miRNAs, from Evolutionary Junk to Possible Prognostic Markers and Therapeutic Targets in COVID-19
Abstract
:1. Introduction
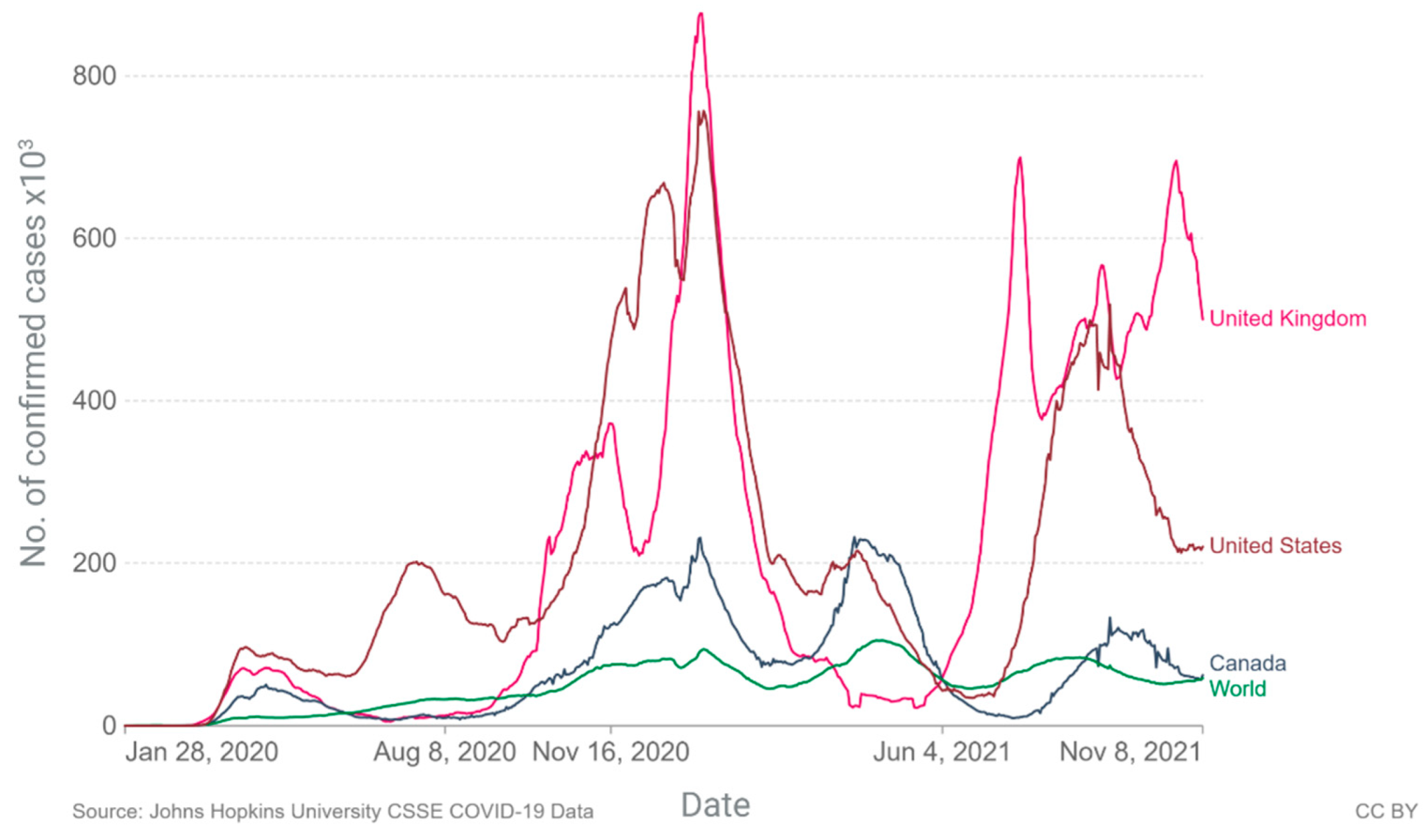
2. Current Epidemiology
3. miRNAs
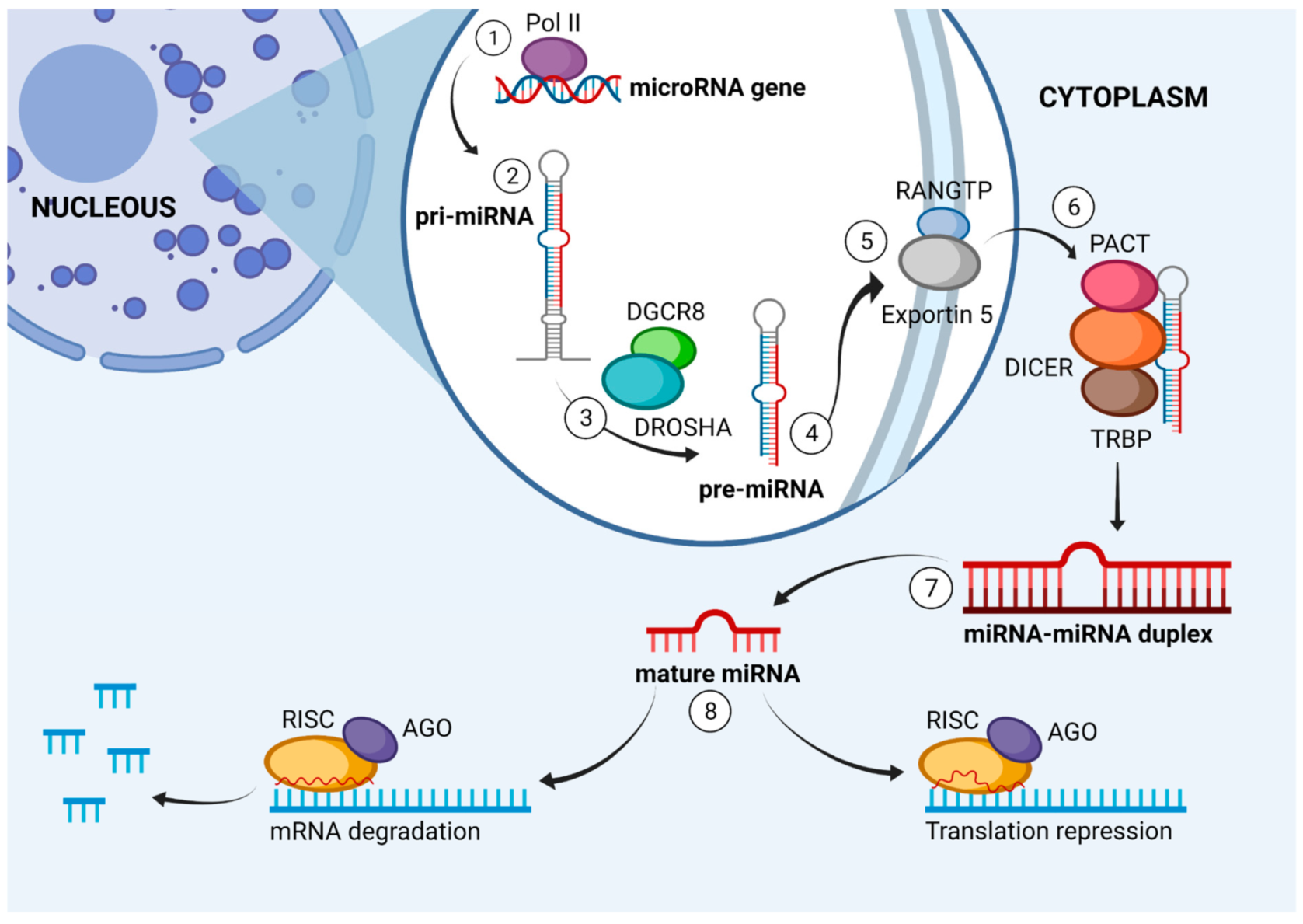
4. miRNA Biogenesis
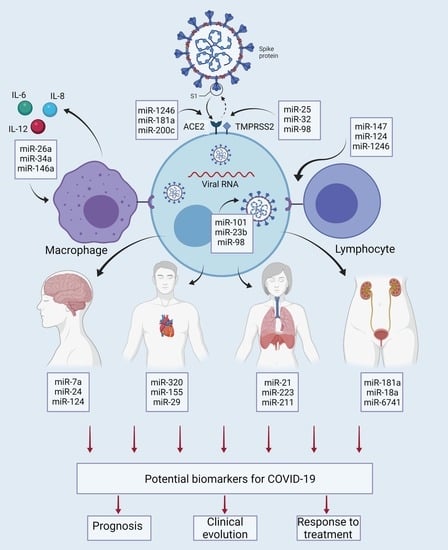
5. Interaction of miRNAs with Molecular and Cellular Elements in COVID-19
6. miRNAs of Viral Origin in COVID-19
7. miRNAs as Drivers of Major Complications in COVID-19
7.1. Coagulopathy
7.2. Lung Damage
7.3. Cardiac Damage
7.4. Kidney Damage
7.5. Neurological Damage
8. miRNAs as Possible Prognostic Markers in COVID-19
9. miRNAs as Predictive Markers of Clinical Course
10. miRNAs Associated with Response to Therapeutic Alternatives for COVID-19
11. miRNAs as Possible Therapeutic Targets
12. microRNA Delivery Systems
13. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Huang, C.; Wang, Y.; Li, X.; Ren, L.; Zhao, J.; Hu, Y.; Zhang, L.; Fan, G.; Xu, J.; Gu, X.; et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020, 395, 497–506. [Google Scholar] [CrossRef] [Green Version]
- Chen, N.; Zhou, M.; Dong, X.; Qu, J.; Gong, F.; Han, Y.; Qiu, Y.; Wang, J.; Liu, Y.; Wei, Y.; et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: A descriptive study. Lancet 2020, 395, 507–513. [Google Scholar] [CrossRef] [Green Version]
- Wang, D.; Hu, B.; Hu, C.; Zhu, F.; Liu, X.; Zhang, J.; Wang, B.; Xiang, H.; Cheng, Z.; Xiong, Y.; et al. Clinical Characteristics of 138 Hospitalized Patients With 2019 Novel Coronavirus—Infected Pneumonia in Wuhan, China. JAMA 2020, 323, 1061. [Google Scholar] [CrossRef] [PubMed]
