Genetic Diversity and Spatiotemporal Dynamics of Chikungunya Infections in Mexico during the Outbreak of 2014–2016
Abstract
:1. Introduction
2. Materials and Methods
2.1. Virus Isolation, Reverse Transcription-Polymerase Chain Reaction Amplification, and Nucleotide Sequencing
2.2. Data Sets
2.3. Phylogenetic Analysis
2.4. Phylogeographic Inference
3. Results
3.1. Nonsynonymous Mutations in Mexican CHIKV Sequences
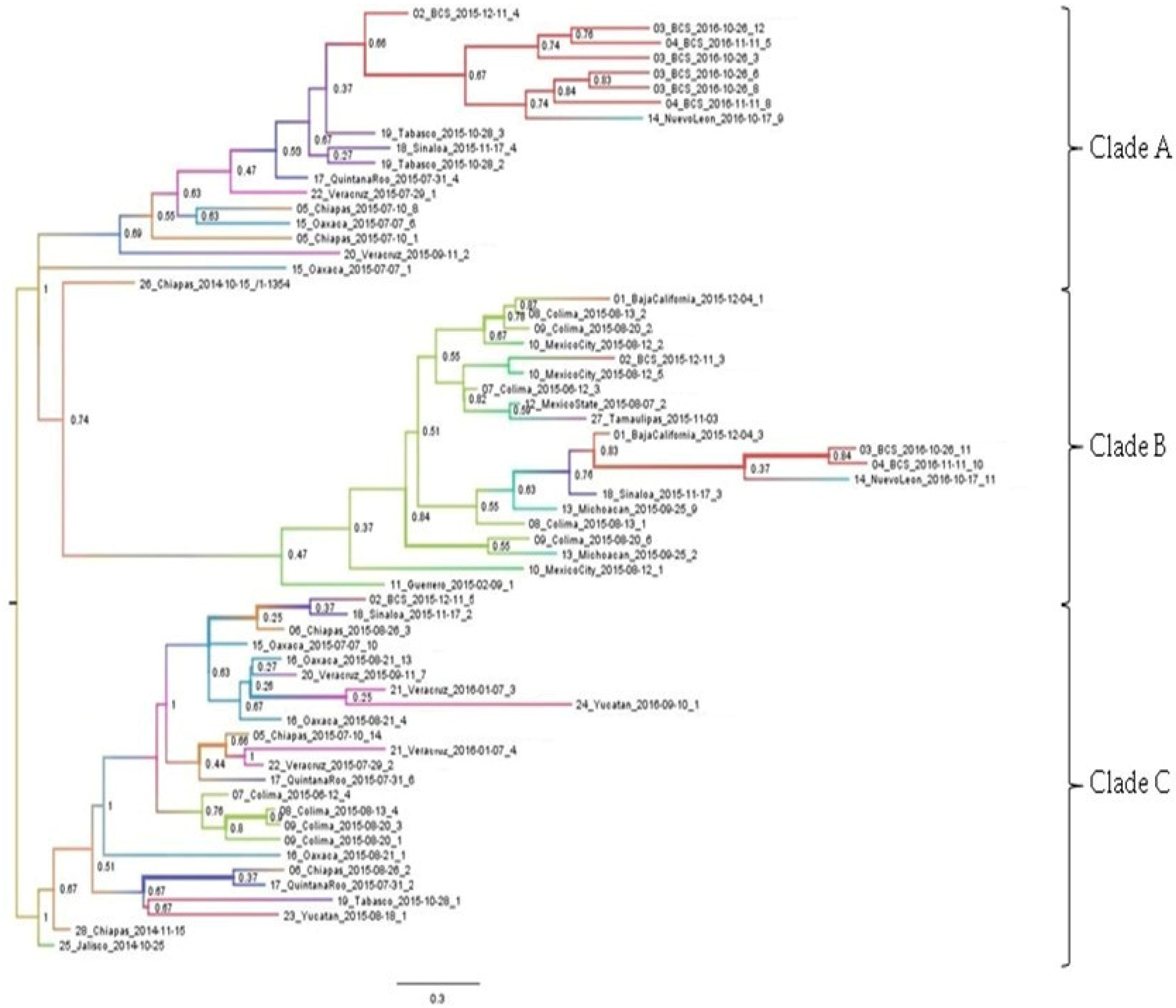
3.2. Phylogenetic Analysis
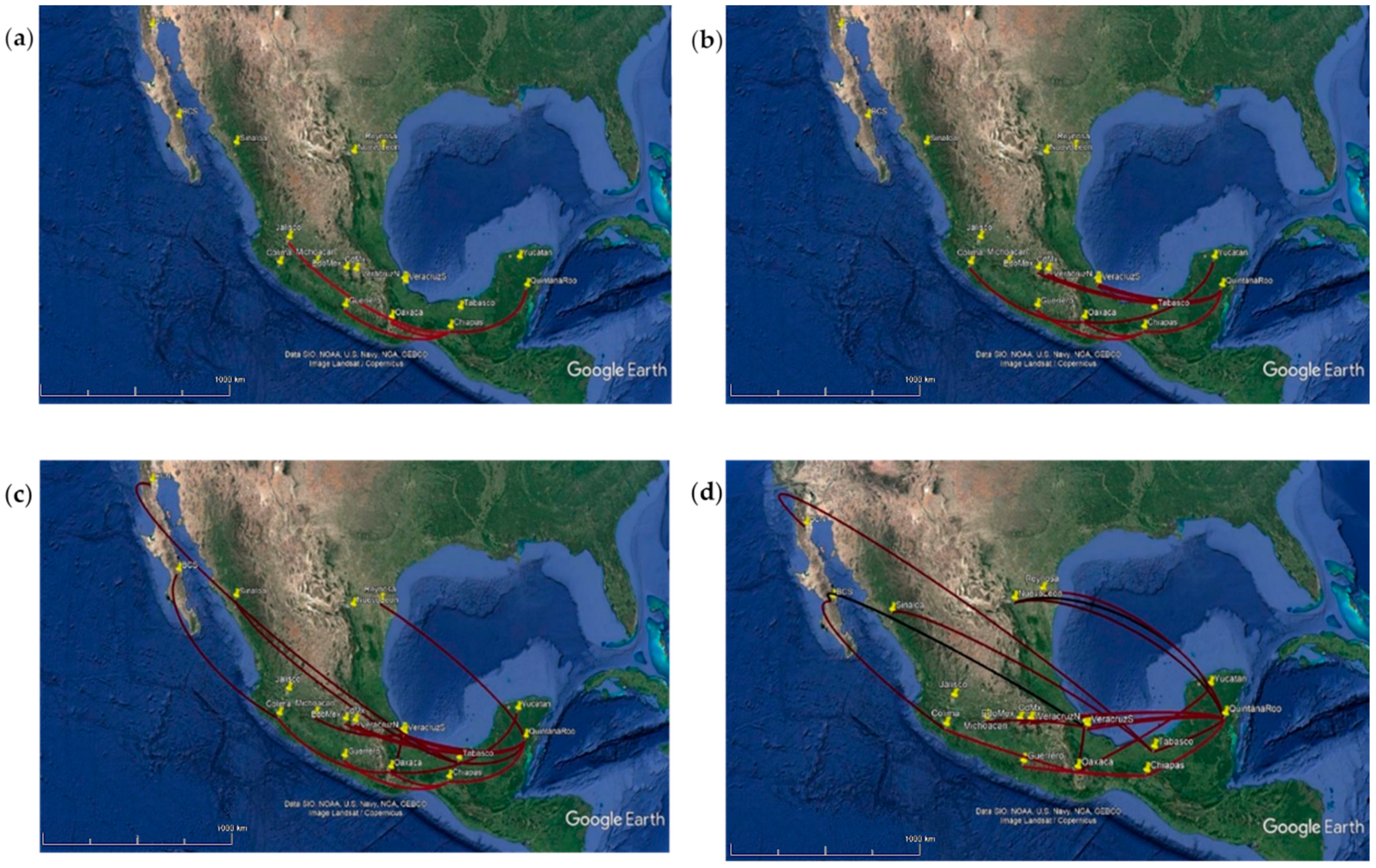
3.3. Phylogeographic Inference
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ramon-Pardo, P.; Cibrelus, L.; Yactayo, S. Chikungunya: Case definitions for acute, atypical and chronic cases. Conclusions of an expert consultation, Managua, Nicaragua, 20–21 May 2015. Wkly. Epidemiol. Rec. 2015, 90, 410–414. [Google Scholar]
- Suhrbier, A. Rheumatic manifestations of chikungunya: Emerging concepts and interventions. Nat. Rev. Rheumatol. 2019, 15, 597–611. [Google Scholar] [CrossRef]
- Economopoulou, A.; Dominguez, M.; Helynck, B.; Sissoko, D.; Wichmann, O.; Quenel, P.; Germonneau, P.; Quatresous, I. Atypical Chikungunya virus infections: Clinical manifestations, mortality and risk factors for severe disease during the 2005-2006 outbreak on Réunion. Epidemiol. Infect. 2009, 137, 534–541. [Google Scholar] [CrossRef] [Green Version]
- Godaert, L.; Najioullah, F.; Bartholet, S.; Colas, S.; Yactayo, S.; Cabié, A.; Fanon, J.-L.; Césaire, R.; Dramé, M. Atypical Clinical Presentations of Acute Phase Chikungunya Virus Infection in Older Adults. J. Am. Geriatr. Soc. 2017, 65, 2510–2515. [Google Scholar] [CrossRef]
- Dorléans, F.; Hoen, B.; Najioullah, F.; Herrmann-Storck, C.; Schepers, K.M.; Abel, S.; Lamaury, I.; Fagour, L.; Césaire, R.; Guyomard, S.; et al. Outbreak of Chikungunya in the French Caribbean Islands of Martinique and Guadeloupe: Findings from a Hospital-Based Surveillance System (2013–2015). Am. J. Trop. Med. Hyg. 2018, 98, 1819–1825. [Google Scholar] [CrossRef] [Green Version]
- Alvarez, M.F.; Bolívar-Mejía, A.; Rodriguez-Morales, A.J.; Ramirez-Vallejo, E. Cardiovascular involvement and manifestations of systemic Chikungunya virus infection: A systematic review. F1000Research 2017, 6, 390. [Google Scholar] [CrossRef]
- Burt, F.J.; Chen, W.; Miner, J.J.; Lenschow, D.J.; Merits, A.; Schnettler, E.; Kohl, A.; Rudd, P.A.; Taylor, A.; Herrero, L.J.; et al. Chikungunya virus: An update on the biology and pathogenesis of this emerging pathogen. Lancet Infect. Dis. 2017, 17, e107–e117. [Google Scholar] [CrossRef]
