Simultaneous Detection of RIG-1, MDA5, and IFIT-1 Expression Is a Convenient Tool for Evaluation of the Interferon-Mediated Response
Abstract
1. Introduction
2. Materials and Methods
2.1. Cells
2.2. WBC Isolation
2.3. In Vitro Transcription
2.4. Cell Transfection
2.5. Primer and Probe Design
2.6. RNA Isolation
2.7. Reverse Transcription
2.8. PCR Analysis
2.9. ELISA
2.10. Statistical Data Processing
3. Results
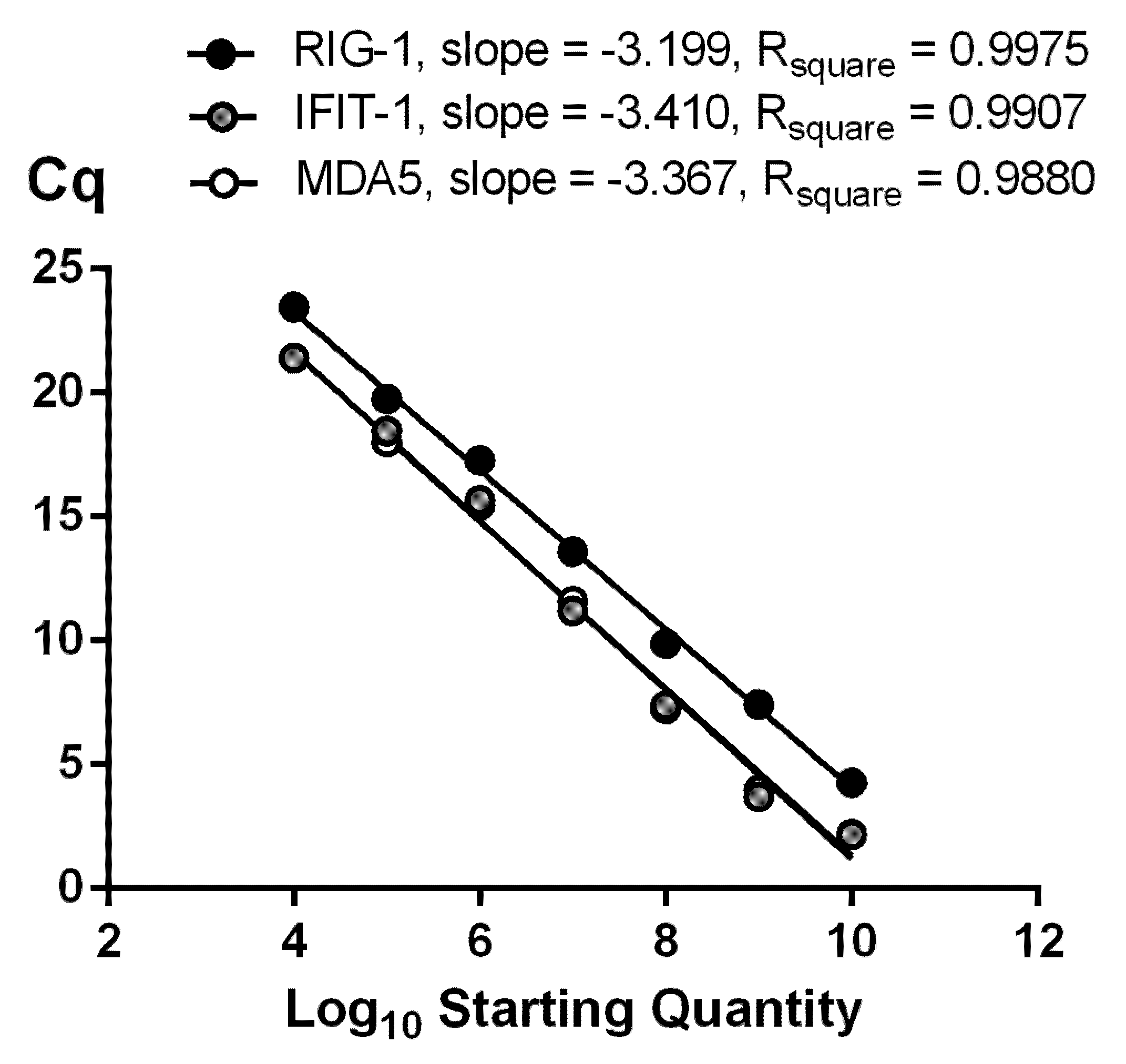
3.1. Multiplex Optimization
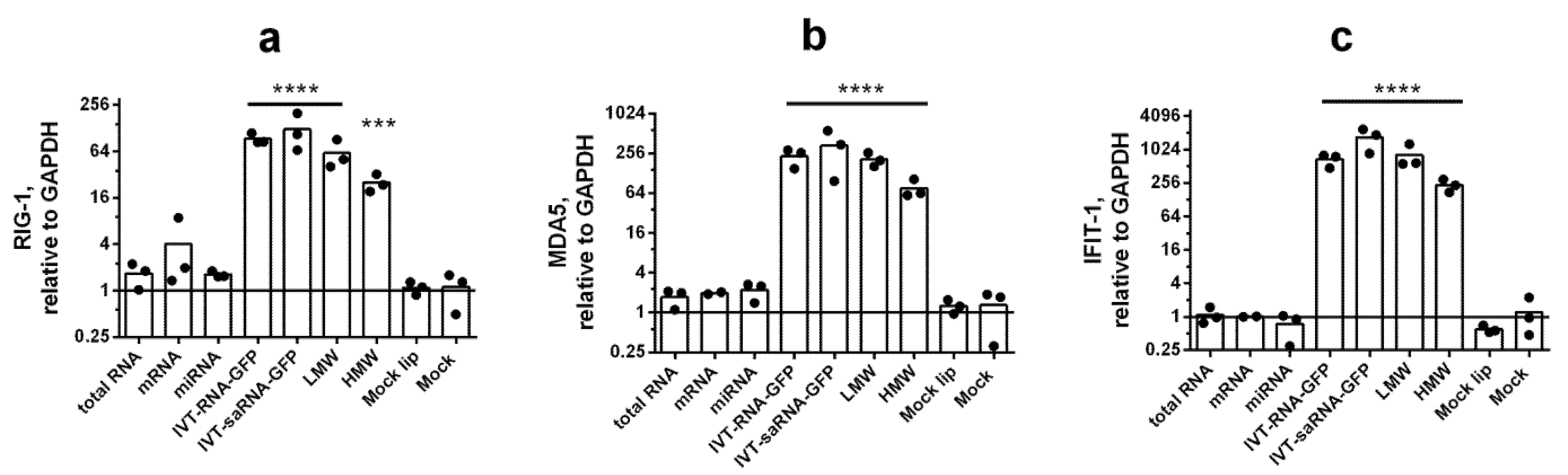
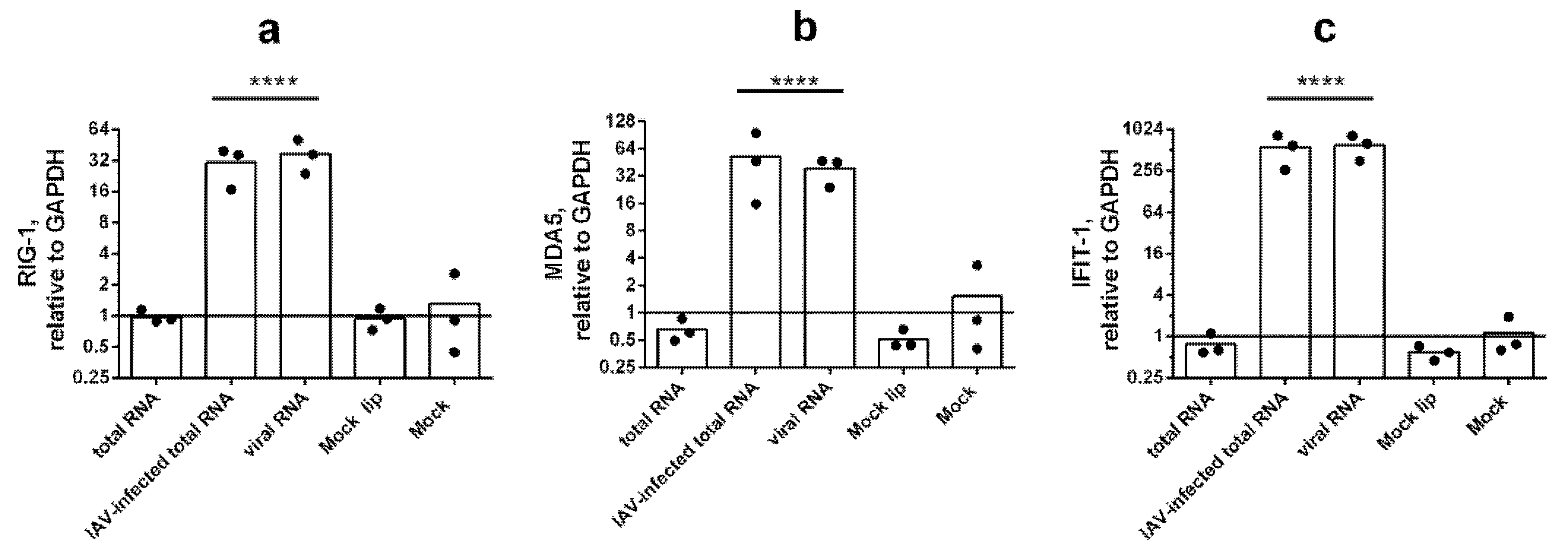
3.2. IFN-Mediated Response to Transfection
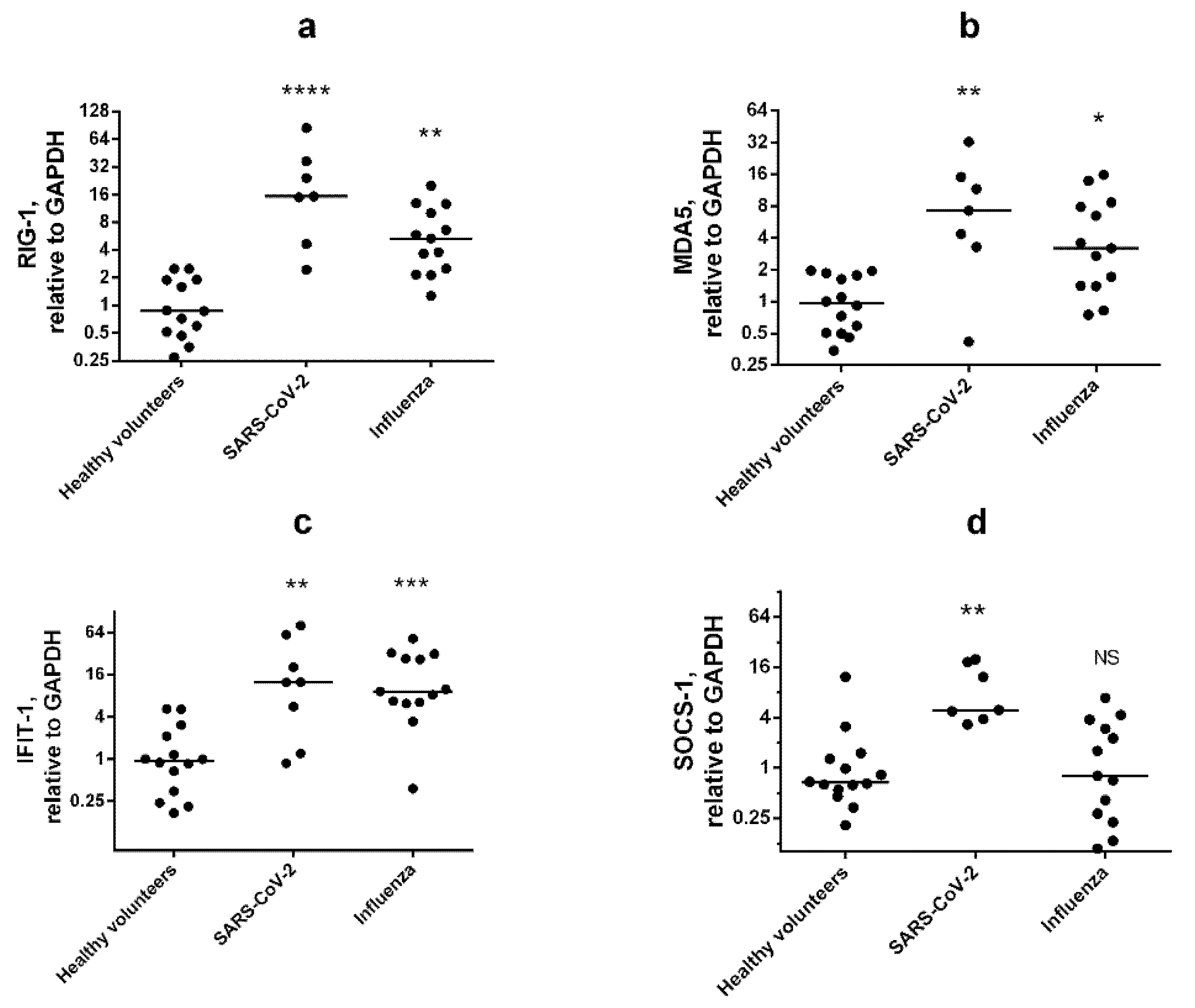
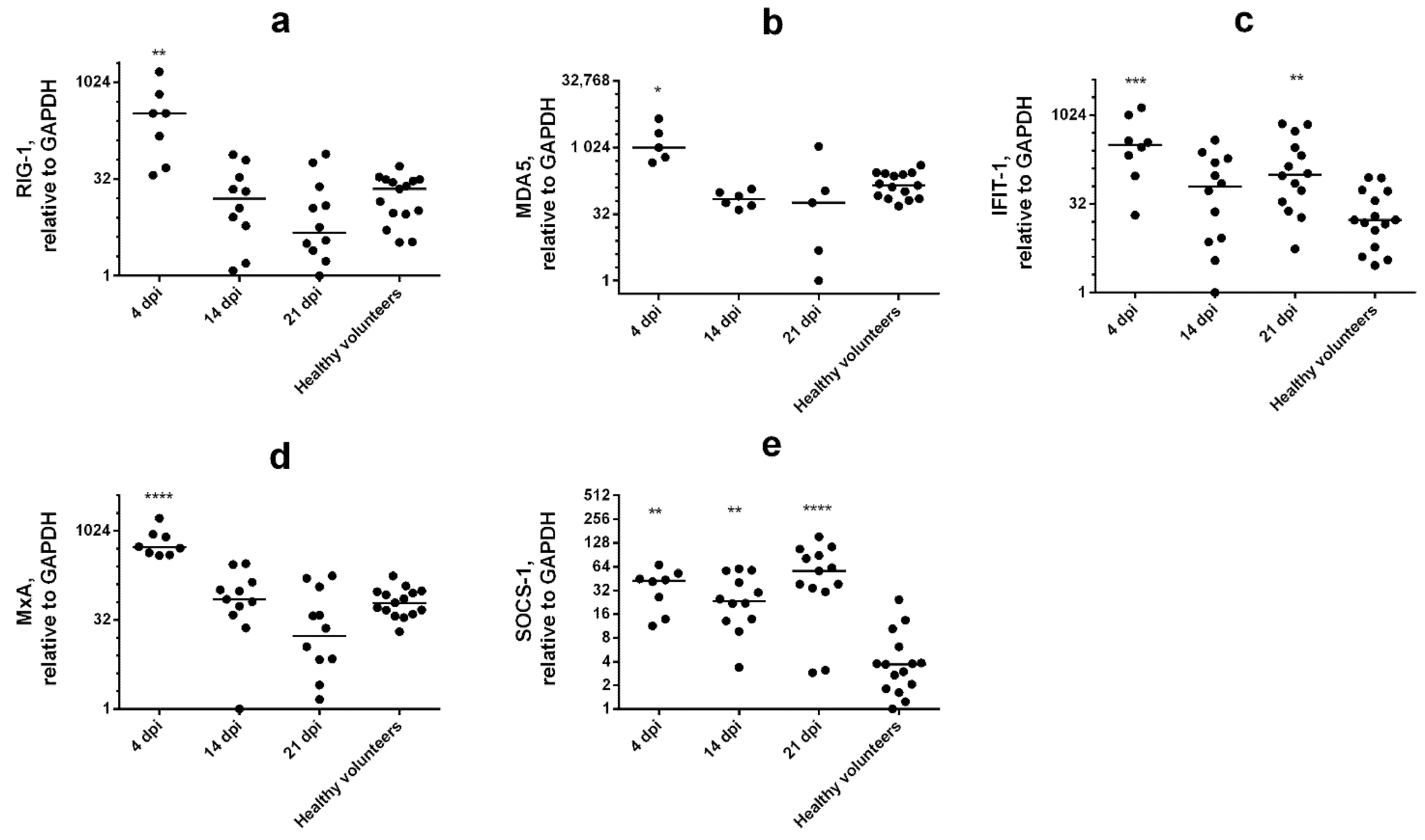
3.3. Multiplex Testing with WBCs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Thompson, M.R.; Kaminski, J.J.; Kurt-Jones, E.A.; Fitzgerald, K.A. Pattern recognition receptors and the innate immune response to viral infection. Viruses 2011, 3, 920–940. [Google Scholar] [CrossRef] [PubMed]
- Rehwinkel, J.; Gack, M.U. RIG-I-like receptors: Their regulation and roles in RNA sensing. Nat. Reviews. Immunol. 2020, 20, 537–551. [Google Scholar] [CrossRef] [PubMed]
- Li, M.M.; MacDonald, M.R.; Rice, C.M. To translate, or not to translate: Viral and host mRNA regulation by interferon-stimulated genes. Trends Cell Biol. 2015, 25, 320–329. [Google Scholar] [CrossRef] [PubMed]