- Coronaviridae Study Group of the International Committee on Taxonomy of Viruses. The species Severe acute respiratory syndrome-related coronavirus: Classifying 2019-nCoV and naming it SARS-CoV-2. Nat. Microbiol. 2020, 5, 536–544. [Google Scholar] [CrossRef] [Green Version]
- Director-General’s Opening Remarks at the Media Briefing on COVID-19—26 April World Health Organization. Available online: https://www.who.int/director-general/speeches/detail/director-general-s-opening-remarks-at-the-media-briefing-on-covid-19-26-april-2021 (accessed on 19 August 2021).
- Chams, N.; Chams, S.; Badran, R.; Shams, A.; Araji, A.; Raad, M.; Mukhopadhyay, S.; Stroberg, E.; Duval, E.J.; Barton, L.M.; et al. COVID-19: A Multidisciplinary Review. Front. Public Health 2020, 8, 383. [Google Scholar] [CrossRef] [PubMed]
- Hariyanto, T.I.; Japar, K.V.; Kwenandar, F.; Damay, V.; Siregar, J.I.; Lugito, N.P.H.; Tjiang, M.M.; Kurniawan, A. Inflammatory and hematologic markers as predictors of severe outcomes in COVID-19 infection: A systematic review and meta-analysis. Am. J. Emerg. Med. 2021, 41, 110–119. [Google Scholar] [CrossRef]
- Malik, P.; Patel, U.; Mehta, D.; Patel, N.; Kelkar, R.; Akrmah, M.; Gabrilove, J.L.; Sacks, H. Biomarkers and outcomes of COVID-19 hospitalisations: Systematic review and meta-analysis. BMJ Evid. Based Med. 2021, 26, 107–108. [Google Scholar] [CrossRef] [PubMed]
- Gobierno de México. Guía Clínica Para el Tratamiento de la COVID-19 en México. Actualización del 02 de Agosto. Available online: https://coronavirus.gob.mx/wpcontent/uploads/2021/08/GuiaTx_COVID19_ConsensoInterinstitucional_2021.08.03.pdf (accessed on 9 August 2021).