- Lo Presti, A.; Cella, E.; Angeletti, S.; Ciccozzi, M. Molecular epidemiology, evolution and phylogeny of Chikungunya virus: An updating review. Infect. Genet. Evol. 2016, 41, 270–278. [Google Scholar] [CrossRef]
- Zeller, H.; Van Bortel, W.; Sudre, B. Chikungunya: Its History in Africa and Asia and Its Spread to New Regions in 2013–2014. J. Infect. Dis. 2016, 214, S436–S440. [Google Scholar] [CrossRef]
- Nimmannitya, S.; Halstead, S.B.; Cohen, S.N.; Margiotta, M.R. Dengue and chikungunya virus infection in man in Thailand, 1962–1964. I. Observations on hospitalized patients with hemorrhagic fever. Am. J. Trop. Med. Hyg. 1969, 18, 954–971. [Google Scholar] [CrossRef]
- Schuffenecker, I.; Iteman, I.; Michault, A.; Murri, S.; Frangeul, L.; Vaney, M.-C.; Lavenir, R.; Pardigon, N.; Reynes, J.-M.; Pettinelli, F.; et al. Genome Microevolution of Chikungunya Viruses Causing the Indian Ocean Outbreak. PLoS Med. 2006, 3, e263. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arankalle, V.A.; Shrivastava, S.; Cherian, S.; Gunjikar, R.S.; Walimbe, A.M.; Jadhav, S.M.; Sudeep, A.B.; Mishra, A.C. Genetic divergence of Chikungunya viruses in India (1963–2006) with special reference to the 2005–2006 explosive epidemic. J. Gen. Virol. 2007, 88, 1967–1976. [Google Scholar] [CrossRef] [PubMed]
- Hapuarachchi, H.C.; Bandara, K.B.; Sumanadasa, S.D.; Hapugoda, M.D.; Lai, Y.L.; Lee, K.S.; Tan, L.K.; Lin, R.T.; Ng, L.F.; Bucht, G.; et al. Re-emergence of Chikungunya virus in South-east Asia: Virological evidence from Sri Lanka and Singapore. J. Gen. Virol. 2010, 91, 1067–1076. [Google Scholar] [CrossRef]
- Peyrefitte, C.N.; Bessaud, M.; Pastorino, B.A.M.; Gravier, P.; Plumet, S.; Merle, O.L.; Moltini, I.; Coppin, E.; Tock, F.; Daries, W.; et al. Circulation of Chikungunya virus in Gabon, 2006–2007. J. Med. Virol. 2008, 80, 430–433. [Google Scholar] [CrossRef]
- Kariuki Njenga, M.; Nderitu, L.; Ledermann, J.P.; Ndirangu, A.; Logue, C.H.; Kelly, C.H.L.; Sang, R.; Sergon, K.; Breiman, R.; Powers, A.M. Tracking epidemic Chikungunya virus into the Indian Ocean from East Africa. J. Gen. Virol. 2008, 89, 2754–2760. [Google Scholar] [CrossRef]
- Niyas, K.P.; Abraham, R.; Unnikrishnan, R.N.; Mathew, T.; Nair, S.; Manakkadan, A.; Issac, A.; Sreekumar, E. Molecular characterization of Chikungunya virus isolates from clinical samples and adult Aedes albopictus mosquitoes emerged from larvae from Kerala, South India. Virol. J. 2010, 7, 189. [Google Scholar] [CrossRef] [Green Version]
- Weaver, S.C.; Forrester, N.L. Chikungunya: Evolutionary history and recent epidemic spread. Antivir. Res. 2015, 120, 32–39. [Google Scholar] [CrossRef]
