Proteome Analysis of Swine Macrophages after Infection with Two Genotype II African Swine Fever Isolates of Different Pathogenicity
Abstract
1. Introduction
2. Materials and Methods
2.1. Isolation of Primary Porcine Monocyte-Derived Macrophages
2.2. Viruses
2.3. Infection of moMΦ with ASFV
2.4. SDS-Polyacrylamide Gel Electrophoresis and Immunoblot Analysis
2.5. Experimental Design of Proteome Analyses
2.6. LC-MS Analysis of Whole Cell Lysates
2.7. Proteome Analysis
2.8. Role of p38 during ASFV Infection
2.9. Cell Viability
2.10. Modelling of Protein Structures
2.11. Calculation of Genome Coverage
2.12. Data Availability
3. Results
3.1. Expression of Viral Proteins
3.2. Host Response to Infection
3.3. Analysis of p38 during ASFV Infection
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Blome, S.; Franzke, K.; Beer, M. African swine fever—A review of current knowledge. Virus Res. 2020, 287, 198099. [Google Scholar] [CrossRef] [PubMed]
- Dixon, L.K.; Islam, M.; Nash, R.; Reis, A.L. African swine fever virus evasion of host defences. Virus Res. 2019, 266, 25–33. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.J.; Ramanathan, P.; Bishop, E.A.; O’Donnell, V.; Gladue, D.P.; Borca, M.V. Mechanisms of African swine fever virus pathogenesis and immune evasion inferred from gene expression changes in infected swine macrophages. PLoS ONE 2019, 14, e0223955. [Google Scholar] [CrossRef]
- Wang, S.; Zhang, J.; Zhang, Y.; Yang, J.; Wang, L.; Qi, Y.; Han, X.; Zhou, X.; Miao, F.; Chen, T.; et al. Cytokine Storm in Domestic Pigs Induced by Infection of Virulent African Swine Fever Virus. Front. Vet. Sci. 2020, 7, 601641. [Google Scholar] [CrossRef]
- Shimmon, G.L.; Hui, J.Y.K.; Wileman, T.E.; Netherton, C.L. Autophagy impairment by African swine fever virus. J. Gen. Virol. 2021, 102, 001637. [Google Scholar] [CrossRef]
- Gaudreault, N.N.; Madden, D.W.; Wilson, W.C.; Trujillo, J.D.; Richt, J.A. African Swine Fever Virus: An Emerging DNA Arbovirus. Front. Vet. Sci. 2020, 7, 215. [Google Scholar] [CrossRef]
- Galindo, I.; Alonso, C. African Swine Fever Virus: A Review. Viruses 2017, 9, 103. [Google Scholar] [CrossRef]
- Revilla, Y.; Pérez-Núñez, D.; Richt, J.A. African Swine Fever Virus Biology and Vaccine Approaches. Adv. Virus Res. 2018, 100, 41–74. [Google Scholar]
- Keßler, C.; Forth, J.H.; Keil, G.M.; Mettenleiter, T.C.; Blome, S.; Karger, A. The intracellular proteome of African swine fever virus. Sci. Rep. 2018, 8, 14714. [Google Scholar] [CrossRef] [PubMed]
- Urzainqui, A.; Tabarés, E.; Carrasco, L. Proteins synthesized in African swine fever virus-infected cells analyzed by two-dimensional gel electrophoresis. Virology 1987, 160, 286–291. [Google Scholar] [CrossRef]
- Esteves, A.; Marques, M.I.; Costa, J.V. Two-dimensional analysis of African swine fever virus proteins and proteins induced in infected cells. Virology 1986, 152, 192–206. [Google Scholar] [CrossRef]
- Cackett, G.; Portugal, R.; Matelska, D.; Dixon, L.; Werner, F. African Swine Fever Virus and host response—Transcriptome profiling of the Georgia 2007/1 strain and porcine macrophages. J. Virol. 2022, 96, jvi0193921. [Google Scholar] [CrossRef]
- Chapman, D.A.G.; Tcherepanov, V.; Upton, C.; Dixon, L.K. Comparison of the genome sequences of non-pathogenic and pathogenic African swine fever virus isolates. J. Gen. Virol. 2008, 89 Pt 2, 397–408. [Google Scholar] [CrossRef]
- Cackett, G.; Matelska, D.; Sýkora, M.; Portugal, R.; Malecki, M.; Bähler, J.; Dixon, L.; Werner, F. The African Swine Fever Virus Transcriptome. J. Virol. 2020, 94, e00119-20. [Google Scholar] [CrossRef]
- Jaing, C.; Rowland, R.R.R.; Allen, J.E.; Certoma, A.; Thissen, J.B.; Bingham, J.; Rowe, B.; White, J.R.; Wynne, J.W.; Johnson, D.; et al. Gene expression analysis of whole blood RNA from pigs infected with low and high pathogenic African swine fever viruses. Sci. Rep. 2017, 7, 10115. [Google Scholar] [CrossRef]