- Kermali, M.; Khalsa, R.K.; Pillai, K.; Ismail, Z.; Harky, A. The role of biomarkers in diagnosis of COVID-19—A systematic review. Life Sci. 2020, 254, 117788. [Google Scholar] [CrossRef]
- Zeng, F.; Huang, Y.; Guo, Y.; Yin, M.; Chen, X.; Xiao, L.; Deng, G. Association of inflammatory markers with the severity of COVID-19: A meta-analysis. Int. J. Infect. Dis. 2020, 96, 467–474. [Google Scholar] [CrossRef] [PubMed]
- Majumder, J.; Minko, T. Recent Developments on Therapeutic and Diagnostic Approaches for COVID-19. AAPS J. 2021, 23, 1–22. [Google Scholar] [CrossRef]
- Ritchie, H.; Mathieu, E.; Rodés-Guirao, L.; Appel, C.; Giattino, C.; Ortiz-Ospina, E.; Hasell, J.; Macdonald, B.; Beltekian, D.; Roser, M. Coronavirus Pandemic (COVID-19). Our World in Data. 2021. Available online: https://ourworldindata.org/coronavirus-data (accessed on 29 September 2021).
- World Health Organization. Listings of WHO’s Response to COVID-19. Available online: https://www.who.int/news/item/29-06-2020-covidtimeline (accessed on 29 September 2021).
- CDC. Cases, Data, and Surveillance. Centers for Disease Control and Prevention. 2021. Available online: https://www.cdc.gov/coronavirus/2019-ncov/covid-data/investigations-discovery/hospitalization-death-by-race-ethnicity.html (accessed on 31 August 2021).
- Selden, T.M.; Berdahl, T.A. COVID-19 And Racial/Ethnic Disparities in Health Risk, Employment, and Household Composition. Health Affairs 2020, 39, 1624–1632. [Google Scholar] [CrossRef]
- Price-Haywood, E.G.; Burton, J.; Fort, D.; Seoane, L. Hospitalization and Mortality among Black Patients and White Patients with COVID-19. N. Engl. J. Med. 2020, 382, 2534–2543. [Google Scholar] [CrossRef]
- Rodriguez, F.; Solomon, N.; de Lemos, J.A.; Das, S.R.; Morrow, D.A.; Bradley, S.M.; Elkind, M.S.V.; Williams, J.H.; Holmes, D.; Matsouaka, R.A.; et al. Racial and Ethnic Differences in Presentation and Outcomes for Patients Hospitalized With COVID-19: Findings from the American Heart Association’s COVID-19 Cardiovascular Disease Registry. Circulation 2021, 143, 2332–2342. [Google Scholar] [CrossRef] [PubMed]
- Mackey, K.; Ayers, C.K.; Kondo, K.K.; Saha, S.; Advani, S.M.; Young, S.; Spencer, H.; Rusek, M.; Anderson, J.; Veazie, S.; et al. Racial and Ethnic Disparities in COVID-19–Related Infections, Hospitalizations, and Deaths: A Systematic Review. Ann. Intern. Med. 2021, 174, 362–373. [Google Scholar] [CrossRef] [PubMed]
- Dluzen, D.F.; Noren Hooten, N.; Zhang, Y.; Kim, Y.; Glover, F.E.; Tajuddin, S.M.; Jacob, K.D.; Zonderman, A.B.; Evans, M.K. Racial Differences in MicroRNA and Gene Expression in Hypertensive Women. Sci. Rep. 2016, 6, 35815. [Google Scholar] [CrossRef]
- Liu, B.; Li, J.; Cairns, M.J. Identifying miRNAs, targets and functions. Brief. Bioinform. 2014, 15, 1–19. [Google Scholar] [CrossRef]
- Beermann, J.; Piccoli, M.-T.; Viereck, J.; Thum, T. Non-coding RNAs in Development and Disease: Background, Mechanisms, and Therapeutic Approaches. Physiol. Rev. 2016, 96, 1297–1325. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.; Zhu, S.; Meng, N.; He, Y.; Lu, R.; Yan, G.-R. ncRNA-Encoded Peptides or Proteins and Cancer. Mol. Ther. 2019, 27, 1718–1725. [Google Scholar] [CrossRef]
- Kabekkodu, S.; Shukla, V.; Varghese, V.K.; Souza, J.D.; Chakrabarty, S.; Satyamoorthy, K. Clustered miRNAs and their role in biological functions and diseases. Biol. Rev. 2018, 93, 1955–1986. [Google Scholar] [CrossRef]
- Li, C.; Hu, X.; Li, L.; Li, J. Differential microRNA expression in the peripheral blood from human patients with COVID-19. J. Clin. Lab. Anal. 2020, 34. [Google Scholar] [CrossRef]
- Chauhan, N.; Jaggi, M.; Chauhan, S.C.; Yallapu, M.M. COVID-19: Fighting the invisible enemy with microRNAs. Expert Rev. Anti-Infect. Ther. 2020, 19, 137–145. [Google Scholar] [CrossRef] [PubMed]
- Benz, F.; Roy, S.; Trautwein, C.; Roderburg, C.; Luedde, T. Circulating MicroRNAs as Biomarkers for Sepsis. Int. J. Mol. Sci. 2016, 17, 78. [Google Scholar] [CrossRef] [Green Version]
- De Sousa, M.C.; Gjorgjieva, M.; Dolicka, D.; Sobolewski, C.; Foti, M. Deciphering miRNAs’ Action through miRNA Editing. Int. J. Mol. Sci. 2019, 20, 6249. [Google Scholar] [CrossRef] [Green Version]
- Girardi, E.; López, P.; Pfeffer, S. On the Importance of Host MicroRNAs during Viral Infection. Front. Genet. 2018, 9, 439. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saliminejad, K.; Khorram Khorshid, H.R.; Soleymani Fard, S.; Ghaffari, S.H. An overview of microRNAs: Biology, functions, therapeutics, and analysis methods. J. Cell Physiol. 2019, 234, 5451–5465. [Google Scholar] [CrossRef]