- Terenzi, F.; Hui, D.J.; Merrick, W.C.; Sen, G.C. Distinct induction patterns and functions of two closely related interferon-inducible human genes, ISG54 and ISG56. J. Biol. Chem. 2006, 281, 34064–34071. [Google Scholar] [CrossRef]
- Guo, J.; Peters, K.L.; Sen, G.C. Induction of the human protein P56 by interferon, double-stranded RNA, or virus infection. Virology 2000, 267, 209–219. [Google Scholar] [CrossRef]
- Wacher, C.; Müller, M.; Hofer, M.J.; Getts, D.R.; Zabaras, R.; Ousman, S.S.; Terenzi, F.; Sen, G.C.; King, N.J.; Campbell, I.L. Coordinated regulation and widespread cellular expression of interferon-stimulated genes (ISG) ISG-49, ISG-54, and ISG-56 in the central nervous system after infection with distinct viruses. J. Virol. 2007, 81, 860–871. [Google Scholar] [CrossRef]
- Raychoudhuri, A.; Shrivastava, S.; Steele, R.; Kim, H.; Ray, R.; Ray, R.B. ISG56 and IFITM1 proteins inhibit hepatitis C virus replication. J. Virol. 2011, 85, 12881–12889. [Google Scholar] [CrossRef]
- Saikia, P.; Fensterl, V.; Sen, G.C. The inhibitory action of P56 on select functions of E1 mediates interferon’s effect on human papillomavirus DNA replication. J Virol. 2010, 84, 13036–13039. [Google Scholar] [CrossRef]
- Kimura, T.; Katoh, H.; Kayama, H.; Saiga, H.; Okuyama, M.; Okamoto, T.; Umemoto, E.; Matsuura, Y.; Yamamoto, M.; Takeda, K. Ifit1 inhibits Japanese encephalitis virus replication through binding to 5’ capped 2’-O unmethylated RNA. J. Virol. 2013, 87, 9997–10003. [Google Scholar] [CrossRef]