- Olesen, A.S.; Kodama, M.; Lohse, L.; Accensi, F.; Rasmussen, T.B.; Lazov, C.M.; Limborg, M.T.; Gilbert, M.T.P.; Bøtner, A.; Belsham, G.J. Identification of African Swine Fever Virus Transcription within Peripheral Blood Mononuclear Cells of Acutely Infected Pigs. Viruses 2021, 13, 2333. [Google Scholar] [CrossRef]
- Wang, N.; Zhao, D.; Wang, J.; Zhang, Y.; Wang, M.; Gao, Y.; Li, F.; Wang, J.; Bu, Z.; Rao, Z.; et al. Architecture of African swine fever virus and implications for viral assembly. Science 2019, 366, 640–644. [Google Scholar] [CrossRef]
- Netherton, C.L.; Connell, S.; Benfield, C.T.O.; Dixon, L.K. The Genetics of Life and Death: Virus-Host Interactions Underpinning Resistance to African Swine Fever, a Viral Hemorrhagic Disease. Front. Genet. 2019, 10, 402. [Google Scholar] [CrossRef]
- Boinas, F.S.; Hutchings, G.H.; Dixon, L.K.; Wilkinson, P.J. Characterization of pathogenic and non-pathogenic African swine fever virus isolates from Ornithodoros erraticus inhabiting pig premises in Portugal. J. Gen. Virol. 2004, 85 Pt 8, 2177–2187. [Google Scholar] [CrossRef] [PubMed]
- Gallardo, C.; Soler, A.; Rodze, I.; Nieto, R.; Cano-Gómez, C.; Fernandez-Pinero, J.; Arias, M. Attenuated and non-haemadsorbing (non-HAD) genotype II African swine fever virus (ASFV) isolated in Europe, Latvia 2017. Transbound. Emerg. Dis. 2019, 66, 1399–1404. [Google Scholar] [CrossRef] [PubMed]
- Zani, L.; Forth, J.H.; Forth, L.; Nurmoja, I.; Leidenberger, S.; Henke, J.; Carlson, J.; Breidenstein, C.; Viltrop, A.; Höper, D.; et al. Deletion at the 5’-end of Estonian ASFV strains associated with an attenuated phenotype. Sci. Rep. 2018, 8, 6510. [Google Scholar] [CrossRef]
- Sun, E.; Zhang, Z.; Wang, Z.; He, X.; Zhang, X.; Wang, L.; Wang, W.; Huang, L.; Xi, F.; Huangfu, H.; et al. Emergence and prevalence of naturally occurring lower virulent African swine fever viruses in domestic pigs in China in 2020. Sci. China Life Sci. 2021, 64, 752–765. [Google Scholar] [CrossRef] [PubMed]
- Vuono, E.A.; Ramirez-Medina, E.; Pruitt, S.; Rai, A.; Espinoza, N.; Velazquez-Salinas, L.; Gladue, D.P.; Borca, M.V. Evaluation of the Function of the ASFV KP177R Gene, Encoding for Structural Protein p22, in the Process of Virus Replication and in Swine Virulence. Viruses 2021, 13, 986. [Google Scholar] [CrossRef] [PubMed]
- Borca, M.V.; O’Donnell, V.; Holinka, L.G.; Ramírez-Medina, E.; Clark, B.A.; Vuono, E.A.; Berggren, K.; Alfano, M.; Carey, L.B.; Richt, J.A.; et al. The L83L ORF of African swine fever virus strain Georgia encodes for a non-essential gene that interacts with the host protein IL-1β. Virus Res. 2018, 249, 116–123. [Google Scholar] [CrossRef]
- Hühr, J.; Schäfer, A.; Schwaiger, T.; Zani, L.; Sehl, J.; Mettenleiter, T.C.; Blome, S.; Blohm, U. Impaired T-cell responses in domestic pigs and wild boar upon infection with a highly virulent African swine fever virus strain. Transbound. Emerg. Dis. 2020, 67, 3016–3032. [Google Scholar] [CrossRef]
- Schäfer, A.; Zani, L.; Pikalo, J.; Hühr, J.; Sehl, J.; Mettenleiter, T.C.; Breithaupt, A.; Blome, S.; Blohm, U. T-cell responses in domestic pigs and wild boar upon infection with the moderately virulent African swine fever virus strain “Estonia2014”. Transbound. Emerg. Dis. 2021, 68, 2733–2749. [Google Scholar] [CrossRef]
- Gil, S.; Sepúlveda, N.; Albina, E.; Leitão, A.; Martins, C. The low-virulent African swine fever virus (ASFV/NH/P68) induces enhanced expression and production of relevant regulatory cytokines (IFNalpha, TNFalpha and IL12p40) on porcine macrophages in comparison to the highly virulent ASFV/L60. Arch. Virol. 2008, 153, 1845–1854. [Google Scholar] [CrossRef]