- Rezza, G.; Nicoletti, L.; Angelini, R.; Romi, R.; Finarelli, A.C.; Panning, M.; Cordioli, P.; Fortuna, C.; Boros, S.; Magurano, F.; et al. Infection with chikungunya virus in Italy: An outbreak in a temperate region. Lancet 2007, 370, 1840–1846. [Google Scholar] [CrossRef]
- AbuBakar, S.; Sam, I.-C.; Wong, P.-F.; MatRahim, N.; Hooi, P.-S.; Roslan, N. Reemergence of endemic Chikungunya, Malaysia. Emerg. Infect. Dis. 2007, 13, 147–149. [Google Scholar] [CrossRef]
- Cassadou, S.; Boucau, S.; Petit-Sinturel, M.; Huc, P.; Leparc-Goffart, I.; Ledrans, M. Emergence of chikungunya fever on the French side of Saint Martin island, October to December 2013. Euro Surveill. 2014, 19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leparc-Goffart, I.; Nougairede, A.; Cassadou, S.; Prat, C.; Lamballerie, X. de Chikungunya in the Americas. Lancet 2014, 383, 514. [Google Scholar] [CrossRef]
- Sahadeo, N.S.D.; Allicock, O.M.; De Salazar, P.M.; Auguste, A.J.; Widen, S.; Olowokure, B.; Gutierrez, C.; Valadere, A.M.; Polson-Edwards, K.; Weaver, S.C.; et al. Understanding the evolution and spread of chikungunya virus in the Americas using complete genome sequences. Virus Evol. 2017, 3, vex010. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Díaz-Quiñonez, J.A.; Ortiz-Alcántara, J.; Fragoso-Fonseca, D.E.; Garcés-Ayala, F.; Escobar-Escamilla, N.; Vázquez-Pichardo, M.; Núñez-León, A.; Torres-Rodríguez, M.D.; Torres-Longoria, B.; López-Martínez, I.; et al. Complete Genome Sequences of Chikungunya Virus Strains Isolated in Mexico: First Detection of Imported and Autochthonous Cases. Genome Announc. 2015, 3, e00300-15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kautz, T.F.; Díaz-González, E.E.; Erasmus, J.H.; Malo-García, I.R.; Langsjoen, R.M.; Patterson, E.I.; Auguste, D.I.; Forrester, N.L.; Sanchez-Casas, R.M.; Hernández-Ávila, M.; et al. Chikungunya Virus as Cause of Febrile Illness Outbreak, Chiapas, Mexico, 2014. Emerg. Infect. Dis. 2015, 21, 2070–2073. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Galan-Huerta, K.A.; Zomosa-Signoret, V.C.; Vidaltamayo, R.; Caballero-Sosa, S.; Fernández-Salas, I.; Ramos-Jiménez, J.; Rivas-Estilla, A.M. Genetic Variability of Chikungunya Virus in Southern Mexico. Viruses 2019, 11, 714. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Díaz-Quiñonez, J.A.; Escobar-Escamilla, N.; Ortíz-Alcántara, J.; Vázquez-Pichardo, M.; de la Luz Torres-Rodríguez, M.; Nuñez-León, A.; Torres-Longoria, B.; López-Martínez, I.; Ruiz-Matus, C.; Kuri-Morales, P.; et al. Identification of Asian genotype of chikungunya virus isolated in Mexico. Virus Genes 2016, 52, 127–129. [Google Scholar] [CrossRef]