- Rabbani, M.A.; Ribaudo, M.; Guo, J.T.; Barik, S. Identification of Interferon-Stimulated Gene Proteins That Inhibit Human Parainfluenza Virus Type 3. J. Virol. 2016, 90, 11145–11156. [Google Scholar] [CrossRef]
- Yap, G.; Sachaphibulkij, K.; Foo, S.L.; Cui, J.; Fairhurst, A.M.; Lim, L. Annexin-A1 promotes RIG-I-dependent signaling and apoptosis via regulation of the IRF3-IFNAR-STAT1-IFIT1 pathway in A549 lung epithelial cells. Cell Death Dis. 2020, 11, 463. [Google Scholar] [CrossRef] [PubMed]
- Hawkins, S.; Guest, P.C. Multiplex Analyses Using Real-Time Quantitative PCR. Methods Mol. Biol. 2017, 1546, 125–133. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Li, C.; Xue, P.; Zhong, B.; Mao, A.P.; Ran, Y.; Chen, H.; Wang, Y.Y.; Yang, F.; Shu, H.B. ISG56 is a negative-feedback regulator of virus-triggered signaling and cellular antiviral response. Proc. Natl. Acad. Sci. USA 2009, 106, 7945–7950. [Google Scholar] [CrossRef] [PubMed]
- Haller, O.; Staeheli, P.; Schwemmle, M.; Kochs, G. Mx GTPases: Dynamin-like antiviral machines of innate immunity. Trends Microbiol. 2015, 23, 154–163. [Google Scholar] [CrossRef]
- Schneider, W.M.; Chevillotte, M.D.; Rice, C.M. Interferon-stimulated genes: A complex web of host defenses. Annu Rev Immunol. 2014, 32, 513–545. [Google Scholar] [CrossRef]
