Activation of Host Cellular Signaling and Mechanism of Enterovirus 71 Viral Proteins Associated with Hand, Foot and Mouth Disease
Abstract
1. Introduction
2. Molecular Mechanisms Associated with Host-Pathogen Interaction in EV71
3. Immune Cells Involvement during Evasion Process
3.1. Study of Animal Pathogenesis Using EV71
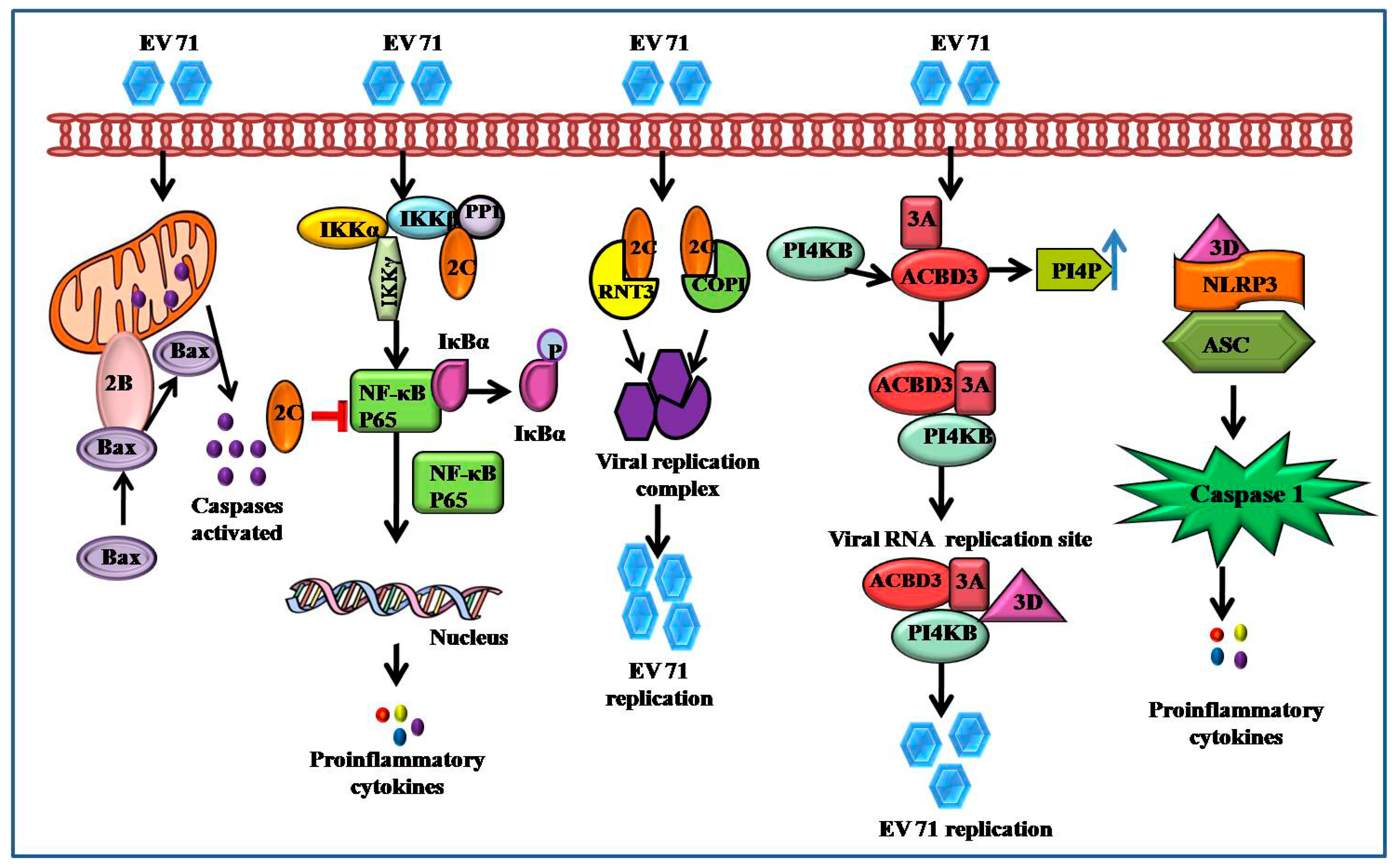
3.2. Apoptosis
3.3. Autophagy
3.4. Innate Immunity
3.5. Acquired Immunity
3.6. Immune Evasion
4. Characterization of Coding and Non-Coding Regions of EV71
4.1. VP1
4.2. VP2–VP3
4.3. VP4
4.4. 2A–2C and 3A–3D
4.5. 2A and 3C Proteases
5. Immune Related Signaling Pathways Associated with EV71
5.1. MAPK Signaling Activated by EV71
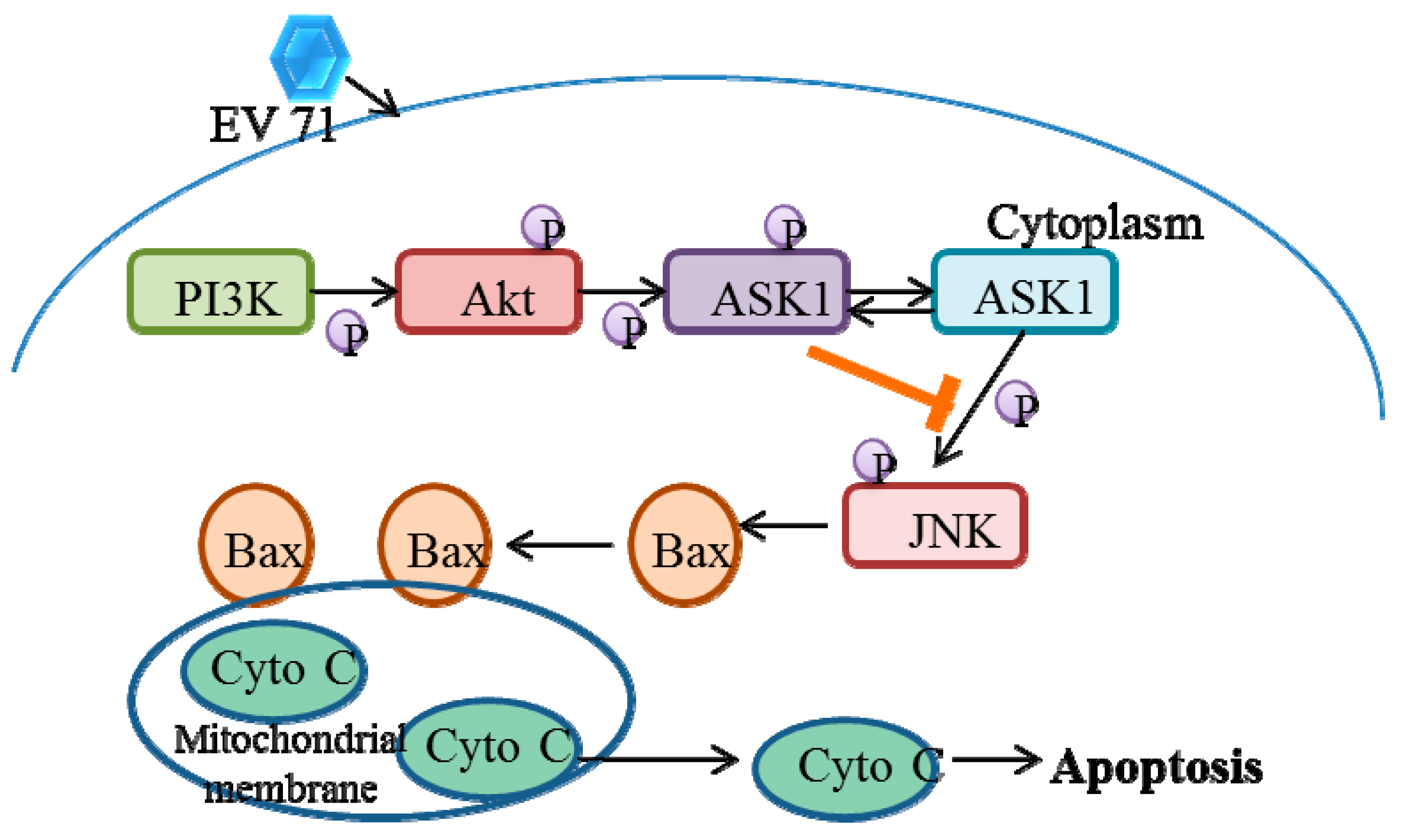
5.2. EV71 Induces Phosphatidylinositol 3-Kinase (PI3K) Signaling
5.3. EV71 Activates Calcium (Ca2+)-Dependent Signaling
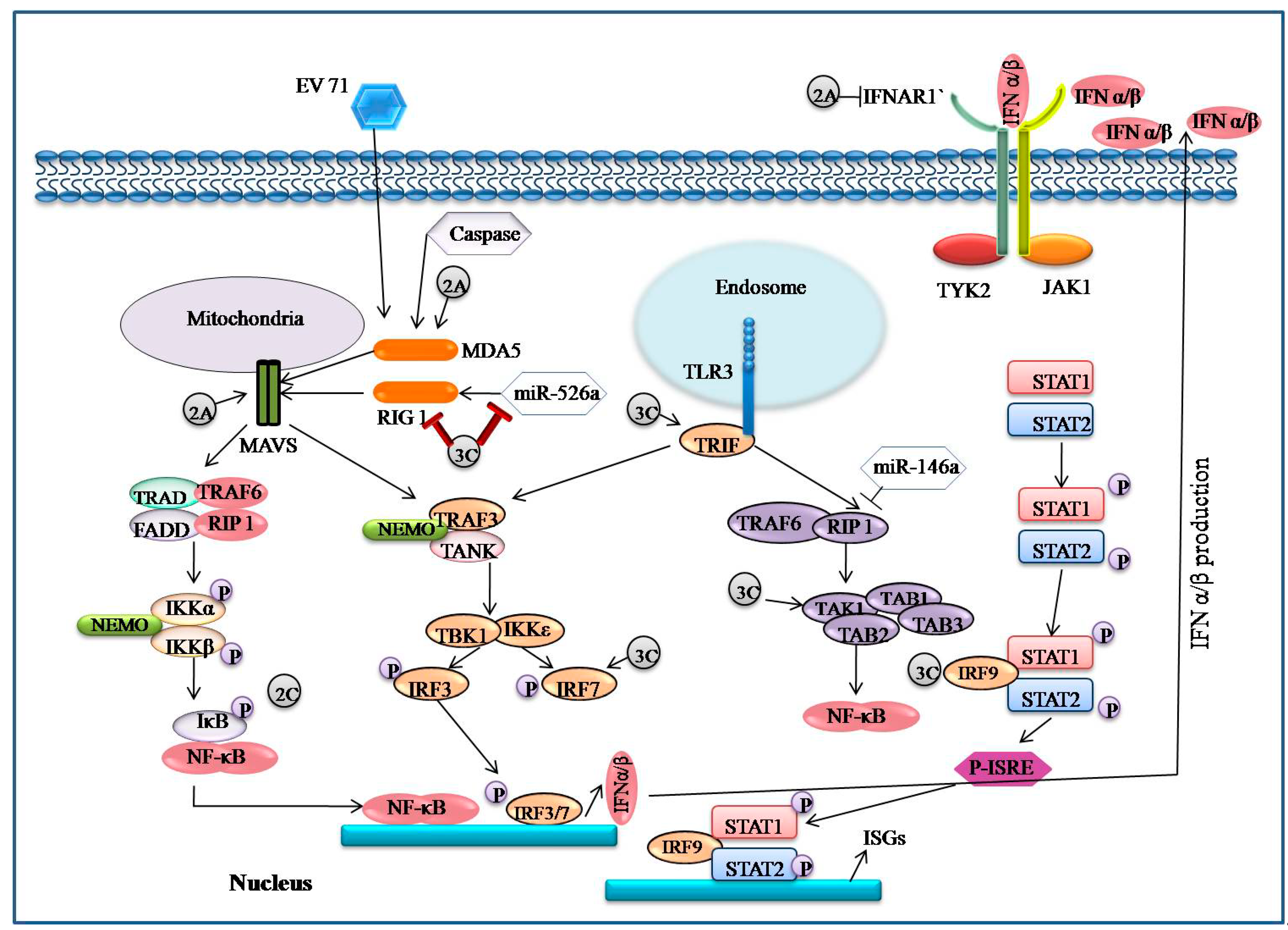
5.4. EV71 Encoded Proteases Inhibit MAVS-Mediated Antiviral Signaling
5.5. EV71 Infection-Associated IFN Signaling
5.6. EV71 Interacts with IRF Signaling
5.7. EV71 Triggers NF-κB Signaling
5.8. EV71 Interacts with TLR Signaling
6. Preventive and Therapeutic Strategiesand Control of EV71 (Pathological Perspective)
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Puenpa, J.; Wanlapakorn, N.; Vongpunsawad, S.; Poovorawan, Y. The history of enterovirus A71 out-breaks and molecular epidemiology in the Asia-Pacific region. J. Biomed. Sci. 2019, 26, 75. [Google Scholar] [CrossRef] [PubMed]
- McMinn, P.C. An overview of the evolution of enterovirus 71 and its clinical and public health significance. FEMS Microbial. Rev. 2002, 26, 91–107. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Qi, M.; Ma, C.; Yang, M.; Huang, P.; Sun, J.; Shi, J.; Hu, Y. Natural intertypic and intratypic re-combinants of enterovirus 71 from mainland China during 2009–2018: A complete genome analysis. Virus Genes 2021, 57, 172–180. [Google Scholar] [CrossRef]
- Lu, G.; Qi, J.; Chen, Z.; Xu, X.; Gao, F.; Lin, D.; Qian, W.; Liu, H.; Jiang, H.; Yan, J.; et al. Enterovirus 71 and Coxsackievirus A16 3C Proteases: Binding to Rupintrivir and Their Substrates and Anti-Hand, Foot, and Mouth Disease Virus Drug Design. J. Virol. 2011, 85, 10319–10331. [Google Scholar] [CrossRef]
- Huang, K.-Y.A.; Chen, M.-F.; Huang, Y.-C.; Shih, S.-R.; Chiu, C.-H.; Lin, J.-J.; Wang, J.-R.; Tsao, K.-C.; Lin, T.-Y. Epitope-associated and specificity-focused features of EV71-neutralizing antibody repertoires from plasmablasts of infected children. Nat. Commun. 2017, 8, 1762. [Google Scholar] [CrossRef]
- Pathinayake, P.S.; Hsu, A.C.-Y.; Wark, P.A.B. Innate Immunity and Immune Evasion by Enterovirus 71. Viruses 2015, 7, 6613–6630. [Google Scholar] [CrossRef]
- Dang, D.; Zhang, C.; Zhang, R.; Wu, W.; Chen, S.; Ren, J.; Zhang, P.; Zhou, G.; Feng, D.; Sun, T. In-volvement of inducible nitric oxide synthase and mitochondrial dysfunction in the pathogenesis of en-terovirus 71 infection. Oncotarget 2017, 8, 81014–81026. [Google Scholar] [CrossRef]
- Duan, G.; Yang, H.; Shi, L.; Sun, W.; Sui, M.; Zhang, R.; Wang, X.; Wang, F.; Zhang, W.; Xi, Y.; et al. Serum Inflammatory Cytokine Levels Correlate with Hand-Foot-Mouth Disease Severity: A Nested Serial Case-Control Study. PLoS ONE 2014, 9, e112676. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Wang, B.; Wu, S.; Wen, Y.; Wang, X.; Song, X.; Zhang, J.; Hou, L.; Chen, W. PD169316, a specific p38 inhibitor, shows antiviral activity against Enterovirus71. Virology 2017, 508, 150–158. [Google Scholar] [CrossRef] [PubMed]