- Guterres, A.; Lima, C.H.D.A.; Miranda, R.; Gadelha, M.R. What is the potential function of microRNAs as biomarkers and therapeutic targets in COVID-19? Infect. Genet. Evol. 2020, 85, 104417. [Google Scholar] [CrossRef] [PubMed]
- Parasher, A. COVID-19: Current understanding of its Pathophysiology, Clinical presentation and Treatment. Postgrad. Med. J. 2020, 97, 312–320. [Google Scholar] [CrossRef] [PubMed]
- Conti, P.; Ronconi, G.; Caraffa, A.; Gallenga, C.; Ross, R.; Frydas, I.; Kritas, S. Induction of pro-inflammatory cytokines (IL-1 and IL-6) and lung inflammation by Coronavirus-19 (COVI-19 or SARS-CoV-2): Anti-inflammatory strategies. J. Biol. Regul. Homeost. Agents 2020, 34, 1. [Google Scholar] [CrossRef]
- Nile, S.H.; Nile, A.; Qiu, J.; Li, L.; Jia, X.; Kai, G. COVID-19: Pathogenesis, cytokine storm and therapeutic potential of interferons. Cytokine Growth Factor Rev. 2020, 53, 66–70. [Google Scholar] [CrossRef]
- Mehta, P.; McAuley, D.; Brown, M.; Sanchez, E.; Tattersall, R.S.; Manson, J.J. COVID-19: Consider cytokine storm syndromes and immunosuppression. Lancet 2020, 395, 1033–1034. [Google Scholar] [CrossRef]
- Marques-Rocha, J.L.; Samblas-García, M.; Milagro, F.; Bressan, J.; Martínez, J.A.; Marti, A. Noncoding RNAs, cytokines, and inflammation-related diseases. FASEB J. 2015, 29, 3595–3611. [Google Scholar] [CrossRef] [Green Version]
- Nemecz, M.; Alexandru, N.; Tanko, G.; Georgescu, A. Role of MicroRNA in Endothelial Dysfunction and Hypertension. Curr. Hypertens. Rep. 2016, 18, 87. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Liang, L.; Zhang, X.-F.; Jia, H.-L.; Qin, Y.; Zhu, X.-C.; Gao, X.-M.; Qiao, P.; Zheng, Y.; Sheng, Y.-Y.; et al. MicroRNA-26a suppresses tumor growth and metastasis of human hepatocellular carcinoma by targeting interleukin-6-Stat3 pathway. Hepatology 2013, 58, 158–170. [Google Scholar] [CrossRef]
- Centa, A.; Fonseca, A.S.; Da Silva Ferreira, S.G.; Azevedo, M.L.V.; de Paula, C.B.V.; Nagashima, S.; Machado-Souza, C.; Dos Santos Miggiolaro, A.F.R.; Baena, C.P.; de Noronha, L.; et al. Deregulated miRNA expression is associated with endothelial dysfunction in post-mortem lung biopsies of COVID-19 patients. Am. J. Physiol. Cell. Mol. Physiol. 2021, 320, L405–L412. [Google Scholar] [CrossRef]
- Hart, M.; Walch-Rückheim, B.; Friedmann, K.S.; Rheinheimer, S.; Tänzer, T.; Glombitza, B.; Sester, M.; Lenhof, H.-P.; Hoth, M.; Schwarz, E.C.; et al. miR-34a: A new player in the regulation of T cell function by modulation of NF-κB signaling. Cell Death Dis. 2019, 10, 1–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tang, H.; Gao, Y.; Li, Z.; Miao, Y.; Huang, Z.; Liu, X.; Xie, L.; Li, H.; Wen, W.; Zheng, Y.; et al. The noncoding and coding transcriptional landscape of the peripheral immune response in patients with COVID-19. Clin. Transl. Med. 2020, 10. [Google Scholar] [CrossRef] [PubMed]
- O’Connell, R.M.; Rao, D.; Baltimore, D. microRNA Regulation of Inflammatory Responses. Annu. Rev. Immunol. 2012, 30, 295–312. [Google Scholar] [CrossRef]
- Cheng, H.; Sivachandran, N.; Lau, A.; Boudreau, E.; Zhao, J.L.; Baltimore, D.; Olguín, P.D.; Cybulsky, M.; Fish, J.E. Micro RNA-146 represses endothelial activation by inhibiting pro-inflammatory pathways. EMBO Mol. Med. 2013, 5, 1017–1034. [Google Scholar] [CrossRef]
- Li, S.; Yue, Y.; Xu, W.; Xiong, S. MicroRNA-146a Represses Mycobacteria-Induced Inflammatory Response and Facilitates Bacterial Replication via Targeting IRAK-1 and TRAF-6. PLoS ONE 2013, 8, e81438. [Google Scholar] [CrossRef]
- Ramkaran, P.; Khan, S.; Phulukdaree, A.; Moodley, D.; Chuturgoon, A.A. miR-146a Polymorphism Influences Levels of miR-146a, IRAK-1, and TRAF-6 in Young Patients with Coronary Artery Disease. Cell Biophys. 2014, 68, 259–266. [Google Scholar] [CrossRef]
- Hou, J.; Wang, P.; Lin, L.; Liu, X.; Ma, F.; An, H.; Wang, Z.; Cao, X. MicroRNA-146a Feedback Inhibits RIG-I-Dependent Type I IFN Production in Macrophages by Targeting TRAF6, IRAK1, and IRAK2. J. Immunol. 2009, 183, 2150–2158. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sheedy, F.J. Turning 21: Induction of miR-21 as a Key Switch in the Inflammatory Response. Front. Immunol. 2015, 6, 19. [Google Scholar] [CrossRef] [Green Version]