- Wang, R.; Yang, X.; Chang, M.; Xue, Z.; Wang, W.; Bai, L.; Zhao, S.; Liu, E. ORF3a Protein of Severe Acute Respiratory Syndrome Coronavirus 2 Inhibits Interferon-Activated Janus Kinase/Signal Transducer and Activator of Transcription Signaling via Elevating Suppressor of Cytokine Signaling 1. Front. Microbiol. 2021, 12, 752597. [Google Scholar] [CrossRef]
- Wei, H.; Wang, S.; Chen, Q.; Chen, Y.; Chi, X.; Zhang, L.; Huang, S.; Gao, G.F.; Chen, J.L. Suppression of interferon lambda signaling by SOCS-1 results in their excessive production during influenza virus infection. PLoS Pathog. 2014, 10, e1003845. [Google Scholar] [CrossRef]
- Pichlmair, A.; Lassnig, C.; Eberle, C.A.; Górna, M.W.; Baumann, C.L.; Burkard, T.R.; Bürckstümmer, T.; Stefanovic, A.; Krieger, S.; Bennett, K.L.; et al. IFIT1 is an antiviral protein that recognizes 5′-triphosphate RNA. Nat. Immunol. 2011, 12, 624–630. [Google Scholar] [CrossRef]
- Cagigi, A.; Loré, K. Immune Responses Induced by mRNA Vaccination in Mice, Monkeys and Humans. Vaccines 2021, 9, 61. [Google Scholar] [CrossRef]
- Edwards, D.K.; Jasny, E.; Yoon, H.; Horscroft, N.; Schanen, B.; Geter, T.; Fotin-Mleczek, M.; Petsch, B.; Wittman, V. Adjuvant effects of a sequence-engineered mRNA vaccine: Translational profiling demonstrates similar human and murine innate response. J. Transl. Med. 2017, 15, 1. [Google Scholar] [CrossRef]