- Hsiao, H.B.; Chou, A.H.; Lin, S.I.; Chen, I.H.; Lien, S.P.; Liu, C.C.; Chong, P.; Liu, S.J. Toll-like receptor 9-mediated protection of enterovirus 71 infection in mice is due to the release of danger-associated molecular patterns. J. Virol. 2014, 88, 11658–11670. [Google Scholar] [CrossRef] [PubMed]
- Zhu, K.; Yang, J.; Luo, K.; Yang, C.; Zhang, N.; Xu, R.; Chen, J.; Jin, M.; Xu, B.; Guo, N.; et al. TLR3 sig-naling in macrophages is indispensable for the protective immunity of invariant natural killer T cells against enterovirus 71 infection. PLoS Pathog. 2015, 11, e1004613. [Google Scholar] [CrossRef]
- Zhao, Q.; Xiong, Y.; Xu, J.; Chen, S.; Li, P.; Huang, Y.; Wang, Y.; Chen, W.X.; Wang, B. Host MicroRNA hsa-miR-494-3p Promotes EV71 Replication by Directly Targeting PTEN. Front. Cell Infect. Microbiol. 2018, 8, 278. [Google Scholar] [CrossRef] [PubMed]
- Pawson, T. Protein modules and signalling networks. Nature 1995, 373, 573–580. [Google Scholar] [CrossRef]
- Yamayoshi, S.; Fujii, K.; Koike, S. Receptors for enterovirus 71. Emerg. Microbes Infect. 2014, 3, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Yuan, M.; Yan, J.; Xun, J.; Chen, C.; Zhang, Y.; Wang, M.; Chu, W.; Song, Z.; Hu, Y.; Zhang, S.; et al. Enhanced human enterovirus 71 infection by endocytosis inhibitors reveals multiple entry pathways by enterovirus causing hand-foot-and-mouth diseases. Virol. J. 2018, 15, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.-L.; Liu, Y.-G.; Zhou, Y.-T.; Zhao, P.; Ren, H.; Xiao, M.; Zhu, Y.-Z.; Qi, Z.-T. Endophilin-A2-mediated endocytic pathway is critical for enterovirus 71 entry into caco-2 cells. Emerg. Microbes Infect. 2019, 8, 773–786. [Google Scholar] [CrossRef]
- Renard, H.-F.; Simunovic, M.; Lemiere, J.; Boucrot, E.; Garcia-Castillo, M.D.; Arumugam, S.; Chambon, V.; Lamaze, C.; Wunder, C.; Kenworthy, A.; et al. Endophilin-A2 functions in membrane scission in clathrin-independent endocytosis. Nature 2014, 517, 493–496. [Google Scholar] [CrossRef]
- Cui, L.; Guo, X.; Qi, Y.; Qi, X.; Ge, Y.; Shi, Z.; Wu, T.; Shan, J.; Shan, Y.; Zhu, Z.; et al. Identification of microRNAs Involved in the Host Response to Enterovirus 71 Infection by a Deep Sequencing Approach. J. Biomed. Biotechnol. 2010, 2010, 425939. [Google Scholar] [CrossRef]
- Liao, Y.-W.; Ho, B.-C.; Chen, M.-H.; Yu, S.-L. Enterovirus 71 Infection Shapes Host T Cell Receptor Repertoire and Presumably Expands VP1-Specific TCRβ CDR3 Cluster. Pathogens 2020, 9, 121. [Google Scholar] [CrossRef] [PubMed]
- Wu, K.X.; Phuektes, P.; Kumar, P.; Goh, G.Y.; Moreau, D.; Chow, V.T.; Bard, F.; Chu, J.J. Human ge-nome-wide RNAi screen reveals host factors required for enterovirus 71 replication. Nat. Commun. 2016, 7, 13150. [Google Scholar] [CrossRef] [PubMed]
- Ho, B.C.; Yu, S.L.; Chen, J.J.; Chang, S.Y.; Yan, B.S.; Hong, Q.S.; Singh, S.; Kao, C.L.; Chen, H.Y.; Su, K.Y.; et al. Enterovirus-induced miR-141 con-tributes to shutoff of host protein translation by targeting the translation initiation factor eIF4E. Cell Host Microbe 2011, 9, 58–69. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, J.; Li, Q. Immune Evasion of Enteroviruses Under Innate Immune Monitoring. Front. Microbiol. 2018, 9, 1866. [Google Scholar] [CrossRef]