- Gil, S.; Spagnuolo-Weaver, M.; Canals, A.; Sepúlveda, N.; Oliveira, J.; Aleixo, A.; Allan, G.; Leitão, A.; Martins, C.L.V. Expression at mRNA level of cytokines and A238L gene in porcine blood-derived macrophages infected in vitro with African swine fever virus (ASFV) isolates of different virulence. Arch. Virol. 2003, 148, 2077–2097. [Google Scholar] [CrossRef]
- Franzoni, G.; Graham, S.P.; Dei Giudici, S.; Bonelli, P.; Pilo, G.; Anfossi, A.G.; Pittau, M.; Nicolussi, P.S.; Laddomada, A.; Oggiano, A. Characterization of the interaction of African swine fever virus with monocytes and derived macrophage subsets. Vet. Microbiol. 2017, 198, 88–98. [Google Scholar] [CrossRef]
- García-Belmonte, R.; Pérez-Núñez, D.; Pittau, M.; Richt, J.A.; Revilla, Y. African Swine Fever Virus Armenia/07 Virulent Strain Controls Interferon Beta Production through the cGAS-STING Pathway. J. Virol. 2019, 93, e02298-18. [Google Scholar] [CrossRef]
- Franzoni, G.; Razzuoli, E.; Dei Giudici, S.; Carta, T.; Galleri, G.; Zinellu, S.; Ledda, M.; Angioi, P.; Modesto, P.; Graham, S.P.; et al. Comparison of Macrophage Responses to African Swine Fever Viruses Reveals that the NH/P68 Strain is Associated with Enhanced Sensitivity to Type I IFN and Cytokine Responses from Classically Activated Macrophages. Pathogens 2020, 9, 209. [Google Scholar] [CrossRef] [PubMed]
- Razzuoli, E.; Franzoni, G.; Carta, T.; Zinellu, S.; Amadori, M.; Modesto, P.; Oggiano, A. Modulation of Type I Interferon System by African Swine Fever Virus. Pathogens 2020, 9, 361. [Google Scholar] [CrossRef] [PubMed]
- Tatoyan, M.R.; Izmailyan, R.A.; Semerjyan, A.B.; Karalyan, N.Y.; Sahakyan, C.T.; Mkrtchyan, G.L.; Ghazaryan, H.K.; Arzumanyan, H.H.; Semerjyan, Z.B.; Karalova, E.M.; et al. Patterns of alveolar macrophage activation upon attenuated and virulent African swine fever viruses in vitro. Comp. Immunol. Microbiol. Infect. Dis. 2020, 72, 101513. [Google Scholar] [CrossRef] [PubMed]
- Riera, E.; Pérez-Núñez, D.; García-Belmonte, R.; Miorin, L.; García-Sastre, A.; Revilla, Y. African Swine Fever Virus Induces STAT1 and STAT2 Degradation to Counteract IFN-I Signaling. Front. Microbiol. 2021, 12, 2467. [Google Scholar] [CrossRef] [PubMed]
- Herrera-Uribe, J.; Jiménez-Marín, Á.; Lacasta, A.; Monteagudo, P.L.; Pina-Pedrero, S.; Rodríguez, F.; Moreno, Á.; Garrido, J.J. Comparative proteomic analysis reveals different responses in porcine lymph nodes to virulent and attenuated homologous African swine fever virus strains. Vet. Res. 2018, 49, 90. [Google Scholar] [CrossRef]
- Gabriel, C.; Blome, S.; Malogolovkin, A.; Parilov, S.; Kolbasov, D.; Teifke, J.P.; Beer, M. Characterization of African swine fever virus Caucasus isolate in European wild boars. Emerg. Infect. Dis. 2011, 17, 2342–2345. [Google Scholar] [CrossRef]
- Wöhnke, E.; Fuchs, W.; Hartmann, L.; Blohm, U.; Blome, S.; Mettenleiter, T.C.; Karger, A. Comparison of the Proteomes of Porcine Macrophages and a Stable Porcine Cell Line after Infection with African Swine Fever Virus. Viruses 2021, 13, 2198. [Google Scholar] [CrossRef]
- Reed, L.J.; Muench, H. A Simple method of estimating fifty per cent endpoints. Am. J. Hyg. 1938, 27, 493–497. [Google Scholar]
- Hübner, A.; Petersen, B.; Keil, G.M.; Niemann, H.; Mettenleiter, T.C.; Fuchs, W. Efficient inhibition of African swine fever virus replication by CRISPR/Cas9 targeting of the viral p30 gene (CP204L). Sci. Rep. 2018, 8, 1449. [Google Scholar] [CrossRef]
- Stoscheck, C.M. Quantitation of protein. Methods Enzymol. 1990, 182, 50–68. [Google Scholar]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef] [PubMed]
- Neuhoff, V.; Arold, N.; Taube, D.; Ehrhardt, W. Improved staining of proteins in polyacrylamide gels including isoelectric focusing gels with clear background at nanogram sensitivity using Coomassie Brilliant Blue G-250 and R-250. Electrophoresis 1988, 9, 255–262. [Google Scholar] [CrossRef] [PubMed]
