen, D.; Goldner, T.; Ruebsamen-Schaeff, H.; Zimmermann, H. In Vitro and In Vivo Activities of the Novel Anticytomegalovirus Compound AIC. Antimicrob. Agents Chemother. 2010, 54, 1290–1297. [Google Scholar] [CrossRef] [PubMed]
- Goldner, T.; Hewlett, G.; Ettischer, N.; Ruebsamen-Schaeff, H.; Zimmermann, H.; Lischka, P. The Novel Anticytomegalovirus Compound AIC246 (Letermovir) Inhibits Human Cytomegalovirus Replication through a Specific Antiviral Mechanism That Involves the Viral Terminase. J. Virol. 2011, 85, 10884–10893. [Google Scholar] [CrossRef]
- Goldner, T.; Hempel, C.; Ruebsamen-Schaeff, H.; Zimmermann, H.; Lischka, P. Geno- and Phenotypic Characterization of Human Cytomegalovirus Mutants Selected In Vitro after Letermovir (AIC246) Exposure. Antimicrob. Agents Chemother. 2014, 58, 610–613. [Google Scholar] [CrossRef] [PubMed]
| Virus | Protomer M.W. (kDa) | Channel Diameter Min–Max (Å) | Number of Subunits | Accession Number(s) | Methodology | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| EMDB | PDB | NMR | X-ray | Cryo-EM | ||||||
| Bacteriophage | Podoviridae | P22 | 18.6 | 20–25 | 9 | 3P9A | + | |||
| Sf6 | 15.5 | ~19 | 8 | 4DYQ | + | |||||
| PaP3 | 16.6 | 9–15 | 9 | 6W7T, 7JOQ | + | |||||
| Siphoviridae | P74-26 | 18.7 | ~30 | 9 | 21,012 | 6V1I | + | |||
| HK97 | 18.4 | ~18 | 9 | 6Z6E | + | |||||
| G20C | 18.8 | ~30 | 9 | 6EJQ, 4XVN | + | |||||
| SF6 (SPP1-like) | 16.0 | 11–29 | 9 | 3ZQM, 3ZQN, 3ZQO, 3ZQP, 3ZQQ, 4ZC3, 2CMP | + | |||||
| Lambda | 20.4 | n/a | 2 * | 1J9I | + | |||||
| Myoviridae | E217 | 21.3 | 22–52 | 10 | 26,858 | 7UXE | + | |||
| 44RR | 17.3 | 24–32 | 11,12 | 3TXQ, 3TXS | + | |||||
| Herpesviruses | HSV-1 | 85.6 | n/a | n/a | nd ** | 6M5S | + | |||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lokareddy, R.K.; Hou, C.-F.D.; Li, F.; Yang, R.; Cingolani, G. Viral Small Terminase: A Divergent Structural Framework for a Conserved Biological Function. Viruses 2022, 14, 2215. https://doi.org/10.3390/v14102215
Lokareddy RK, Hou C-FD, Li F, Yang R, Cingolani G. Viral Small Terminase: A Divergent Structural Framework for a Conserved Biological Function. Viruses. 2022; 14(10):2215. https://doi.org/10.3390/v14102215
Chicago/Turabian StyleLokareddy, Ravi K., Chun-Feng David Hou, Fenglin Li, Ruoyu Yang, and Gino Cingolani. 2022. "Viral Small Terminase: A Divergent Structural Framework for a Conserved Biological Function" Viruses 14, no. 10: 2215. https://doi.org/10.3390/v14102215
APA StyleLokareddy, R. K., Hou, C.-F. D., Li, F., Yang, R., & Cingolani, G. (2022). Viral Small Terminase: A Divergent Structural Framework for a Conserved Biological Function. Viruses, 14(10), 2215. https://doi.org/10.3390/v14102215