- Sonda, N.; Simonato, F.; Peranzoni, E.; Calì, B.; Bortoluzzi, S.; Bisognin, A.; Wang, E.; Marincola, F.M.; Naldini, L.; Gentner, B.; et al. miR-142-3p Prevents Macrophage Differentiation during Cancer-Induced Myelopoiesis. Immunity 2013, 38, 1236–1249. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aslani, M.; Mortazavi-Jahromi, S.S.; Mirshafiey, A. Cytokine storm in the pathophysiology of COVID-19: Possible functional disturbances of miRNAs. Int. Immunopharmacol. 2021, 101, 108172. [Google Scholar] [CrossRef]
- Salvi, V.; Gianello, V.; Tiberio, L.; Sozzani, S.; Bosisio, D. Cytokine Targeting by miRNAs in Autoimmune Diseases. Front. Immunol. 2019, 10, 15. [Google Scholar] [CrossRef] [PubMed]
- Bartoszewski, R.; Dabrowski, M.; Jakiela, B.; Matalon, S.; Harrod, K.S.; Sanak, M.; Collawn, J.F. SARS-CoV-2 may regulate cellular responses through depletion of specific host miRNAs. Am. J. Physiol. Cell. Mol. Physiol. 2020, 319, L444–L455. [Google Scholar] [CrossRef]
- Gatti, P.; Ilamathi, H.S.; Todkar, K.; Germain, M. Mitochondria Targeted Viral Replication and Survival Strategies-Prospective on SARS-CoV-2. Front. Pharmacol. 2020, 11, 578599. [Google Scholar] [CrossRef]
- McDonald, J.T.; Enguita, F.J.; Taylor, D.; Griffin, R.J.; Priebe, W.; Emmett, M.R.; Sajadi, M.M.; Harris, A.D.; Clement, J.; Dybas, J.M.; et al. Role of miR-2392 in driving SARS-CoV-2 infection. Cell Rep. 2021, 37, 109839. [Google Scholar] [CrossRef]
- Burtscher, J.; Cappellano, G.; Omori, A.; Koshiba, T.; Millet, G.P. Mitochondria: In the Cross Fire of SARS-CoV-2 and Immunity. iScience 2020, 23, 101631. [Google Scholar] [CrossRef]
- Yasukawa, K.; Kinoshita, D.; Yaku, K.; Nakagawa, T.; Koshiba, T. The microRNAs miR-302b and miR-372 regulate mitochondrial metabolism via the SLC25A12 transporter, which controls MAVS-mediated antiviral innate immunity. J. Biol. Chem. 2020, 295, 444–457. [Google Scholar] [CrossRef]
- Dutta, S.; Das, N.; Mukherjee, P. Picking up a Fight: Fine Tuning Mitochondrial Innate Immune Defenses against RNA Viruses. Front. Microbiol. 2020, 11, 1990. [Google Scholar] [CrossRef]
- Mishra, R.; Kumar, A.; Ingle, H.; Kumar, H. The Interplay between Viral-Derived miRNAs and Host Immunity During Infection. Front. Immunol. 2020, 10, 3079. [Google Scholar] [CrossRef] [Green Version]
- Aydemir, M.N.; Aydemir, H.B.; Korkmaz, E.M.; Budak, M.; Cekin, N.; Pinarbasi, E. Computationally predicted SARS-CoV-2 encoded microRNAs target NFKB, JAK/STAT and TGFB signaling pathways. Gene Rep. 2021, 22, 101012. [Google Scholar] [CrossRef]
- Liu, Z.; Wang, J.; Xu, Y.; Guo, M.; Mi, K.; Xu, R.; Pei, Y.; Zhang, Q.; Luan, X.; Liu, X.; et al. Implications of the virus-encoded miRNA and host miRNA in the pathogenicity of SARS-CoV-2. arXiv 2020, arXiv:2004.04874. [Google Scholar]
- Riolo, G.; Cantara, S.; Marzocchi, C.; Ricci, C. miRNA Targets: From Prediction Tools to Experimental Validation. Methods Protoc. 2020, 4, 1. [Google Scholar] [CrossRef] [PubMed]
- Meng, F.; Siu, G.K.-H.; Mok, B.W.-Y.; Sun, J.; Fung, K.S.C.; Lam, J.Y.-W.; Wong, N.K.; Gedefaw, L.; Luo, S.; Lee, T.M.H.; et al. Viral MicroRNAs Encoded by Nucleocapsid Gene of SARS-CoV-2 Are Detected during Infection, and Targeting Metabolic Pathways in Host Cells. Cells 2021, 10, 1762. [Google Scholar] [CrossRef] [PubMed]
- Mao, L.; Jin, H.; Wang, M.; Hu, Y.; Chen, S.; He, Q.; Chang, J.; Hong, C.; Zhou, Y.; Wang, D.; et al. Neurologic Manifestations of Hospitalized Patients with Coronavirus Disease 2019 in Wuhan, China. JAMA Neurol. 2020, 77, 683. [Google Scholar] [CrossRef] [Green Version]
- Helms, J.; Kremer, S.; Merdji, H.; Clere-Jehl, R.; Schenck, M.; Kummerlen, C.; Collange, O.; Boulay, C.; Fafi-Kremer, S.; Ohana, M.; et al. Neurologic Features in Severe SARS-CoV-2 Infection. New Engl. J. Med. 2020, 382, 2268–2270. [Google Scholar] [CrossRef] [PubMed]
- Levi, M.; Thachil, J.; Iba, T.; Levy, J.H. Coagulation abnormalities and thrombosis in patients with COVID-19. Lancet Haematol. 2020, 7, e438–e440. [Google Scholar] [CrossRef]
- Middeldorp, S.; Coppens, M.; Van Haaps, T.F.; Foppen, M.; Vlaar, A.P.; Müller, M.C.A.; Bouman, C.C.S.; Beenen, L.F.M.; Kootte, R.S.; Heijmans, J.; et al. Incidence of venous thromboembolism in hospitalized patients with COVID-19. J. Thromb. Haemost. 2020, 18, 1995–2002. [Google Scholar] [CrossRef] [PubMed]
- Grasselli, G.; Zangrillo, A.; Zanella, A.; Antonelli, M.; Cabrini, L.; Castelli, A.; Cereda, D.; Coluccello, A.; Foti, G.; Fumagali, R.; et al. Baseline Characteristics and Outcomes of 1591 Patients Infected with SARS-CoV-2 Admitted to ICUs of the Lombardy Region, Italy. JAMA 2020, 323, 1574. [Google Scholar] [CrossRef] [Green Version]