- Towbin, H.; Staehelin, T.; Gordon, J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: Procedure and some applications. Proc. Natl. Acad. Sci. USA 1979, 76, 4350–4354. [Google Scholar] [CrossRef] [PubMed]
- Hübner, A.; Keßler, C.; Pannhorst, K.; Forth, J.H.; Kabuuka, T.; Karger, A.; Mettenleiter, T.C.; Fuchs, W. Identification and characterization of the 285L and K145R proteins of African swine fever virus. J. Gen. Virol. 2019, 100, 1303–1314. [Google Scholar] [CrossRef]
- Wisniewski, J.R.; Zougman, A.; Nagaraj, N.; Mann, M. Universal sample preparation method for proteome analysis. Nat. Methods 2009, 6, 359–362. [Google Scholar] [CrossRef]
- Cox, J.; Mann, M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008, 26, 1367–1372. [Google Scholar] [CrossRef]
- Prianichnikov, N.; Koch, H.; Koch, S.; Lubeck, M.; Heilig, R.; Brehmer, S.; Fischer, R.; Cox, J. MaxQuant Software for Ion Mobility Enhanced Shotgun Proteomics. Mol. Cell. Proteom. MCP 2020, 19, 1058–1069. [Google Scholar] [CrossRef]
- Aken, B.L.; Achuthan, P.; Akanni, W.; Amode, M.R.; Bernsdorff, F.; Bhai, J.; Billis, K.; Carvalho-Silva, D.; Cummins, C.; Clapham, P.; et al. Ensembl 2017. Nucleic Acids Res. 2017, 45, D635–D642. [Google Scholar] [CrossRef]
- R_Core_Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2020. [Google Scholar]
- Tyanova, S.; Temu, T.; Sinitcyn, P.; Carlson, A.; Hein, M.Y.; Geiger, T.; Mann, M.; Cox, J. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods 2016, 13, 731–740. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef] [PubMed]
- Kolberg, L.; Raudvere, U.; Kuzmin, I.; Vilo, J.; Peterson, H. gprofiler2- an R package for gene list functional enrichment analysis and namespace conversion toolset g:Profiler. F1000Research 2020, 9, 709. [Google Scholar] [CrossRef] [PubMed]
- Tweedie, S.; Braschi, B.; Gray, K.; Jones, T.E.M.; Seal, R.L.; Yates, B.; Bruford, E.A. Genenames.org: The HGNC and VGNC resources in 2021. Nucleic Acids Res. 2021, 49, D939–D946. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Bindea, G.; Mlecnik, B.; Hackl, H.; Charoentong, P.; Tosolini, M.; Kirilovsky, A.; Fridman, W.-H.; Pagès, F.; Trajanoski, Z.; Galon, J. ClueGO: A Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 2009, 25, 1091–1093. [Google Scholar] [CrossRef]
- Bindea, G.; Galon, J.; Mlecnik, B. CluePedia Cytoscape plugin: Pathway insights using integrated experimental and in silico data. Bioinformatics 2013, 29, 661–663. [Google Scholar] [CrossRef]
- Wickham, H.; Chang, W.; Henry, L.; Pedersen, T.L.; Takahashi, K.; Wilke, C.; Woo, K.; Yutani, H.; Dunnington, D. ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016. [Google Scholar]
- Tunyasuvunakool, K.; Adler, J.; Wu, Z.; Green, T.; Zielinski, M.; Žídek, A.; Bridgland, A.; Cowie, A.; Meyer, C.; Laydon, A.; et al. Highly accurate protein structure prediction for the human proteome. Nature 2021, 596, 590–596. [Google Scholar] [CrossRef]
- Mészáros, B.; Erdos, G.; Dosztányi, Z. IUPred2A: Context-dependent prediction of protein disorder as a function of redox state and protein binding. Nucleic Acids Res. 2018, 46, W329–W337. [Google Scholar] [CrossRef]
- Ward, J.J.; McGuffin, L.J.; Bryson, K.; Buxton, B.F.; Jones, D.T. The DISOPRED server for the prediction of protein disorder. Bioinformatics 2004, 20, 2138–2139. [Google Scholar] [CrossRef]
- Mitchell, A.; Chang, H.-Y.; Daugherty, L.; Fraser, M.; Hunter, S.; Lopez, R.; McAnulla, C.; McMenamin, C.; Nuka, G.; Pesseat, S.; et al. The InterPro protein families database: The classification resource after 15 years. Nucleic Acids Res. 2015, 43, D213–D221. [Google Scholar] [CrossRef]
- Kall, L.; Krogh, A.; Sonnhammer, E.L. A combined transmembrane topology and signal peptide prediction method. J. Mol. Biol. 2004, 338, 1027–1036. [Google Scholar] [CrossRef] [PubMed]