- Cifuente, J.O.; Moratorio, G. Evolutionary and Structural Overview of Human Picornavirus Capsid An-tibody Evasion. Front. Cell Infect Microbiol. 2019, 9, 283. [Google Scholar] [CrossRef] [PubMed]
- Sim, A.C.N.; Luhur, A.; Tan, T.M.C.; Chow, V.T.K.; Poh, C.L. RNA interference against Enterovirus 71 infection. Virology 2005, 341, 72–79. [Google Scholar] [CrossRef]
- Chan, L.G.; Parashar, U.D.; Lye, M.S.; Ong, F.G.L.; Zaki, S.R.; Alexander, J.P.; Ho, K.K.; Han, L.L.; Pallansch, M.A.; Suleiman, A.B.; et al. Deaths of Children during an Outbreak of Hand, Foot, and Mouth Disease in Sarawak, Malaysia: Clinical and Pathological Characteristics of the Disease. Clin. Infect. Dis. 2000, 31, 678–683. [Google Scholar] [CrossRef] [PubMed]
- Nagata, N.; Iwasaki, T.; Ami, Y. Differential localization of neurons susceptible to enterovirus 71 and po-liovirus type 1 in the central nervous system of cynomolgus monkeys after intravenous inoculation. J. Gen. Virol. 2004, 85, 2981–2989. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Cui, W.; Liu, L.; Wang, J.; Zhao, H.; Liao, Y.; Na, R.; Dong, C.; Wang, L.; Xie, Z.; et al. Pathogenesis study of enterovirus 71 infection in rhesus monkeys. Lab. Investig. 2011, 91, 1337–1350. [Google Scholar] [CrossRef]
- Arit, C.M.; Shimizu, H.; Nagata, N.; Ami, Y.; Suzaki, Y.; Sata, T.; Iwasaki, T.; Miyamura, T. Temperature-sensitive mutants of enterovirus 71 show attenuation in cynomolgus monkeys. J. Gen. Virol. 2005, 86, 1391–1401. [Google Scholar] [CrossRef]
- Yamayoshi, S.; Koike, S. Identification of a Human SCARB2 Region That Is Important for Enterovirus 71 Binding and Infection. J. Virol. 2011, 85, 4937–4946. [Google Scholar] [CrossRef] [PubMed]
- Nishimura, Y.; Shimojima, M.; Tano, Y.; Miyamura, T.; Wakita, T.; Shimizu, H. Human P-selectin glyco-protein ligand-1 is a functional receptor for enterovirus 71. Nat. Med. 2009, 15, 794–797. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.H.; Liang, C.T.; Jiang, S.T.; Chen, K.H.; Yang, C.C.; Cheng, M.L.; Ho, H.Y. A novel murine model expressing a chimeric mSCARB2/hSCARB2 receptor is highly susceptible to oral infection with clinical isolates of enterovirus 71. J. Virol. 2019, 93, e00183-19. [Google Scholar] [CrossRef]
- Jin, Y.; Sun, T.; Zhou, G.; Li, D.; Chen, S.; Zhang, W.; Li, X.; Zhang, R.; Yang, H.; Duan, G. Pathogenesis Study of Enterovirus 71 Using a Novel Human SCARB2 Knock-In Mouse Model. mSphere 2021, 6, e01048-20. [Google Scholar] [CrossRef]
- Huang, S.-C.; Chang, C.-L.; Wang, P.-S.; Tsai, Y.; Liu, H.-S. Enterovirus 71-induced autophagy detected in vitro and in vivo promotes viral replication. J. Med. Virol. 2009, 81, 1241–1252. [Google Scholar] [CrossRef]
- Li, M.-L.; Hsu, T.-A.; Chen, T.-C.; Chang, S.-C.; Lee, J.-C.; Chen, C.-C.; Stollar, V.; Shih, S.-R. The 3C Protease Activity of Enterovirus 71 Induces Human Neural Cell Apoptosis. Virology 2002, 293, 386–395. [Google Scholar] [CrossRef]
- Chang, S.-C.; Lin, J.-Y.; Lo, L.Y.-C.; Li, M.-L.; Shih, S.-R. Diverse apoptotic pathways in enterovirus 71–infected cells. J. NeuroVirology 2004, 10, 338–349. [Google Scholar] [CrossRef]
- Liang, C.-C.; Sun, M.-J.; Lei, H.-Y.; Chen, S.-H.; Yu, C.-K.; Liu, C.-C.; Wang, J.-R.; Yeh, T.-M. Human endothelial cell activation and apoptosis induced by enterovirus 71 infection. J. Med. Virol. 2004, 74, 597–603. [Google Scholar] [CrossRef]