- Daffis, S.; Szretter, K.J.; Schriewer, J.; Li, J.; Youn, S.; Errett, J.; Lin, T.Y.; Schneller, S.; Zust, R.; Dong, H.; et al. 2’-O methylation of the viral mRNA cap evades host restriction by IFIT family members. Nature 2010, 468, 452–456. [Google Scholar] [CrossRef] [PubMed]
- Hoagland, D.A.; Møller, R.; Uhl, S.A.; Oishi, K.; Frere, J.; Golynker, I.; Horiuchi, S.; Panis, M.; Blanco-Melo, D.; Sachs, D.; et al. Leveraging the antiviral type I interferon system as a first line of defense against SARS-CoV-2 pathogenicity. Immunity 2021, 54, 557–570. [Google Scholar] [CrossRef] [PubMed]
- Mantlo, E.; Bukreyeva, N.; Maruyama, J.; Paessler, S.; Huang, C. Antiviral activities of type I interferons to SARS-CoV-2 infection. Antivir. Res. 2020, 179, 104811. [Google Scholar] [CrossRef]
- Plotnikova, M.; Lozhkov, A.; Romanovskaya-Romanko, E.; Baranovskaya, I.; Sergeeva, M.; Kàà, K.; Klotchenko, S.; Vasin, A. IFN-λ1Displays Various Levels of AntiviralActivity In Vitro in a Select Panel ofRNA Viruses. Viruses 2021, 13, 1602. [Google Scholar] [CrossRef]
- Hadjadj, J.; Yatim, N.; Barnabei, L.; Corneau, A.; Boussier, J.; Smith, N.; Péré, H.; Charbit, B.; Bondet, V.; Chenevier-Gobeaux, C.; et al. Impaired type I interferon activity and inflammatory responses in severe COVID-19 patients. Science 2020, 369, 718–724. [Google Scholar] [CrossRef] [PubMed]