- McGuffin, L.J.; Bryson, K.; Jones, D.T. The PSIPRED protein structure prediction server. Bioinformatics 2000, 16, 404–405. [Google Scholar] [CrossRef]
- Nugent, T.; Jones, D.T. Transmembrane protein topology prediction using support vector machines. BMC Bioinform. 2009, 10, 159. [Google Scholar] [CrossRef] [PubMed]
- Kahsay, R.Y.; Gao, G.; Liao, L. An improved hidden Markov model for transmembrane protein detection and topology prediction and its applications to complete genomes. Bioinformatics 2005, 21, 1853–1858. [Google Scholar] [CrossRef] [PubMed]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef]
- Quinlan, A.R.; Hall, I.M. BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 2010, 26, 841–842. [Google Scholar] [CrossRef] [PubMed]
- Perez-Riverol, Y.; Csordas, A.; Bai, J.; Bernal-Llinares, M.; Hewapathirana, S.; Kundu, D.J.; Inuganti, A.; Griss, J.; Mayer, G.; Eisenacher, M.; et al. The PRIDE database and related tools and resources in 2019: Improving support for quantification data. Nucleic Acids Res. 2019, 47, D442–D450. [Google Scholar] [CrossRef] [PubMed]
- Muñoz, M.; Freije, J.M.; Salas, M.L.; Viñuela, E.; López-Otín, C. Structure and expression in E. coli of the gene coding for protein p10 of African swine fever virus. Arch. Virol. 1993, 130, 93–107. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Ma, B.; Qian, N.; Zhang, F.; Tan, X.; Lei, J.; Xiang, Y. Structure of the African swine fever virus major capsid protein p72. Cell Res. 2019, 29, 953–955. [Google Scholar] [CrossRef] [PubMed]
- Andres, G.; Charro, D.; Matamoros, T.; Dillard, R.S.; Abrescia, N.G.A. The cryo-EM structure of African swine fever virus unravels a unique architecture comprising two icosahedral protein capsids and two lipoprotein membranes. J. Biol. Chem. 2019, 295, jbc.AC119.011196. [Google Scholar] [CrossRef]
- Guo, F.; Shi, Y.; Yang, M.; Guo, Y.; Shen, Z.; Li, M.; Chen, Y.; Liang, R.; Yang, Y.; Chen, H.; et al. The structural basis of African swine fever virus core shell protein p15 binding to DNA. FASEB J. 2021, 35, e21350. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Luo, Y.; Wang, Y.; Li, S.; Zhao, Z.; Bi, Y.; Sun, J.; Peng, R.; Song, H.; Zhu, D.; et al. Cryo-EM Structure of the African Swine Fever Virus. Cell Host Microbe 2019, 26, 836–843.e3. [Google Scholar] [CrossRef]
- Lee, T.H.; Huang, Q.; Oikemus, S.; Shank, J.; Ventura, J.-J.; Cusson, N.; Vaillancourt, R.R.; Su, B.; Davis, R.J.; Kelliher, M.A. The death domain kinase RIP1 is essential for tumor necrosis factor alpha signaling to p38 mitogen-activated protein kinase. Mol. Cell. Biol. 2003, 23, 8377–8385. [Google Scholar] [CrossRef][Green Version]
- Antonio Alcamí, A.A.; Eladio, V. Mapping and sequence of the gene encoding the African swine fever virion protein of M r 11500. J. Gen. Virol. 1993, 74, 2317–2324. [Google Scholar] [CrossRef] [PubMed]
- Alejo, A.; Matamoros, T.; Guerra, M.; Andrés, G. A Proteomic Atlas of the African Swine Fever Virus Particle. J. Virol. 2018, 92, e01293-18. [Google Scholar] [CrossRef]
- Nagaraj, N.; Wisniewski, J.R.; Geiger, T.; Cox, J.; Kircher, M.; Kelso, J.; Pääbo, S.; Mann, M. Deep proteome and transcriptome mapping of a human cancer cell line. Mol. Syst. Biol. 2011, 7, 548. [Google Scholar] [CrossRef]
- Ai, Q.; Lin, X.; Xie, H.; Li, B.; Liao, M.; Fan, H. Proteome Analysis in PAM Cells Reveals That African Swine Fever Virus Can Regulate the Level of Intracellular Polyamines to Facilitate Its Own Replication through ARG1. Viruses 2021, 13, 1236. [Google Scholar] [CrossRef]
- Nunes-Correia, I.; Rodríguez, J.M.; Eulálio, A.; Carvalho, A.L.; Citovsky, V.; Simões, S.; Faro, C.; Salas, M.L.; Pedroso de Lima, M.C. African swine fever virus p10 protein exhibits nuclear import capacity and accumulates in the nucleus during viral infection. Vet. Microbiol. 2008, 130, 47–59. [Google Scholar] [CrossRef][Green Version]