- Chen, L.-C.; Shyu, H.-W.; Chen, S.-H.; Lei, H.-Y.; Yu, C.-K.; Yeh, T.-M. Enterovirus 71 infection induces Fas ligand expression and apoptosis of Jurkat cells. J. Med. Virol. 2006, 78, 780–786. [Google Scholar] [CrossRef]
- Wong, W.-R.; Chen, Y.-Y.; Yang, S.-M.; Chen, Y.-L.; Horng, J.-T. Phosphorylation of PI3K/Akt and MAPK/ERK in an early entry step of enterovirus 71. Life Sci. 2005, 78, 82–90. [Google Scholar] [CrossRef]
- Tung, W.H.; Lee, I.T.; Hsieh, H.L.; Yang, C.M. EV71 induces COX-2 expres-sion via c-Src/PDGFR/PI3K/Akt/p42/p44 MAPK/AP-1 and NF-kappaB in rat brainastrocytes. J. Cell. Physiol. 2010, 224, 376–386. [Google Scholar] [CrossRef]
- Wang, B.; Zhang, H.; Zhu, M.; Luo, Z.J.; Peng, Y.H. MEK1-ERKs signal cascade isrequired for the repli-cation of Enterovirus 71 (EV71). Antivir. Res. 2012, 93, 110–117. [Google Scholar] [CrossRef]
- Zhang, H.; Li, F.; Pan, Z.; Wu, Z.; Wang, Y.; Cui, Y. Activation of PI3K/Akt pathway limits JNK-mediated apoptosis during EV71 infection. Virus Res. 2014, 192, 74–84. [Google Scholar] [CrossRef]
- Yorimitsu, T.; Nair, U.; Yang, Z.; Klionsky, D.J. Endoplasmic reticulum stress triggers autophagy. J. Biol. Chem. 2006, 281, 30299–30304. [Google Scholar] [CrossRef]
- Jheng, J.-R.; Lau, K.S.; Tang, W.-F.; Wu, M.-S.; Horng, J.-T. Endoplasmic reticulum stress is induced and modulated by enterovirus 71. Cell. Microbiol. 2010, 12, 796–813. [Google Scholar] [CrossRef]
- Brennan, K.; Bowie, A.G. Activation of host pattern recognition receptors by viruses. Curr. Opin. Microbiol. 2010, 13, 503–507. [Google Scholar] [CrossRef]
- Lei, X.; Sun, Z.; Liu, X.; Jin, Q.; He, B.; Wang, J. Cleavage of the Adaptor Protein TRIF by Enterovirus 71 3C Inhibits Antiviral Responses Mediated by Toll-Like Receptor 3. J. Virol. 2011, 85, 8811–8818. [Google Scholar] [CrossRef]
- Hsia, S.H.; Wu, C.T.; Chang, J. Predictors of unfavorable outcomes in enterovirus 71-related cardiopul-monary failure in children. Pediatr. Infect. Dis. J. 2005, 24, 331–334. [Google Scholar] [CrossRef]
- Chang, L.-Y.; Lee, C.-Y.; Kao, C.-L.; Fang, T.-Y.; Lu, C.-Y.; Lee, P.-I.; Huang, L.-M. Hand, Foot and Mouth Disease Complicated with Central Nervous System Involvement in Taiwan in 1980–1981. J. Formos. Med Assoc. 2007, 106, 173–176. [Google Scholar] [CrossRef]
- Diedrich, S.; Weinbrecht, A.; Schreier, E. Seroprevalence and molecular epidemiology of enterovirus 71 in Germany. Arch. Virol. 2009, 154, 1139–1142. [Google Scholar] [CrossRef]
- Yang, C.; Deng, C.; Wan, J.; Zhu, L.; Leng, Q. Neutralizing antibody response in the patients with hand, foot and mouth disease to enterovirus 71 and its clinical implications. Virol. J. 2011, 8, 306. [Google Scholar] [CrossRef]
- Chang, L.-Y.; Hsiung, C.A.; Lu, C.-Y.; Lin, T.-Y.; Huang, F.-Y.; Lai, Y.-H.; Chiang, Y.-P.; Chiang, B.-L.; Lee, C.-Y.; Huang, L.-M. Status of Cellular Rather Than Humoral Immunity is Correlated with Clinical Outcome of Enterovirus 71. Pediatr. Res. 2006, 60, 466–471. [Google Scholar] [CrossRef]
- Chen, X.; Zhang, Q.; Li, J.; Cao, W.; Zhang, J.-X.; Zhang, L.; Zhang, W.; Shao, Z.-J.; Yan, Y. Analysis of recombination and natural selection in human enterovirus 71. Virology 2010, 398, 251–261. [Google Scholar] [CrossRef]
- Kiener, T.K.; Jia, Q.; Lim, X.F.; He, F.; Meng, T.; Chow, V.T.K.; Kwang, J. Characterization and specificity of the linear epitope of the enterovirus 71 VP2 protein. Virol. J. 2012, 9, 55. [Google Scholar] [CrossRef]
- Rowlands, D.J.; Tuthill, T.J.; Groppelli, E.; Rowlands, D.J. Picornaviruses. Curr. Top Microbiol Immunol. 2010, 343, 43–89. [Google Scholar]