- Laredo-Tiscareño, S.V.; Machain-Williams, C.; Rodríguez-Pérez, M.A.; Garza-Hernandez, J.A.; Doria-Cobos, G.L.; Cetina-Trejo, R.C.; Bacab-Cab, L.A.; Tangudu, C.S.; Charles, J.; De Luna-Santillana, E.J.; et al. Arbovirus Surveillance near the Mexico–U.S. Border: Isolation and Sequence Analysis of Chikungunya Virus from Patients with Dengue-like Symptoms in Reynosa, Tamaulipas. Am. J. Trop. Med. Hyg. 2018, 99, 191–194. [Google Scholar] [CrossRef] [Green Version]
- Galán-Huerta, K.A.; Martínez-Landeros, E.; Delgado-Gallegos, J.L.; Caballero-Sosa, S.; Malo-García, I.R.; Fernández-Salas, I.; Ramos-Jiménez, J.; Rivas-Estilla, A.M. Molecular and Clinical Characterization of Chikungunya Virus Infections in Southeast Mexico. Viruses 2018, 10, 248. [Google Scholar] [CrossRef] [Green Version]
- Muñoz-Medina, J.E.; Garcia-Knight, M.A.; Sanchez-Flores, A.; Monroy-Muñoz, I.E.; Grande, R.; Esbjörnsson, J.; Santacruz-Tinoco, C.E.; González-Bonilla, C.R. Evolutionary analysis of the Chikungunya virus epidemic in Mexico reveals intra-host mutational hotspots in the E1 protein. PLoS ONE 2018, 13, e0209292. [Google Scholar] [CrossRef] [Green Version]
- Danis-Lozano, R.; Díaz-González, E.E.; Malo-García, I.R.; Rodríguez, M.H.; Ramos-Castañeda, J.; Juárez-Palma, L.; Ramos, C.; López-Ordóñez, T.; Mosso-González, C.; Fernández-Salas, I. Vertical transmission of dengue virus in Aedes aegypti and its role in the epidemiological persistence of dengue in Central and Southern Mexico. Trop. Med. Int. Health 2019, 24, 1311–1319. [Google Scholar] [CrossRef]
- Yang, B.; Wang, Y.; Qian, P.-Y. Sensitivity and correlation of hypervariable regions in 16S rRNA genes in phylogenetic analysis. BMC Bioinform. 2016, 17, 135. [Google Scholar] [CrossRef] [Green Version]
- Dong, W.; Liu, J.; Yu, J.; Wang, L.; Zhou, S. Highly Variable Chloroplast Markers for Evaluating Plant Phylogeny at Low Taxonomic Levels and for DNA Barcoding. PLoS ONE 2012, 7, e35071. [Google Scholar] [CrossRef] [PubMed]
- Pichler, M.; Coskun, Ö.K.; Ortega-Arbulú, A.; Conci, N.; Wörheide, G.; Vargas, S.; Orsi, W.D. A 16S rRNA gene sequencing and analysis protocol for the Illumina MiniSeq platform. Microbiologyopen 2018, 7, e00611. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [Green Version]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [Green Version]
- Rognes, T.; Flouri, T.; Nichols, B.; Quince, C.; Mahé, F. VSEARCH: A versatile open source tool for metagenomics. PeerJ 2016, 4, e2584. [Google Scholar] [CrossRef]
- Illingworth, C.J.R.; Roy, S.; Beale, M.A.; Tutill, H.; Williams, R.; Breuer, J. On the effective depth of viral sequence data. Virus Evol. 2017, 3, vex030. [Google Scholar] [CrossRef] [Green Version]
- Kosakovsky Pond, S.L.; Posada, D.; Gravenor, M.B.; Woelk, C.H.; Frost, S.D.W. Automated Phylogenetic Detection of Recombination Using a Genetic Algorithm. Mol. Biol. Evol. 2006, 23, 1891–1901. [Google Scholar] [CrossRef] [PubMed]