- van der Lee, R.; Buljan, M.; Lang, B.; Weatheritt, R.J.; Daughdrill, G.W.; Dunker, A.K.; Fuxreiter, M.; Gough, J.; Gsponer, J.; Jones, D.T.; et al. Classification of intrinsically disordered regions and proteins. Chem. Rev. 2014, 114, 6589–6631. [Google Scholar] [CrossRef]
- Wallmann, A.; Kesten, C. Common Functions of Disordered Proteins across Evolutionary Distant Organisms. Int. J. Mol. Sci. 2020, 21, 2105. [Google Scholar] [CrossRef] [PubMed]
- Andrés, G.; Simón-Mateo, C.; Viñuela, E. Characterization of two African swine fever virus 220-kDa proteins: A precursor of the major structural protein p150 and an oligomer of phosphoprotein p32. Virology 1993, 194, 284–293. [Google Scholar] [CrossRef]
- Andrés, G.; Alejo, A.; Simón-Mateo, C.; Salas, M.L. African swine fever virus protease, a new viral member of the SUMO-1-specific protease family. J. Biol. Chem. 2001, 276, 780–787. [Google Scholar] [CrossRef]
- Heath, C.M.; Windsor, M.; Wileman, T. Membrane association facilitates the correct processing of pp220 during production of the major matrix proteins of African swine fever virus. J. Virol. 2003, 77, 1682–1690. [Google Scholar] [CrossRef]
- Tabata, K.; Neufeldt, C.J.; Bartenschlager, R. Hepatitis C Virus Replication. Cold Spring Harb. Perspect. Med. 2020, 10, a037093. [Google Scholar] [CrossRef]
- Tews, B.A.; Meyers, G. The pestivirus glycoprotein Erns is anchored in plane in the membrane via an amphipathic helix. J. Biol. Chem. 2007, 282, 32730–32741. [Google Scholar] [CrossRef]
- Varkey, J.; Zhang, J.; Kim, J.; George, G.; He, G.; Belov, G.; Langen, R.; Wang, X. An Amphipathic Alpha-Helix Domain from Poliovirus 2C Protein Tubulate Lipid Vesicles. Viruses 2020, 12, 1466. [Google Scholar] [CrossRef]
- Portugal, R.; Leitão, A.; Martins, C. Apoptosis in porcine macrophages infected in vitro with African swine fever virus (ASFV) strains with different virulence. Arch. Virol. 2009, 154, 1441–1450. [Google Scholar] [CrossRef]
- Neilan, J.G.; Lu, Z.; Afonso, C.L.; Kutish, G.F.; Sussmann, M.D.; Rock, D.L. An African Swine Fever Viurs Gene with Similarity to the Proto-Oncogene bcl-2 and EBV Gene BHRF1. J. Virol. 1993, 67, 4391–4394. [Google Scholar] [CrossRef]
- Chacón, M.R.; Almazán, F.; Nogal, M.L.; Viñuela, E.; Rodríguez, J.F. The African Swine Fever Virus IAP Homolog Is a Late Structural Polypeptide. Virology 1995, 214, 670–674. [Google Scholar] [CrossRef]
- Banjara, S.; Caria, S.; Dixon, L.K.; Hinds, M.G.; Kvansakul, M. Structural Insight into African Swine Fever Virus A179L-Mediated Inhibition of Apoptosis. J. Virol. 2017, 91, e02228-16. [Google Scholar] [CrossRef]
- Tummers, B.; Green, D.R. Caspase-8: Regulating life and death. Immunol. Rev. 2017, 277, 76–89. [Google Scholar] [CrossRef]
- Orning, P.; Lien, E. Multiple roles of caspase-8 in cell death, inflammation, and innate immunity. J. Leukoc. Biol. 2021, 109, 121–141. [Google Scholar] [CrossRef]
- Parrish, A.B.; Freel, C.D.; Kornbluth, S. Cellular mechanisms controlling caspase activation and function. Cold Spring Harb. Perspect. Biol. 2013, 5, a008672. [Google Scholar] [CrossRef] [PubMed]
- Franzoni, G.; Graham, S.P.; Sanna, G.; Angioi, P.; Fiori, M.S.; Anfossi, A.; Amadori, M.; Dei Giudici, S.; Oggiano, A. Interaction of porcine monocyte-derived dendritic cells with African swine fever viruses of diverse virulence. Vet. Microbiol. 2018, 216, 190–197. [Google Scholar] [CrossRef]
- Kumar, R.; Khandelwal, N.; Thachamvally, R.; Tripathi, B.N.; Barua, S.; Kashyap, S.K.; Maherchandani, S.; Kumar, N. Role of MAPK/MNK1 signaling in virus replication. Virus Res. 2018, 253, 48–61. [Google Scholar] [CrossRef]
- Mathieson, T.; Franken, H.; Kosinski, J.; Kurzawa, N.; Zinn, N.; Sweetman, G.; Poeckel, D.; Ratnu, V.S.; Schramm, M.; Becher, I.; et al. Systematic analysis of protein turnover in primary cells. Nat. Commun. 2018, 9, 689. [Google Scholar] [CrossRef]