- Tan, Y.W.; Hong, W.J.; Chu, J.J.H. Inhibition of enterovirus VP4 myristoylation is a potential antiviral strategy for hand, foot and mouth disease. Antivir. Res. 2016, 133, 191–195. [Google Scholar] [CrossRef]
- Hsu, Y.Y.; Liu, Y.N.; Wang, W.; Kao, F.J.; Kung, S.H. Invivodynamics of enterovirus protease revealed by fluorescence resonance emissiontransfer (FRET) based on a novel FRET pair. Biochem. Biophys. Res. Commun. 2007, 353, 939–945. [Google Scholar] [CrossRef]
- Ventoso, I.; Carrasco, L. A poliovirus 2A(pro) mutant unable to cleave 3CD shows inefficient viral protein synthesis and transactivation defects. J. Virol. 1995, 69, 6280–6288. [Google Scholar] [CrossRef]
- Wang, B.; Xi, X.; Lei, X.; Zhang, X.; Cui, S.; Wang, J.; Jin, Q.; Zhao, Z. Enterovirus 71 Protease 2Apro Targets MAVS to Inhibit Anti-Viral Type I Interferon Responses. PLoS Pathog. 2013, 9, e1003231. [Google Scholar] [CrossRef]
- Kuo, R.-L.; Kao, L.-T.; Lin, S.-J.; Wang, R.Y.-L.; Shih, S.-R. MDA5 Plays a Crucial Role in Enterovirus 71 RNA-Mediated IRF3 Activation. PLoS ONE 2013, 8, e63431. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Yi, L.; Zhao, J.; Yu, J.; Chen, Y.; Lin, M.C.; Kung, H.-F.; He, M.-L. Enterovirus 71 Disrupts Interferon Signaling by Reducing the Level of Interferon Receptor 1. J. Virol. 2012, 86, 3767–3776. [Google Scholar] [CrossRef] [PubMed]
- Xie, S.; Wang, K.; Yu, W.; Lu, W.; Xu, K.; Wang, J.; Ye, B.; Schwarz, W.; Jin, Q.; Sun, B. DIDS blocks a chloride-dependent current that is mediated by the 2B protein of enterovirus 71. Cell Res. 2011, 21, 1271–1275. [Google Scholar] [CrossRef]
- Cong, H.; Du, N.; Yang, Y.; Song, L.; Zhang, W.; Tien, P. Enterovirus 71 2B Induces Cell Apoptosis by Directly Inducing the Conformational Activation of the Proapoptotic Protein Bax. J. Virol. 2016, 90, 9862–9877. [Google Scholar] [CrossRef]
- Guan, H.; Tian, J.; Qin, B.; Wojdyla, J.A.; Wang, B.; Zhao, Z.; Wang, M.; Cui, S. Crystal structure of 2C helicase from enterovirus 71. Sci. Adv. 2017, 3, e1602573. [Google Scholar] [CrossRef]
- Hober, D.; Sané, F.; Riedweg, K.; Moumna, I.; Goffard, A.; Choteau, L.; Kazali, E.; Desaillou, R. Viruses and Type 1 Diabetes: Focus on the Enteroviruses. InType 1 Diabetes; IntechOpen: London, UK, 2013. [Google Scholar]
- Laitinen, O.H.; Svedin, E.; Kapell, S.; Nurminen, A.; Hytönen, V.P.; Flodström-Tullberg, M. Enteroviral proteases: Structure, host interactions and pathogenicity. Rev. Med. Virol. 2016, 26, 251–267. [Google Scholar] [CrossRef] [PubMed]
- Weng, K.-F.; Li, M.-L.; Hung, C.-T.; Shih, S.-R. Enterovirus 71 3C Protease Cleaves a Novel Target CstF-64 and Inhibits Cellular Polyadenylation. PLoS Pathog. 2009, 5, e1000593. [Google Scholar] [CrossRef]
- Guo, Y.; Wang, Y.; Cao, L.; Wang, P.; Qing, J.; Zheng, Q.; Shang, L.; Yin, Z.; Sun, Y. A Conserved Inhibitory Mechanism of a Lycorine Derivative against Enterovirus and Hepatitis C Virus. Antimicrob. Agents Chemother. 2016, 60, 913–924. [Google Scholar] [CrossRef] [PubMed]
- Bogoyevitch, M.A.; Kobe, B. Uses for JNK: The Many and Varied Substrates of the c-Jun N-Terminal Kinases. Microbiol. Mol. Biol. Rev. 2006, 70, 1061–1095. [Google Scholar] [CrossRef] [PubMed]
- Krishna, M.; Narang, H. The complexity of mitogen-activated protein kinases (MAPKs) made simple. Experientia 2008, 65, 3525–3544. [Google Scholar] [CrossRef]