- Sievers, F.; Wilm, A.; Dineen, D.; Gibson, T.J.; Karplus, K.; Li, W.; Lopez, R.; McWilliam, H.; Remmert, M.; Söding, J.; et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011, 7, 539. [Google Scholar] [CrossRef]
- Posada, D.; Crandall, K.A. MODELTEST: Testing the model of DNA substitution. Bioinformatics 1998, 14, 817–818. [Google Scholar] [CrossRef] [Green Version]
- Bouckaert, R.; Vaughan, T.G.; Barido-Sottani, J.; Duchêne, S.; Fourment, M.; Gavryushkina, A.; Heled, J.; Jones, G.; Kühnert, D.; Maio, N.D.; et al. BEAST 2.5: An advanced software platform for Bayesian evolutionary analysis. PLOS Comput. Biol. 2019, 15, e1006650. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Drummond, A.J.; Ho, S.Y.W.; Phillips, M.J.; Rambaut, A. Relaxed Phylogenetics and Dating with Confidence. PLOS Biol. 2006, 4, e88. [Google Scholar] [CrossRef]
- Suchard, M.A.; Weiss, R.E.; Sinsheimer, J.S. Bayesian Selection of Continuous-Time Markov Chain Evolutionary Models. Mol. Biol. Evol. 2001, 18, 1001–1013. [Google Scholar] [CrossRef] [Green Version]
- Parker, J.; Rambaut, A.; Pybus, O.G. Correlating viral phenotypes with phylogeny: Accounting for phylogenetic uncertainty. Infect. Genet. Evol. 2008, 8, 239–246. [Google Scholar] [CrossRef] [PubMed]
- Drummond, A.J.; Suchard, M.A.; Xie, D.; Rambaut, A. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol. Biol. Evol. 2012, 29, 1969–1973. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rambaut, A.; Drummond, A.J.; Xie, D.; Baele, G.; Suchard, M.A. Posterior Summarization in Bayesian Phylogenetics Using Tracer 1.7. Syst. Biol. 2018, 67, 901–904. [Google Scholar] [CrossRef] [Green Version]
- Lemey, P.; Rambaut, A.; Drummond, A.J.; Suchard, M.A. Bayesian Phylogeography Finds Its Roots. PLOS Comput. Biol. 2009, 5, e1000520. [Google Scholar] [CrossRef] [Green Version]
- Bielejec, F.; Rambaut, A.; Suchard, M.A.; Lemey, P. SPREAD: Spatial phylogenetic reconstruction of evolutionary dynamics. Bioinformatics 2011, 27, 2910–2912. [Google Scholar] [CrossRef] [Green Version]
- Shrinet, J.; Jain, S.; Sharma, A.; Singh, S.S.; Mathur, K.; Rana, V.; Bhatnagar, R.K.; Gupta, B.; Gaind, R.; Deb, M.; et al. Genetic Characterization of Chikungunya Virus from New Delhi Reveal Emergence of a New Molecular Signature in Indian Isolates. Virol. J. 2012, 9, 100. [Google Scholar] [CrossRef] [Green Version]
- Tandel, K.; Kumar, M.; Shergill, S.P.S.; Sahai, K.; Gupta, R.M. Molecular Characterization and Phylogenetic Analysis of Chikungunya Virus from Delhi, India. Med. J. Armed Forces India 2019, 75, 266–273. [Google Scholar] [CrossRef] [PubMed]