- Blanco-Melo, D.; Nilsson-Payant, B.E.; Liu, W.C.; Uhl, S.; Hoagland, D.; Møller, R.; Jordan, T.X.; Oishi, K.; Panis, M.; Sachs, D.; et al. Imbalanced Host Response to SARS-CoV-2 Drives Development of COVID-19. Cell 2020, 181, 1036–1045.e9. [Google Scholar] [CrossRef]
- Zhang, J.; Zhao, C.; Zhao, W. Virus Caused Imbalance of Type I IFN Responses and Inflammation in COVID-19. Front. Immunol. 2021, 12, 633769. [Google Scholar] [CrossRef]
- Combes, A.J.; Courau, T.; Kuhn, N.F.; Hu, K.H.; Ray, A.; Chen, W.S.; Chew, N.W.; Cleary, S.J.; Kushnoor, D.; Reeder, G.C.; et al. Global absence and targeting of protective immune states in severe COVID-19. Nature 2021, 591, 124–130. [Google Scholar] [CrossRef]
- Han, H.; Ma, Q.; Li, C.; Liu, R.; Zhao, L.; Wang, W.; Zhang, P.; Liu, X.; Gao, G.; Liu, F.; et al. Profiling serum cytokines in COVID-19 patients reveals IL-6 and IL-10 are disease severity predictors. Emerg. Microbes Infect. 2020, 9, 1123–1130. [Google Scholar] [CrossRef]
- Taraskin, A.S.; Semenov, K.K.; Lozhkov, A.A.; Baranovskaya, I.L.; Protasov, A.V.; Ramsay, E.S.; Tyulin, A.A.; Mirgorodskaya, O.A.; Vasin, A.V.; Klotchenko, S.A.; et al. A novel method for multiplex protein biomarker analysis of human serum using quantitative MALDI mass spectrometry. J. Pharm. Biomed. Anal. 2022, 210, 114575. [Google Scholar] [CrossRef]