- Leong, S.Y.; Ong, B.K.T.; Chu, J.J.H. The Role of Misshapen NCK-related kinase (MINK), a Novel Ste20 Family Kinase, in the IRES-Mediated Protein Translation of Human Enterovirus 71. PLoS Pathog. 2015, 11, e1004686. [Google Scholar] [CrossRef] [PubMed]
- Ono, K.; Ohtomo, T.; Sato, S.; Sugamata, Y.; Suzuki, M.; Hisamoto, N.; Ninomiya-Tsuji, J.; Tsuchiya, M.; Matsumoto, K. An evolutionarily conserved motif in the TAB1 C-terminal region is necessary for interaction with and activation of TAK1 MAPKKK. J. Biol. Chem. 2001, 276, 24396–24400. [Google Scholar] [CrossRef] [PubMed]
- Lei, X.; Han, N.; Xiao, X.; Jin, Q.; He, B.; Wang, J. Enterovirus 71 3C inhibits cytokine expression through cleavage of the TAK1/TAB1/TAB2/TAB3 complex. J. Virol. 2014, 88, 9830–9841. [Google Scholar] [CrossRef]
- Wei, L.; Zhu, Z.; Wang, J.; Liu, J. JNK and p38 Mitogen-Activated Protein Kinase Pathways Contribute to Porcine Circovirus Type 2 Infection. J. Virol. 2009, 83, 6039–6047. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ceballos-Olvera, I.; Chávez-Salinas, S.; Medina, F.; Ludert, J.E.; del Angel, R.M. JNK phosphorylation, induced during dengue virus infection, is important for viral infection and requires the presence of cholesterol. J. Virol. 2010, 396, 30–36. [Google Scholar] [CrossRef]
- Bryk, D.; Olejarz, W.; Zapolska-Downar, D. Mitogen-activated protein kinases in atherosclerosis. Postepy Hig. Med. Dosw. 2014, 68, 10–22. [Google Scholar] [CrossRef]
- Waetzig, V.; Czeloth, K.; Hidding, U.; Mielke, K.; Kanzow, M.; Brecht, S.; Goetz, M.; Lucius, R.; Herdegen, T.; Hanisch, U.-K. c-Jun N-terminal kinases (JNKs) mediate pro-inflammatory actions of microglia. Glia 2005, 50, 235–246. [Google Scholar] [CrossRef] [PubMed]
- Sukhumavasi, W.; Egan, C.E.; Denkers, E.Y. Mouse neutrophils require JNK2 MAPK for Toxoplasma gondii-induced IL-12p40 and CCL2/MCP-1 release. J. Immunol. 2007, 179, 3570–3577. [Google Scholar] [CrossRef]
- Peng, H.; Shi, M.; Zhang, L.; Li, Y.; Sun, J.; Zhang, L.; Wang, X.; Xu, X.; Zhang, X.; Mao, Y.; et al. Activation of JNK1/2 and p38 MAPK signaling pathways promotes enterovirus 71 infection in immature dendritic cells. BMC Microbiol. 2014, 14, 147. [Google Scholar] [CrossRef] [PubMed]
- Turner, N.A.; Warburton, P.; O’Regan, D.J.; Ball, S.G.; Porter, K.E. Modulatory effect of interleukin-1α on expression of structural matrix proteins, MMPs and TIMPs in human cardiac myofibroblasts: Role of p38 MAP kinase. Matrix Bio. 2010, 29, 613–620. [Google Scholar] [CrossRef]
- Bardwell, A.J.; Frankson, E.; Bardwell, L. Selectivity of Docking Sites in MAPK Kinases. J. Biol. Chem. 2009, 284, 13165–13173. [Google Scholar] [CrossRef]
- Mastruzzo, C.; Crimi, N.; Vancheri, C. Role of oxidative stress in pulmonary fibrosis. Monaldi Arch. Chest Dis. Arch. Per Le Mal. Del Torace 2002, 57, 173–176. [Google Scholar]
- Kim, A.H.; Khursigara, G.; Sun, X.; Franke, T.F.; Chao, M.V. Akt Phosphorylates and Negatively Regulates Apoptosis Signal-Regulating Kinase 1. Mol. Cell. Biol. 2001, 21, 893–901. [Google Scholar] [CrossRef]
- Madrid, L.V.; Wang, C.Y.; Guttridge, D.C.; Schottelius, A.J.; Baldwin Jr, A.S.; Mayo, M.W. Akt suppresses apoptosis by stimulating the transactivation potential of the RelA/p65 subunit of NF-κB. Mol. Cell Bio. 2000, 20, 1626–1638. [Google Scholar] [CrossRef]
- Goll, D.E.; Thompson, V.F.; Li, H.; Wei, W.; Cong, J. The calpain system. Physiol. Rev. 2003, 83, 731–801. [Google Scholar] [CrossRef] [PubMed]