- Sam, I.-C.; Loong, S.-K.; Michael, J.C.; Chua, C.-L.; Sulaiman, W.Y.W.; Vythilingam, I.; Chan, S.-Y.; Chiam, C.-W.; Yeong, Y.-S.; AbuBakar, S.; et al. Genotypic and Phenotypic Characterization of Chikungunya Virus of Different Genotypes from Malaysia. PLoS ONE 2012, 7, e50476. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chua, C.-L.; Sam, I.-C.; Merits, A.; Chan, Y.-F. Antigenic Variation of East/Central/South African and Asian Chikungunya Virus Genotypes in Neutralization by Immune Sera. PLoS Negl. Trop. Dis. 2016, 10, e0004960. [Google Scholar] [CrossRef] [Green Version]
- Suhana, O.; Nazni, W.A.; Apandi, Y.; Farah, H.; Lee, H.L.; Sofian-Azirun, M. Insight into the Origin of Chikungunya Virus in Malaysian Non-Human Primates via Sequence Analysis. Heliyon 2019, 5, e02682. [Google Scholar] [CrossRef] [Green Version]
- Tan, K.-K.; Sy, A.K.D.; Tandoc, A.O.; Khoo, J.-J.; Sulaiman, S.; Chang, L.-Y.; AbuBakar, S. Independent Emergence of the Cosmopolitan Asian Chikungunya Virus, Philippines 2012. Sci. Rep. 2015, 5, 12279. [Google Scholar] [CrossRef] [Green Version]
- Tsetsarkin, K.A.; Chen, R.; Yun, R.; Rossi, S.L.; Plante, K.S.; Guerbois, M.; Forrester, N.; Perng, G.C.; Sreekumar, E.; Leal, G.; et al. Multi-Peaked Adaptive Landscape for Chikungunya Virus Evolution Predicts Continued Fitness Optimization in Aedes Albopictus Mosquitoes. Nat. Commun. 2014, 5, 4084. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rokas, A.; Williams, B.L.; King, N.; Carroll, S.B. Genome-scale approaches to resolving incongruence in molecular phylogenies. Nature 2003, 425, 798–804. [Google Scholar] [CrossRef]
- Chen, R.; Puri, V.; Fedorova, N.; Lin, D.; Hari, K.L.; Jain, R.; Rodas, J.D.; Das, S.R.; Shabman, R.S.; Weaver, S.C. Comprehensive Genome Scale Phylogenetic Study Provides New Insights on the Global Expansion of Chikungunya Virus. J. Virol. 2016, 90, 10600–10611. [Google Scholar] [CrossRef] [Green Version]
- Gontcharov, A.A.; Marin, B.; Melkonian, M. Are Combined Analyses Better Than Single Gene Phylogenies? A Case Study Using SSU rDNA and rbcL Sequence Comparisons in the Zygnematophyceae (Streptophyta). Mol. Biol. Evol. 2004, 21, 612–624. [Google Scholar] [CrossRef] [PubMed]
- Tsetsarkin, K.A.; Vanlandingham, D.L.; McGee, C.E.; Higgs, S. A single mutation in chikungunya virus affects vector specificity and epidemic potential. PLoS Pathog. 2007, 3, e201. [Google Scholar] [CrossRef]
- Martin, E.; Moutailler, S.; Madec, Y.; Failloux, A.-B. Differential responses of the mosquito Aedes albopictus from the Indian Ocean region to two chikungunya isolates. BMC Ecol. 2010, 10, 8. [Google Scholar] [CrossRef] [Green Version]