- Yang, X.; Yu, Y.; Xu, J.; Shu, H.; Xia, J.; Liu, H.; Wu, Y.; Zhang, L.; Yu, Z.; Fang, M.; et al. Clinical course and outcomes of critically ill patients with SARS-CoV-2 pneumonia in Wuhan, China: A single-centered, retrospective, observational study. Lancet Respir. Med. 2020, 8, 475–481. [Google Scholar] [CrossRef] [Green Version]
- Wang, F.; Kream, R.M.; Stefano, G.B. Long-Term Respiratory and Neurological Sequelae of COVID-19. Med. Sci. Monit. 2020, 26, e928996. [Google Scholar] [CrossRef]
- Chen, Y.-T.; Shao, S.-C.; Hsu, C.-K.; Wu, I.-W.; Hung, M.-J.; Chen, Y.-C. Incidence of acute kidney injury in COVID-19 infection: A systematic review and meta-analysis. Crit. Care 2020, 24, 1–4. [Google Scholar] [CrossRef] [PubMed]
- Han, X.; Ye, Q. Kidney involvement in COVID-19 and its treatments. J. Med. Virol. 2021, 93, 1387–1395. [Google Scholar] [CrossRef]
- Mao, R.; Qiu, Y.; He, J.S.; Tan, J.Y.; Li, X.H.; Liang, J.; Shen, J.; Zhu, L.-R.; Chen, Y.; Iacucci, M.; et al. Manifestations and prognosis of gastrointestinal and liver in-volvement in patients with COVID-19: A systematic review and meta-analysis. Lancet Gastroenterol. Hepatol. 2020, 5, 667–678. [Google Scholar] [CrossRef]
- Alqahtani, S.A.; Schattenberg, J.M. Liver injury in COVID-19: The current evidence. United Eur. Gastroenterol. J. 2020, 8, 509–519. [Google Scholar] [CrossRef]
- Iba, T.; Levy, J.H.; Levi, M.; Thachil, J. Coagulopathy in COVID-19. J. Thromb. Haemost. 2020, 18, 2103–2109. [Google Scholar] [CrossRef]
- Asakura, H.; Ogawa, H. COVID-19-associated coagulopathy and disseminated intravascular coagulation. Int. J. Hematol. 2021, 113, 45–57. [Google Scholar] [CrossRef] [PubMed]
- Zhou, F.; Yu, T.; Du, R.; Fan, G.; Liu, Y.; Liu, Z.; Xiang, J.; Wang, Y.; Song, B.; Gu, X.; et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: A retrospective cohort study. Lancet 2020, 395, 1054–1062. [Google Scholar] [CrossRef]
- Gambardella, J.; Sardu, C.; Morelli, M.B.; Messina, V.; Marfella, R.; Maggi, P.; Paolisso, G.; Wang, X.; Santulli, G. Abstract 221: Exosomal MicroRNAs Drive Tromboembolism in COVID-19. Circulation 2020, 142. [Google Scholar] [CrossRef]
- Di Minno, A.; Ambrosino, P.; Calcaterra, I.; Di Minno, M.N.D. COVID-19 and Venous Thromboembolism: A Meta-analysis of Literature Studies. Semin. Thromb. Hemost. 2020, 46, 763–771. [Google Scholar] [CrossRef]
- Damiani, S.; Fiorentino, M.; De Palma, A.; Foschini, M.P.; Lazzarotto, T.; Gabrielli, L.; Viale, P.L.; Attard, L.; Riefolo, M.; D’Errico, A. Pathological post-mortem findings in lungs infected with SARS-CoV-2. J. Pathol. 2020, 253, 31–40. [Google Scholar] [CrossRef] [PubMed]
- Miró, O.; Jiménez, S.; Mebazaa, A.; Freund, Y.; Burillo-Putze, G.; Martín, A.; Martín-Sánchez, F.J.; García-Lamberechts, E.J.; Alquézar-Arbé, A.; Jacob, J.; et al. Pulmonary embolism in patients with COVID-19: Incidence, risk factors, clinical characteristics, and outcome. Eur. Heart J. 2021, 42, 3127–3142. [Google Scholar] [CrossRef]
- Khurana, P.; Gupta, A.; Sugadev, R.; Sharma, Y.K.; Varshney, R.; Ganju, L.; Kumar, B. nSARS-CoV-2, pulmonary edema and thrombosis: Possible molecular insights using miRNA-gene circuits in regulatory networks. ExRNA 2020, 2, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Idell, S.; Zwieb, C.; Boggaram, J.; Holiday, D.; Johnson, A.R.; Raghu, G. Mechanisms of fibrin formation and lysis by human lung fibroblasts: Influence of TGF-beta and TNF-alpha. Am. J. Physiol. 1992, 263, L487–L494. [Google Scholar] [CrossRef]
- Whyte, C.S.; Mutch, N.J. uPA-mediated plasminogen activation is enhanced by polyphosphate. Haematologica 2021, 106, 522–531. [Google Scholar] [CrossRef] [Green Version]
- Chow, J.; Salmena, L. Prediction and Analysis of SARS-CoV-2-Targeting MicroRNA in Human Lung Epithelium. Genes 2020, 11, 1002. [Google Scholar] [CrossRef]
- Wang, M.; Li, J.; Cai, J.; Cheng, L.; Wang, X.; Xu, P.; Li, G.; Liang, X. Overexpression of MicroRNA-16 Alleviates Atherosclerosis by Inhibition of Inflammatory Pathways. BioMed Res. Int. 2020, 2020, 1–12. [Google Scholar] [CrossRef]
- Jiang, C.; Guo, Y.; Yu, H.; Lu, S.; Meng, L. Pleiotropic microRNA-21 in pulmonary remodeling: Novel insights for molecular mechanism and present advancements. Allergy Asthma Clin. Immunol. 2019, 15, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Dingsdag, S.A.; Clay, O.K.; Quintero, G.A. COVID-19 severity, miR -21 targets, and common human genetic variation. Letter regarding the article ‘Circulating cardiovascular microRNAs in critically ill COVID-19 patients. Eur. J. Heart Fail. 2021. [Google Scholar] [CrossRef]