- Wu, Y.; Ma, L.; Zhuang, Z.; Cai, S.; Zhao, Z.; Zhou, L.; Zhang, J.; Wang, P.H.; Zhao, J.; Cui, J. Main protease of SARS-CoV-2 serves as a bifunctional molecule in restricting type I interferon antiviral signaling. Signal Transduct. Target. Ther. 2020, 5, 221. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lozhkov, A.A.; Plotnikova, M.A.; Egorova, M.A.; Baranovskaya, I.L.; Elpaeva, E.A.; Klotchenko, S.A.; Vasin, A.V. Simultaneous Detection of RIG-1, MDA5, and IFIT-1 Expression Is a Convenient Tool for Evaluation of the Interferon-Mediated Response. Viruses 2022, 14, 2090. https://doi.org/10.3390/v14102090
Lozhkov AA, Plotnikova MA, Egorova MA, Baranovskaya IL, Elpaeva EA, Klotchenko SA, Vasin AV. Simultaneous Detection of RIG-1, MDA5, and IFIT-1 Expression Is a Convenient Tool for Evaluation of the Interferon-Mediated Response. Viruses. 2022; 14(10):2090. https://doi.org/10.3390/v14102090
Chicago/Turabian StyleLozhkov, Alexey A., Marina A. Plotnikova, Marya A. Egorova, Irina L. Baranovskaya, Ekaterina A. Elpaeva, Sergey A. Klotchenko, and Andrey V. Vasin. 2022. "Simultaneous Detection of RIG-1, MDA5, and IFIT-1 Expression Is a Convenient Tool for Evaluation of the Interferon-Mediated Response" Viruses 14, no. 10: 2090. https://doi.org/10.3390/v14102090
APA StyleLozhkov, A. A., Plotnikova, M. A., Egorova, M. A., Baranovskaya, I. L., Elpaeva, E. A., Klotchenko, S. A., & Vasin, A. V. (2022). Simultaneous Detection of RIG-1, MDA5, and IFIT-1 Expression Is a Convenient Tool for Evaluation of the Interferon-Mediated Response. Viruses, 14(10), 2090. https://doi.org/10.3390/v14102090