- Smith, M.A.; Schnellmann, R.G. Calpains, mitochondria, and apoptosis. Cardiovasc. Res. 2012, 96, 32–37. [Google Scholar] [CrossRef]
- Storr, S.J.; Carragher, N.O.; Frame, M.C.; Parr, T.; Martin, S.G. The calpain system and cancer. Nat. Cancer 2011, 11, 364–374. [Google Scholar] [CrossRef]
- Kontsek, P. Human type I interferons: Structure and function. Acta Virol. 1994, 38, 345–360. [Google Scholar]
- Parmar, S.; Platanias, L.C. Interferons: Mechanisms of action and clinical applications. Curr. Opin. Oncol. 2003, 15, 431–439. [Google Scholar] [CrossRef]
- Sadler, A.J.; WilFams, B.R. Interferon-inducible antiviral effectors. Nat. Rev. Immunol. 2008, 8, 559–568. [Google Scholar] [CrossRef]
- Wang, L.C.; Chen, S.O.; Chang, S.P.; Lee, Y.P.; Yu, C.K.; Chen, C.L.; Tseng, P.C.; Hsieh, C.Y.; Chen, S.H.; Lin, C.F. Enterovirus 71 proteins 2A and 3D antagonize the antiviral activity of interferon via signalin-gattenuation. J. Virol. 2015, 89, 7028–7037. [Google Scholar] [CrossRef]
- Liao, C.C.; Liou, A.T.; Chang, Y.S.; Wu, S.Y.; Chang, C.S.; Lee, C.K.; Kung, J.T.; Tu, P.H.; Yu, Y.Y.; Lin, C.Y.; et al. Immunodeficient mouse models with different disease profiles by in vivo infection with the sameclinical isolate of enterovirus 71. J. Virol. 2014, 88, 12485–12499. [Google Scholar] [CrossRef]
- Tamura, T.; Yanai, H.; Savitsky, D.; Taniguchi, T. The IRF Family Transcription Factors in Immunity and Oncogenesis. Annu. Rev. Immunol. 2008, 26, 535–584. [Google Scholar] [CrossRef]
- Lei, X.; Liu, X.; Ma, Y.; Sun, Z.; Yang, Y.; Jin, Q.; He, B.; Wang, J. The 3C protein of enterovirus 71 inhibits retinoid acid-inducible gene I-mediated interferon regulatory factor 3 activation and type i interferon re-sponses. J. Virol. 2010, 84, 8051–8061. [Google Scholar] [CrossRef]
- Lei, X.; Xiao, X.; Xue, Q.; Jin, Q.; He, B.; Wang, J. Cleavage of Interferon Regulatory Factor 7 by Enterovirus 71 3C Suppresses Cellular Responses. J. Virol. 2013, 87, 1690–1698. [Google Scholar] [CrossRef]
- Baeuerle, P.A.; Baltimore, D. NF-_B: Ten years after. Cell 1996, 87, 13–20. [Google Scholar] [CrossRef]
- Hiscott, J.; Nguyen, T.L.; Arguello, M.; Nakhaei, P.; Paz, S. Manipulation of the nuclear factor-_B pathwayand the innate immune response by viruses. Oncogene 2006, 25, 6844–6867. [Google Scholar] [CrossRef]
- Zheng, Z.; Li, H.; Zhang, Z.; Meng, J.; Mao, D.; Bai, B.; Lu, B.; Mao, P.; Hu, Q.; Wang, H. Enterovirus 712C protein inhibits TNF-α-mediated activation of NF-κB by suppressing IκB kinase β phosphorylation. J. Immunol. 2011, 187, 2202–2212. [Google Scholar] [CrossRef]
- Du, H.; Yin, P.; Yang, X.; Zhang, L.; Jin, Q.; Zhu, G. Enterovirus 71 2C protein inhibits NF-κB activation bybinding to RelA(p65). Sci. Rep. 2015, 5, 14302. [Google Scholar] [CrossRef]
- Tartey, S.; Takeuchi, O. Pathogen recognition and Toll-likereceptor targeted therapeutics in innate immune cells. Int. Rev. Immunol. 2017, 36, 57–73. [Google Scholar] [CrossRef]
- Honda, K.; Taniguchi, T. IRFs: Master regulators of signaling by Toll-like receptors and cytosolic pat-tern-recognition receptors. Nat. Rev. Immunol. 2006, 6, 644–658. [Google Scholar] [CrossRef]
- Mishra, S.; Kumar, H. Balancing anti-viral innate immunityand immune homeostasis. Cell Mol. Immunol. 2018, 15, 408–410. [Google Scholar] [CrossRef]