- Garg, A.; Seeliger, B.; Derda, A.A.; Xiao, K.; Gietz, A.; Scherf, K.; Sonnenschein, K.; Pink, I.; Hoeper, M.M.; Welte, T.; et al. Circulating cardiovascular microRNAs in critically ill COVID-19 patients. Eur. J. Heart Fail. 2021, 23, 468–475. [Google Scholar] [CrossRef]
- Ferruelo, A.; Peñuelas, O.; Lorente, J.A. MicroRNAs as biomarkers of acute lung injury. Ann. Transl. Med. 2018, 6, 34. [Google Scholar] [CrossRef]
- Khan, A.T.; Khalid, Z.; Zahid, H.; Yousaf, M.A.; Shakoori, A.R. A computational and bioinformatic analysis of ACE2: An elucidation of its dual role in COVID-19 pathology and finding its associated partners as potential therapeutic targets. J. Biomol. Struct. Dyn. 2020, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Mahesh, G.; Biswas, R. MicroRNA-155: A Master Regulator of Inflammation. J. Interf. Cytokine Res. 2019, 39, 321–330. [Google Scholar] [CrossRef] [PubMed]
- Kim, C.; Jin, J.; Ye, Z.; Jadhav, R.R.; Gustafson, C.E.; Hu, B.; Cao, W.; Tian, L.; Weyand, C.M.; Goronzy, J.J. Histone deficiency and accelerated replication stress in T cell aging. J. Clin. Investig. 2021, 131. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Amahong, K.; Sun, X.; Lian, X.; Liu, J.; Sun, H.; Lou, Y.; Zhu, F.; Qiu, Y. The miRNA: A small but powerful RNA for COVID-19. Brief. Bioinform. 2021, 22, 1137–1149. [Google Scholar] [CrossRef] [PubMed]
- Knarr, M.; Avelar, R.A.; Sekhar, S.C.; Kwiatkowski, L.J.; Dziubinski, M.L.; McAnulty, J.; Skala, S.; Avril, S.; Drapkin, R.; DiFeo, A. miR-181a initiates and perpetuates oncogenic transformation through the regulation of innate immune signaling. Nat. Commun. 2020, 11, 1–21. [Google Scholar] [CrossRef]
- Pierce, J.B.; Simion, V.; Icli, B.; Pérez-Cremades, D.; Cheng, H.S.; Feinberg, M.W. Computational Analysis of Targeting SARS-CoV-2, Viral Entry Proteins ACE2 and TMPRSS2, and Interferon Genes by Host MicroRNAs. Genes 2020, 11, 1354. [Google Scholar] [CrossRef]
- Mirzaei, R.; Mahdavi, F.; Badrzadeh, F.; Hosseini-Fard, S.R.; Heidary, M.; Jeda, A.S.; Mohammadi, T.; Roshani, M.; Yousefimashouf, R.; Keyvani, H.; et al. The emerging role of microRNAs in the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection. Int. Immunopharmacol. 2021, 90, 107204. [Google Scholar] [CrossRef]
- Demiray, A.; Sarı, T.; Çalışkan, A.; Nar, R.; Aksoy, L.; Akbubak, I.H. Serum microRNA signature is capable of predictive and prognostic factor for SARS-CoV-2 virulence. Turk. J. Biochem. 2021, 46, 245–253. [Google Scholar] [CrossRef]
- Marchi, R.; Sugita, B.; Centa, A.; Fonseca, A.S.; Bortoletto, S.; Fiorentin, K.; Ferreira, S.; Cavalli, L.R. The role of microRNAs in modulating SARS-CoV-2 infection in human cells: A systematic review. Infect. Genet. Evol. 2021, 91, 104832. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Li, Z.; Chen, Q.; Wang, L.; Zheng, J.; Lin, Z.; Li, W. NF-κB-Induced MicroRNA-211 Inhibits Interleukin-10 in Macrophages of Rats with Lipopolysaccharide-Induced Acute Respiratory Distress Syndrome. Cell. Physiol. Biochem. 2018, 45, 332–342. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gangemi, S.; Tonacci, A. AntagomiRs: A novel therapeutic strategy for challenging COVID-19 cytokine storm. Cytokine Growth Factor Rev. 2021, 58, 111–113. [Google Scholar] [CrossRef] [PubMed]
- Cordero, A.; Escribano, D.; Bertomeu-González, V. Complicaciones cardiovasculares y pronóstico en pacientes con COVID-19. Rev. Española Cardiol. Supl. 2020, 20, 9–13. [Google Scholar] [CrossRef]
- Zhou, S.S.; Jin, J.P.; Wang, J.Q.; Zhang, Z.G.; Freedman, J.H.; Zheng, Y.; Cai, L. miRNAS in cardiovascular diseases: Potential biomarkers, therapeutic targets and challenges. Acta Pharmacol. Sinica 2018, 39, 1073–1084. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kukreja, R.C.; Yin, C.; Salloum, F. MicroRNAs: New Players in Cardiac Injury and Protection. Mol. Pharmacol. 2011, 80, 558–564. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fayyad-Kazan, M.; Makki, R.; Skafi, N.; El Homsi, M.; Hamade, A.; El Majzoub, R.; Hamade, E.; Fayyad-Kazan, H.; Badran, B. Circulating miRNAs: Potential diagnostic role for coronavirus disease 2019 (COVID-19). Infect. Genet. Evol. 2021, 94, 105020. [Google Scholar] [CrossRef]
- Modin, D.; Claggett, B.; Sindet-Pedersen, C.; Lassen, M.C.H.; Skaarup, K.G.; Jensen, J.U.S.; Fralick, M.; Schou, M.; Lamberts, M.; Gerds, T.; et al. Acute COVID-19 and the Incidence of Is-chemic Stroke and Acute Myocardial Infarction. Circulation 2020, 142, 2080–2082. [Google Scholar] [CrossRef]