- Canovas, B.; Nebreda, A.R. Diversity and versatility of p38 kinase signalling in health and disease. Nat. Rev. Mol. Cell Biol. 2021, 22, 346–366. [Google Scholar] [CrossRef]
- Xue, Q.; Liu, H.; Zhu, Z.; Yang, F.; Song, Y.; Li, Z.; Xue, Z.; Cao, W.; Liu, X.; Zheng, H. African swine fever virus regulates host energy and amino acid metabolism to promote viral replication. J. Virol. 2021, 96, e01919-21. [Google Scholar] [CrossRef]
- Franzoni, G.; Dei Giudici, S.; Oggiano, A. Infection, modulation and responses of antigen-presenting cells to African swine fever viruses. Virus Res. 2018, 258, 73–80. [Google Scholar] [CrossRef]
- Yang, B.; Shen, C.; Zhang, D.; Zhang, T.; Shi, X.; Yang, J.; Hao, Y.; Zhao, D.; Cui, H.; Yuan, X.; et al. Mechanism of interaction between virus and host is inferred from the changes of gene expression in macrophages infected with African swine fever virus CN/GS/2018 strain. Virol. J. 2021, 18, 170. [Google Scholar] [CrossRef]
- Da Silva, T.A.; Zorzetto-Fernandes, A.L.V.; Cecílio, N.T.; Sardinha-Silva, A.; Fernandes, F.F.; Roque-Barreira, M.C. CD14 is critical for TLR2-mediated M1 macrophage activation triggered by N-glycan recognition. Sci. Rep. 2017, 7, 7083. [Google Scholar] [CrossRef] [PubMed]
- Oliveira-Nascimento, L.; Massari, P.; Wetzler, L.M. The Role of TLR2 in Infection and Immunity. Front. Immunol. 2012, 3, 79. [Google Scholar] [CrossRef] [PubMed]
- Henriques, E.S.; Brito, R.M.M.; Soares, H.; Ventura, S.; Oliveira, V.L.; Parkhouse, R.M.E. Modeling of the Toll-like receptor 3 and a putative Toll-like receptor 3 antagonist encoded by the African swine fever virus. Protein Sci. A Publ. Protein Soc. 2011, 20, 247–255. [Google Scholar] [CrossRef]
- Oliveira, V.L.; Almeida, S.C.P.; Soares, H.R.; Crespo, A.; Marshall-Clarke, S.; Parkhouse, R.M.E. A novel TLR3 inhibitor encoded by African swine fever virus (ASFV). Arch. Virol. 2011, 156, 597–609. [Google Scholar] [CrossRef]
- Li, J.; Song, J.; Kang, L.; Huang, L.; Zhou, S.; Hu, L.; Zheng, J.; Li, C.; Zhang, X.; He, X.; et al. pMGF505-7R determines pathogenicity of African swine fever virus infection by inhibiting IL-1β and type I IFN production. PLoS Pathog. 2021, 17, e1009733. [Google Scholar] [CrossRef] [PubMed]
- Bonney, E.A. Mapping out p38MAPK. Am. J. Reprod. Immunol. 2017, 77, e12652. [Google Scholar] [CrossRef]
- Chander, Y.; Kumar, R.; Khandelwal, N.; Singh, N.; Shringi, B.N.; Barua, S.; Kumar, N. Role of p38 mitogen-activated protein kinase signalling in virus replication and potential for developing broad spectrum antiviral drugs. Rev. Med. Virol. 2021, 31, 1–16. [Google Scholar] [CrossRef]
- Rouiller, I.; Brookes, S.M.; Hyatt, A.D.; Windsor, M.; Wileman, T. African swine fever virus is wrapped by the endoplasmic reticulum. J. Virol. 1998, 72, 2373–2387. [Google Scholar] [CrossRef]
- Netherton, C.; Rouiller, I.; Wileman, T. The subcellular distribution of multigene family 110 proteins of African swine fever virus is determined by differences in C-terminal KDEL endoplasmic reticulum retention motifs. J. Virol. 2004, 78, 3710–3721. [Google Scholar] [CrossRef]
- Keil, E.; Hocker, R.; Schuster, M.; Essmann, F.; Ueffing, N.; Hoffman, B.; Liebermann, D.A.; Pfeffer, K.; Schulze-Osthoff, K.; Schmitz, I. Phosphorylation of Atg5 by the Gadd45beta-MEKK4-p38 pathway inhibits autophagy. Cell Death Differ. 2013, 20, 321–332. [Google Scholar] [CrossRef]
- Henson, S.M.; Lanna, A.; Riddell, N.E.; Franzese, O.; Macaulay, R.; Griffiths, S.J.; Puleston, D.J.; Watson, A.S.; Simon, A.K.; Tooze, S.A.; et al. p38 signaling inhibits mTORC1-independent autophagy in senescent human CD8(+) T cells. J. Clin. Investig. 2014, 124, 4004–4016. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; She, H.; Zhang, T.; Xu, H.; Cheng, L.; Yepes, M.; Zhao, Y.; Mao, Z. p38 MAPK inhibits autophagy and promotes microglial inflammatory responses by phosphorylating ULK1. J. Cell Biol. 2018, 217, 315–328. [Google Scholar] [CrossRef] [PubMed]