- Agarwal, A.; Sharma, A.K.; Sukumaran, D.; Parida, M.; Dash, P.K. Two novel epistatic mutations (E1:K211E and E2:V264A) in structural proteins of Chikungunya virus enhance fitness in Aedes aegypti. Virology 2016, 497, 59–68. [Google Scholar] [CrossRef]
- Kawashima, K.D.; Suarez, L.-A.C.; Labayo, H.K.M.; Liles, V.R.; Salvoza, N.C.; Klinzing, D.C.; Daroy, M.L.G.; Matias, R.R.; Natividad, F.F. Complete Genome Sequence of Chikungunya Virus Isolated in the Philippines. Genome Announc. 2014, 2, e00336-14. [Google Scholar] [CrossRef] [Green Version]
- De Maio, N.; Wu, C.-H.; O’Reilly, K.M.; Wilson, D. New Routes to Phylogeography: A Bayesian Structured Coalescent Approximation. PLoS Genet. 2015, 11, e1005421. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fischer, M.; Staples, J.E. Chikungunya Virus Spreads in the Americas—Caribbean and South America, 2013–2014. MMWR Morb. Mortal. Wkly Rep. 2014, 63, 500–501. [Google Scholar]
- Souza, T.M.A.; Azeredo, E.L.; Badolato-Corrêa, J.; Damasco, P.V.; Santos, C.; Petitinga-Paiva, F.; Nunes, P.C.G.; Barbosa, L.S.; Cipitelli, M.C.; Chouin-Carneiro, T.; et al. First Report of the East-Central South African Genotype of Chikungunya Virus in Rio de Janeiro, Brazil. PLoS Curr. 2017, 9. [Google Scholar] [CrossRef] [PubMed]
- Tan, Y.; Pickett, B.E.; Shrivastava, S.; Gresh, L.; Balmaseda, A.; Amedeo, P.; Hu, L.; Puri, V.; Fedorova, N.B.; Halpin, R.A.; et al. Differing epidemiological dynamics of Chikungunya virus in the Americas during the 2014–2015 epidemic. PLoS Negl. Trop. Dis. 2018, 12, e0006670. [Google Scholar] [CrossRef] [PubMed]

| Location | 2015 | 2016 | Total |
|---|---|---|---|
| Baja California (B.C.) | 5 | 5 | |
| Baja California Sur (B.C.S.) | 14 | 24 | 38 |
| Chiapas | 23 | 23 | |
| Colima | 39 | 39 | |
| Ciudad de Mexico (CdMx) | 3 | 3 | |
| Guerrero | 16 | 16 | |
| Estado de Mexico (EdoMex) | 9 | 1 | 10 |
| Michoacan | 16 | 1 | 17 |
| Nuevo Leon | 4 | 4 | |
| Oaxaca | 41 | 41 | |
| Quintana Roo | 15 | 2 | 17 |
| Sinaloa | 9 | 2 | 11 |
| Tabasco | 5 | 5 | |
| Veracruz | 28 | 29 | 57 |
| Yucatan | 21 | 2 | 23 |
| Total | 244 | 65 | 309 |
| Gene | Mutation | Genotype/Lineage | Effect | References | ||||
|---|---|---|---|---|---|---|---|---|
| ECSA | IOL | Asian | Caribbean | Mexico | ||||
| E1 | A226V | A | V | A | A | A | Leads to increased fitness, dissemination to the salivary glands and transmissibility of the virus by Aedes albopictus | Schuffenecker et al., 2006 [11] |
| K211E | E | K | E | E | E | Increases fitness for Ae. aegypti, increase in virus infectivity (13 fold), dissemination (15 fold) and transmission (62 fold) compared to E1:226A virus. | Shrinet et al., 2012 [49] | |