- Song, J.; Hu, Y.; Li, J.; Zheng, H.; Wang, J.; Guo, L.; Shi, H.; Liu, L. Suppression of the toll-like receptor 7-dependent type I interferon production pathway by autophagy resulting from enterovirus 71 and cox-sackievirus A16 infections facilitates their replication. Arch. Virol. 2018, 163, 135–144. [Google Scholar] [CrossRef]
- Luo, Z.; Ge, M.; Chen, J.; Geng, Q.; Tian, M.; Qiao, Z.; Bai, L.; Zhang, Q.; Zhu, C.; Xiong, Y.; et al. HRS playsan important role for TLR7 signaling to orchestrate inflammation and innate immunity upon EV71 infection. PLoS Pathog. 2017, 13, e1006585. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; Li, W.; Qi, G.; Liu, N.; Sheng, L.; Shang, L.; Qi, B. The immune mechanism of intestinal tracttoll-like receptor in mediating EV71 virus type severe hand-foot-and-mouth disease and the mapk pathway. Exp. Ther. Med. 2017, 13, 2263–2266. [Google Scholar] [CrossRef] [PubMed]
- Gong, X.; Zhou, J.; Zhu, W.; Liu, N.; Li, J.; Li, L.; Jin, Y.; Duan, Z. Excessive proinflammatory cytokine and chemokine responses of human monocyte-derived macrophages to enterovirus 71 infection. BMC Infect. Dis. 2012, 12, 224. [Google Scholar] [CrossRef]
- Abzug, M.J.; Michaels, M.G.; Wald, E.; Jacobs, R.F.; Romero, J.R.; Sánchez, P.J.; Wilson, G.; Krogstad, P.; Storch, G.A.; Lawrence, R.; et al. A Randomized, Double-Blind, Placebo-Controlled Trial of Pleconaril for the Treatment of Neonates with Enterovirus Sepsis. J. Pediatr. Infect. Dis. Soc. 2015, 5, 53–62. [Google Scholar] [CrossRef]
- Puenpa, J.; Chieochansin, T.; Linsuwanon, P.; Korkong, S.; Thongkomplew, S.; Vichaiwattana, P.; Theamboonlers, A.; Poovorawan, Y. Hand, Foot, and Mouth Disease Caused by Coxsackievirus A6, Thailand, 2012. Emerg. Infect. Dis. 2013, 19, 641–643. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.-R.; Ling, P. Interplays between Enterovirus A71 and the innate immune system. J. Biomed. Sci. 2019, 26, 95. [Google Scholar] [CrossRef] [PubMed]
- Shih, C.; Liao, C.C.; Chang, Y.S.; Wu, S.Y.; Chang, C.S.; Liou, A.T. Immunocompetent and immunodefi-cient mouse models for enterovirus 71 pathogenesis and therapy. Viruses 2018, 10, 674. [Google Scholar] [CrossRef]
- Koh, W.M.; Bogich, T.; Siegel, K.; Jin, J.; Chong, E.Y.; Tan, C.Y.; Chen, M.I.; Horby, P.; Cook, A.R. The epidemiology of hand, foot, and mouth disease in Asia: A systematic review and analysis. Pediatr. Infect. Dis. J. 2016, 35, e285. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Swain, S.K.; Panda, S.; Sahu, B.P.; Sarangi, R. Activation of Host Cellular Signaling and Mechanism of Enterovirus 71 Viral Proteins Associated with Hand, Foot and Mouth Disease. Viruses 2022, 14, 2190. https://doi.org/10.3390/v14102190
Swain SK, Panda S, Sahu BP, Sarangi R. Activation of Host Cellular Signaling and Mechanism of Enterovirus 71 Viral Proteins Associated with Hand, Foot and Mouth Disease. Viruses. 2022; 14(10):2190. https://doi.org/10.3390/v14102190
Chicago/Turabian StyleSwain, Subrat Kumar, Subhasmita Panda, Basanta Pravas Sahu, and Rachita Sarangi. 2022. "Activation of Host Cellular Signaling and Mechanism of Enterovirus 71 Viral Proteins Associated with Hand, Foot and Mouth Disease" Viruses 14, no. 10: 2190. https://doi.org/10.3390/v14102190
APA StyleSwain, S. K., Panda, S., Sahu, B. P., & Sarangi, R. (2022). Activation of Host Cellular Signaling and Mechanism of Enterovirus 71 Viral Proteins Associated with Hand, Foot and Mouth Disease. Viruses, 14(10), 2190. https://doi.org/10.3390/v14102190