- Nixon, R.A. The role of autophagy in neurodegenerative disease. Nat. Med. 2013, 19, 983–997. [Google Scholar] [CrossRef] [PubMed]

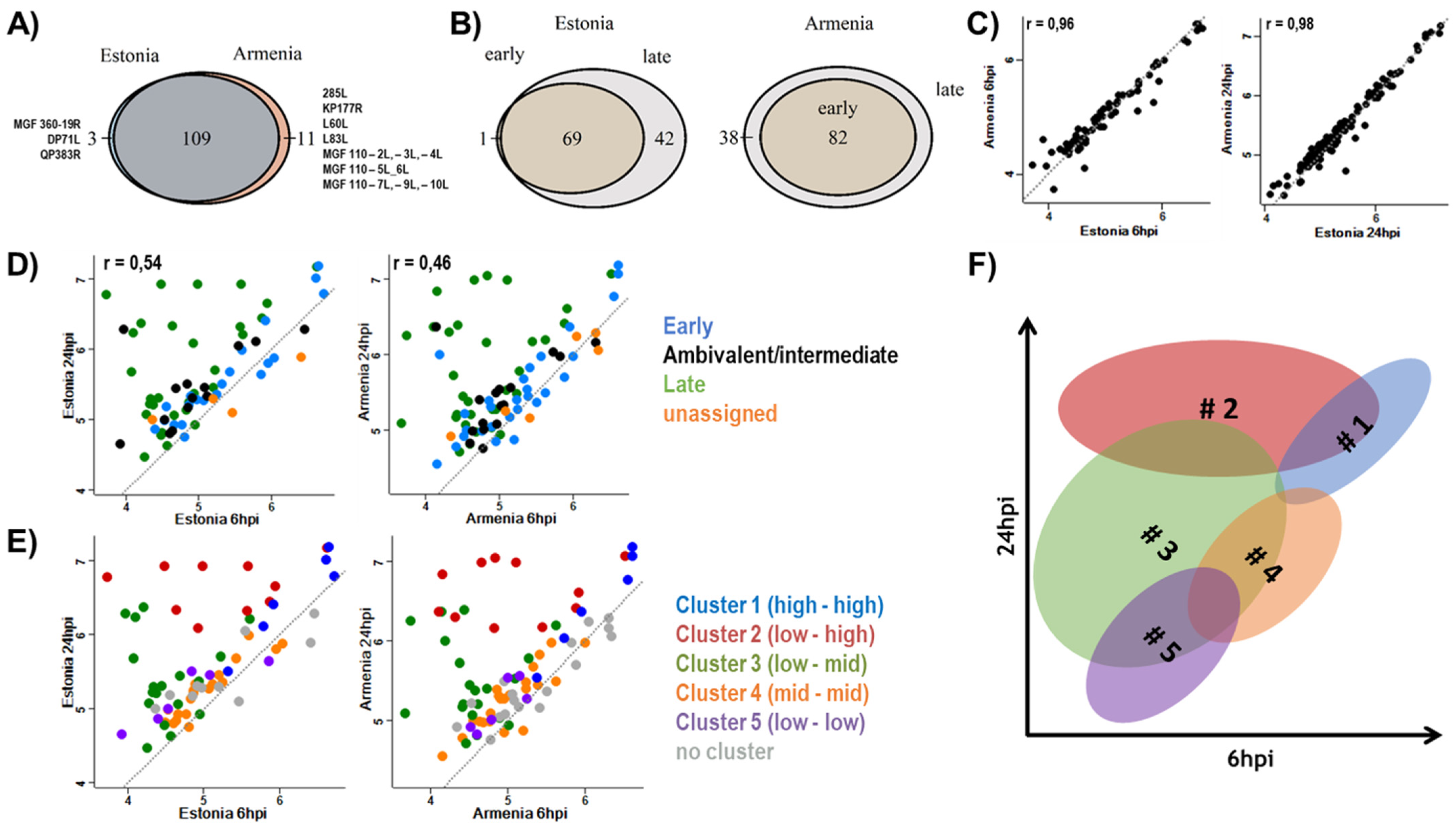
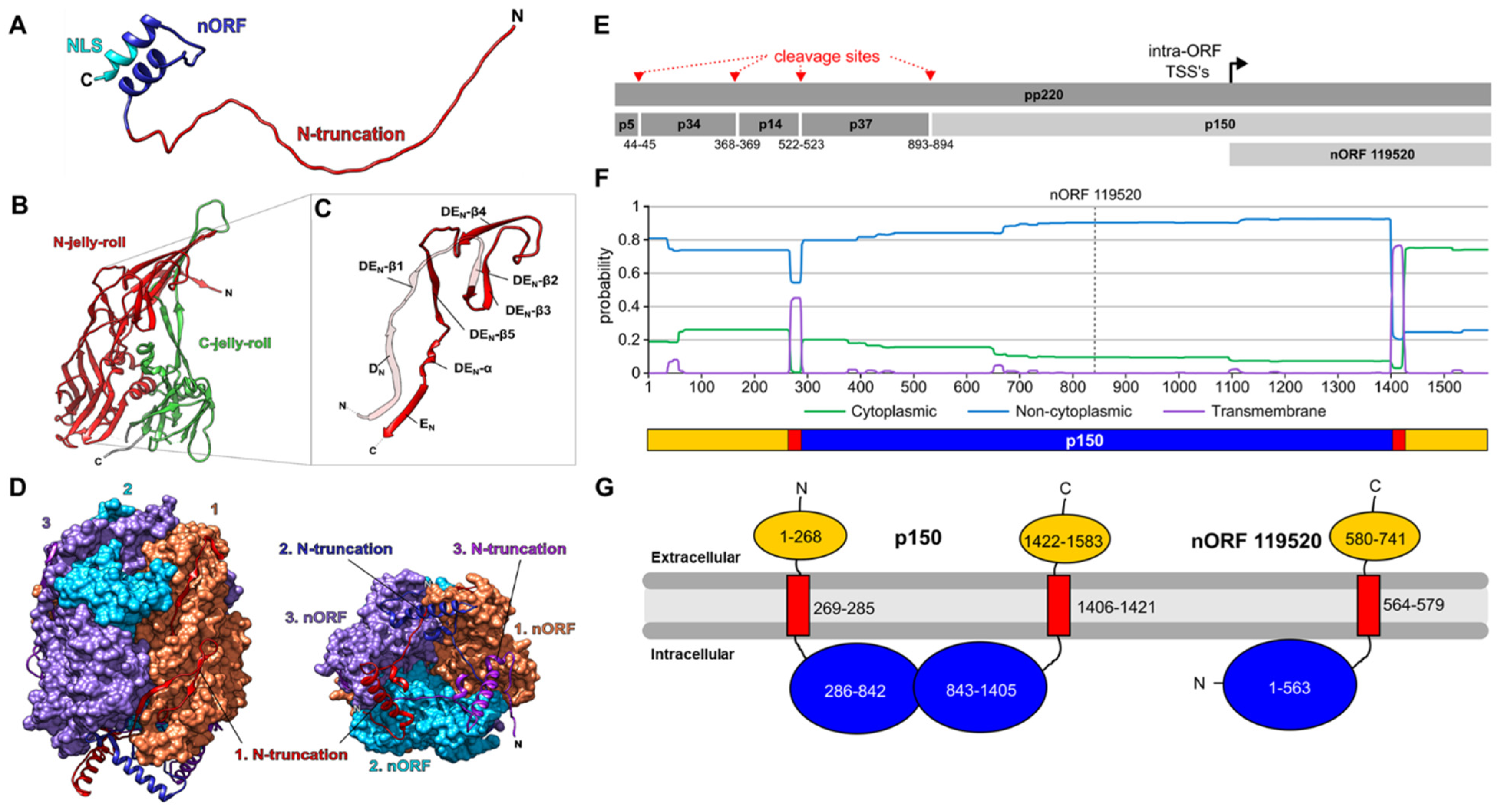

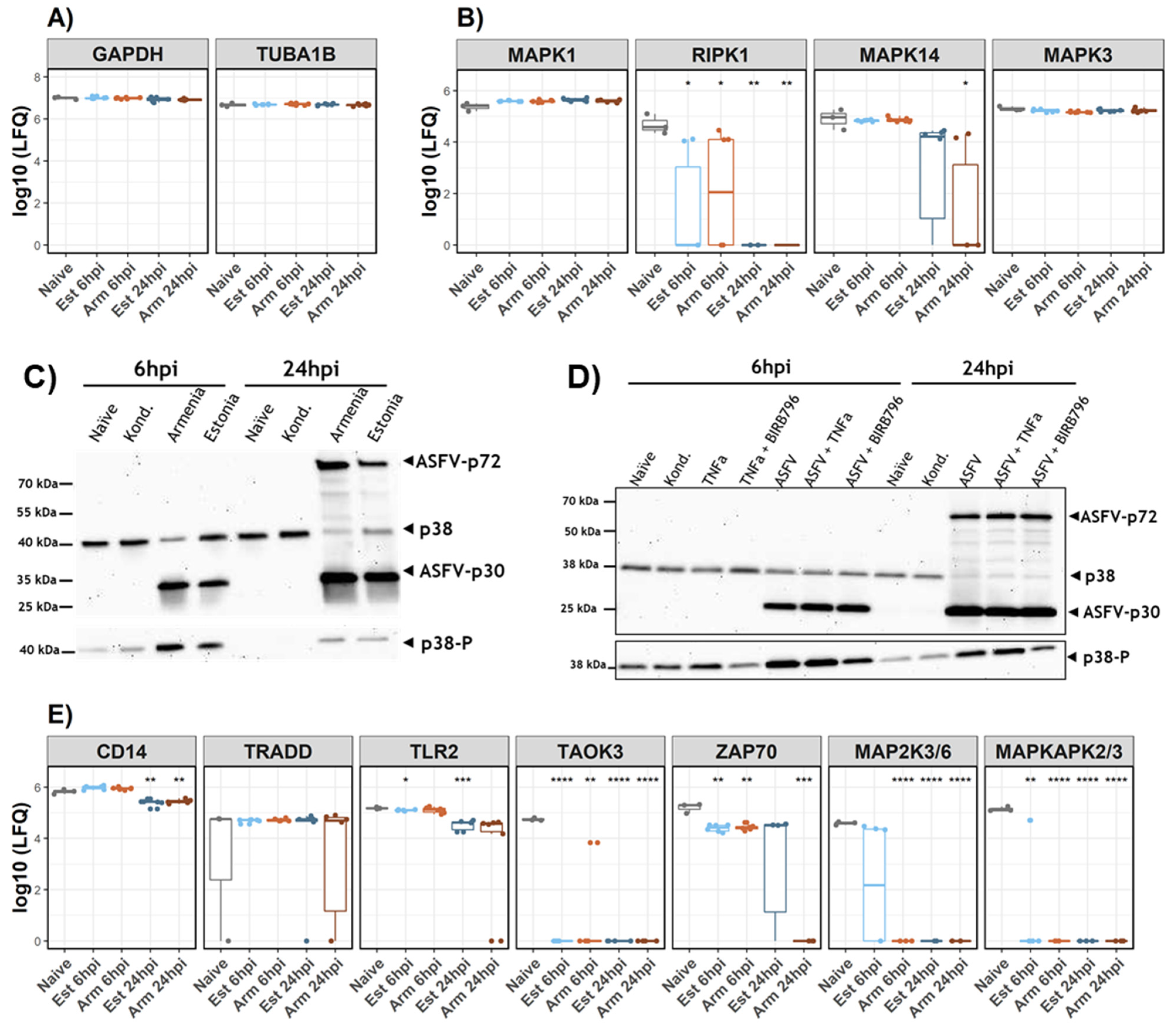

| nORF | Annotated ORF | # Unique Peptides | Detections (Replicates) | Kinetics | |||
|---|---|---|---|---|---|---|---|
| Estonia 2014 | Armenia 2008 | ||||||
| 6 hpi | 24 hpi | 6 hpi | 24 hpi | ||||
| nORF_18417 | 1 | 3 | - | 2 | - | early | |
| nORF_176208 | DP238L (e) | 1 | 6 | 6 | 6 | 6 | early |
| nORF_186513 | 2 | 3 | 3 | 1 | 1 | early | |
| nORF_125163 | CP204L (e) * | 2 | 3 | 4 | 2 | 1 | early |
| nORF_180573 | 4 | 6 | 6 | 6 | 6 | ambivalent | |
| nORF_63288 | K205R (e) | 2 | 6 | 6 | 6 | 6 | ambivalent |
| nORF_105178 | B646L (l) * | 2 | - | 5 | - | 6 | late |
| nORF_143942 | 1 | - | 4 | - | 6 | late | |
| nORF_119520 | CP2475L (l) * | 1 | - | 5 | - | 5 | late |
| nORF_63974 | K78R (l) * | 2 | - | 6 | - | 6 | late |
| nORF_188532 | 1 | - | - | - | 2 | late | |
| Enriched GO and KEGG Terms and Descriptions | Armenia 2008 | Estonia 2014 | ||
|---|---|---|---|---|
| 6 hpi | 24 hpi | 6 hpi | 24 hpi | |
| Phagosome KEGG:04145 | ||||
| Lysosome KEGG:04142 | ||||
| Citrate cycle (TCA cycle) KEGG:00020 | ||||
| Autophagy related diseases KEGG:05010, KEGG:05014, KEGG:05016, KEGG:05012 | ||||
| cell death GO:0008219, GO:0010941, GO:0012501, GO:0006915 | ||||
| regulation of programmed cell death GO:0043067, GO:0042981 | ||||
| intrinsic apoptotic signaling pathway GO:0097193 | ||||
| regulation of intrinsic apoptotic signaling pathway GO:2001242 | ||||
| positive regulation of apoptotic process GO:0043065, GO:2001235, GO:0010942, GO:2001238, GO:0043068, GO:2001236, GO:0043281 | ||||
| Protein processing in endoplasmic reticulum KEGG:04141 | ||||
| Spliceosome KEGG:03040, GO:0043484, GO:0048024 | ||||
| endoplasmic reticulum to Golgi vesicle-mediated transport GO:0006888 | ||||
| response to endoplasmic reticulum stress (ERAD pathway) GO:0036503, GO:0030433, GO:0034976 | ||||
| Proteasome KEGG:03050 | ||||
| immune response and regulation GO:0006955, GO:0002376, GO:0045087, GO:0050778, GO:0002684, GO:0050776, GO:0002682 | ||||
| regulation of innate immune response GO:0045088 | ||||
| Antigen processing and presentation KEGG:04612, GO:0019884, GO:0002478, GO:0002474, GO:0002504 | ||||
| Response to and production of cytokines GO:0071345, GO:0001816, GO:0001817, GO:0034097, GO:0001819, GO:0023056 | ||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wöhnke, E.; Cackett, G.; Werner, F.; Blome, S.; Mettenleiter, T.C.; Karger, A. Proteome Analysis of Swine Macrophages after Infection with Two Genotype II African Swine Fever Isolates of Different Pathogenicity. Viruses 2022, 14, 2140. https://doi.org/10.3390/v14102140
Wöhnke E, Cackett G, Werner F, Blome S, Mettenleiter TC, Karger A. Proteome Analysis of Swine Macrophages after Infection with Two Genotype II African Swine Fever Isolates of Different Pathogenicity. Viruses. 2022; 14(10):2140. https://doi.org/10.3390/v14102140
Chicago/Turabian StyleWöhnke, Elisabeth, Gwenny Cackett, Finn Werner, Sandra Blome, Thomas C. Mettenleiter, and Axel Karger. 2022. "Proteome Analysis of Swine Macrophages after Infection with Two Genotype II African Swine Fever Isolates of Different Pathogenicity" Viruses 14, no. 10: 2140. https://doi.org/10.3390/v14102140
APA StyleWöhnke, E., Cackett, G., Werner, F., Blome, S., Mettenleiter, T. C., & Karger, A. (2022). Proteome Analysis of Swine Macrophages after Infection with Two Genotype II African Swine Fever Isolates of Different Pathogenicity. Viruses, 14(10), 2140. https://doi.org/10.3390/v14102140