| T145A | T | T | A | A | A | Unknown | Tandel et al., 2019 [50] | |
| S225A | A | A | S | S | S | I-Ching Sam et al., 2012 [51] | ||
| E2 | G118S | G | G | S | S | S | Unknown | Chong-Long Chua et al., 2016 [52] |
| S194G | G | G | S | S | S | |||
| V255I | I | I | V | V | V | |||
| D205G | G | G | D | D | D | Unknown | O. Suhana et al., 2019 [53] | |
| S207N | N | N | S | S | S | |||
| S248L | L | L | S | F * | F * | I-Ching Sam et al., 2012 [51]/*Kim-Kee Tan et al., 2015 [54] | ||
| K252Q | Q | K | K | K | K | Increased adaptation to A. albopictus. | Konstantin A. Tsetsarkin et al., 2014 [55] | |
| NSP3 | I383T | T | T | I | T | T | Unknown | O. Suhana et al., 2019 [53] |
| I413T | T | T | I | I | I | |||
| Q434L | L | L | Q | Q | Q | |||
| A437V | V | V | A | T | T | Kim-Kee Tan et al., 2015 [54] | ||
| I449M | M | M | I | I | I | I-Ching Sam et al., 2012 [51] | ||
| L451F | L | L | L | F | F | Kim-Kee Tan et al., 2015 [54] | ||
| R452Q | Q | Q | R | R | R | I-Ching Sam et al., 2012 [51] | ||
| I457T | T | T | I | I | I | |||
| T458A | A | A | T | T | T | |||
| V459T | T | T | V | V | V | |||
| L461P | P | P | L | L | L | |||
| S462N | N | N | S | S | S | |||
| P471S | S | S | P | P | P | |||
| D483N | N | N | D | N | N | Kim-Kee Tan et al., 2015 [54] | ||
| D484E | E | E | D | D | D | I-Ching Sam et al., 2012 [51] | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rodríguez-Aguilar, E.D.; Martínez-Barnetche, J.; González-Bonilla, C.R.; Tellez-Sosa, J.M.; Argotte-Ramos, R.; Rodríguez, M.H. Genetic Diversity and Spatiotemporal Dynamics of Chikungunya Infections in Mexico during the Outbreak of 2014–2016. Viruses 2022, 14, 70. https://doi.org/10.3390/v14010070
Rodríguez-Aguilar ED, Martínez-Barnetche J, González-Bonilla CR, Tellez-Sosa JM, Argotte-Ramos R, Rodríguez MH. Genetic Diversity and Spatiotemporal Dynamics of Chikungunya Infections in Mexico during the Outbreak of 2014–2016. Viruses. 2022; 14(1):70. https://doi.org/10.3390/v14010070
Chicago/Turabian StyleRodríguez-Aguilar, Eduardo D., Jesús Martínez-Barnetche, Cesar R. González-Bonilla, Juan M. Tellez-Sosa, Rocío Argotte-Ramos, and Mario H. Rodríguez. 2022. "Genetic Diversity and Spatiotemporal Dynamics of Chikungunya Infections in Mexico during the Outbreak of 2014–2016" Viruses 14, no. 1: 70. https://doi.org/10.3390/v14010070
APA StyleRodríguez-Aguilar, E. D., Martínez-Barnetche, J., González-Bonilla, C. R., Tellez-Sosa, J. M., Argotte-Ramos, R., & Rodríguez, M. H. (2022). Genetic Diversity and Spatiotemporal Dynamics of Chikungunya Infections in Mexico during the Outbreak of 2014–2016. Viruses, 14(1), 70. https://doi.org/10.3390/v14010070