- Dou, Q.; Wei, X.; Zhou, K.; Yang, S.; Jia, P. Cardiovascular Manifestations and Mechanisms in Patients with COVID-19. Trends Endocrinol. Metab. 2020, 31, 893–904. [Google Scholar] [CrossRef] [PubMed]
- Ren, X.-P.; Wu, J.; Wang, X.; Sartor, M.A.; Qian, J.; Jones, K.; Nicolaou, P.; Pritchard, T.J.; Fan, G.-C. MicroRNA-320 Is Involved in the Regulation of Cardiac Ischemia/Reperfusion Injury by Targeting Heat-Shock Protein 20. Circulation 2009, 119, 2357–2366. [Google Scholar] [CrossRef] [Green Version]
- Wang, B.; Wang, Z.-M.; Ji, J.-L.; Gan, W.; Zhang, A.; Shi, H.-J.; Wang, H.; Lv, L.; Li, Z.; Tang, T.; et al. Macrophage-Derived Exosomal Mir-155 Regulating Cardiomyocyte Pyroptosis and Hypertrophy in Uremic Cardiomyopathy. JACC Basic Transl. Sci. 2020, 5, 148–166. [Google Scholar] [CrossRef]
- Li, J.; Su, H.; Zhu, Y.; Cao, Y.; Ma, X. ETS2 and microRNA-155 regulate the pathogenesis of heart failure through targeting and regulating GPR18 expression. Exp. Ther. Med. 2020, 19, 3469–3478. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, B.; Li, B.; Qin, F.; Bai, F.; Sun, C.; Liu, Q. Expression of serum microRNA-155 and its clinical importance in patients with heart failure after myocardial infarction. J. Int. Med. Res. 2019, 47, 6294–6302. [Google Scholar] [CrossRef]
- Lin, B.; Zhao, H.; Li, L.; Zhang, Z.; Jiang, N.; Yang, X.; Zhang, T.; Lian, B.; Liu, Y.; Zhang, C.; et al. Sirt1 improves heart failure through modulating the NF-κB p65/microRNA-155/BNDF signaling cascade. Aging 2020, 13, 14482–14498. [Google Scholar] [CrossRef] [PubMed]
- Demirci, Y.M.; Demirci, M.D.S. Circular RNA–MicroRNA–MRNA interaction predictions in SARS-CoV-2 infection. J. Integr. Bioinform. 2021, 18, 45–50. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Zhang, C.; Liu, L.; Xi, A.; Chen, B.; Li, Y.; Du, J. Macrophage-Derived mir-155-Containing Exosomes Suppress Fibroblast Proliferation and Promote Fibroblast Inflammation during Cardiac Injury. Mol. Ther. 2017, 25, 192–204. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Yang, L.; Yin, J.; Su, D.; Pan, Z.; Li, P.; Wang, X. MicroRNA-15b deteriorates hypoxia/reoxygenation-induced cardiomyocyte apoptosis by downregulating Bcl-2 and MAPK3. J. Investig. Med. 2018, 66, 39–45. [Google Scholar] [CrossRef] [PubMed]
- Niu, S.; Xu, L.; Yuan, Y.; Yang, S.; Ning, H.; Qin, X.; Xin, P.; Yuan, D.; Jiao, J.; Zhao, Y. Effect of down-regulated miR-15b-5p expression on arrhythmia and myocardial apoptosis after myocardial ischemia reperfusion injury in mice. Biochem. Biophys. Res. Commun. 2020, 530, 54–59. [Google Scholar] [CrossRef] [PubMed]
- Beyerstedt, S.; Casaro, E.B.; Rangel, E.B. COVID-19: Angiotensin-converting enzyme 2 (ACE2) expression and tissue susceptibility to SARS-CoV-2 infection. Eur. J. Clin. Microbiol. Infect. Dis. 2021, 40, 905–919. [Google Scholar] [CrossRef] [PubMed]
- Lu, D.; Chatterjee, S.; Xiao, K.; Riedel, I.; Wang, Y.; Foo, R.; Bär, C.; Thum, T. MicroRNAs targeting the SARS-CoV-2 entry receptor ACE2 in cardiomyocytes. J. Mol. Cell. Cardiol. 2020, 148, 46–49. [Google Scholar] [CrossRef] [PubMed]
- Van Rooij, E.; Sutherland, L.B.; Thatcher, J.E.; DiMaio, J.M.; Naseem, R.H.; Marshall, W.S.; Hill, J.A.; Olson, E.N. Dysregulation of microRNAs after myocardial infarction reveals a role of miR-29 in cardiac fibrosis. Proc. Natl. Acad. Sci. USA 2008, 105, 13027–13032. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Keikha, R.; Hashemi-Shahri, S.M.; Jebali, A. The relative expression of miR-31, miR-29, miR-126, and miR-17 and their mRNA targets in the serum of COVID-19 patients with different grades during hospitalization. Eur. J. Med. Res. 2021, 26, 1–8. [Google Scholar] [CrossRef]
- Widiasta, A.; Sribudiani, Y.; Nugrahapraja, H.; Hilmanto, D.; Sekarwana, N.; Rachmadi, D. Potential role of ACE2-related microRNAs in COVID-19-associated nephropathy. Non-Coding RNA Res. 2020, 5, 153–166. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Chung, A.C.; Dong, Y.; Yang, W.; Zhong, X.; Lan, H.Y. The microRNA miR-433 promotes renal fibrosis by amplifying the TGF-β/Smad3-Azin1 pathway. Kidney Int. 2013, 84, 1129–1144. [Google Scholar] [CrossRef] [Green Version]
- Cheng, Y.; Luo, R.; Wang, K.; Zhang, M.; Wang, Z.; Dong, L.; Li, J.; Yao, Y.; Ge, S.; Xu, G. Kidney disease is associated with in-hospital death of patients with COVID-19. Kidney Int. 2020, 97, 829–838. [Google Scholar] [CrossRef] [PubMed]
- Gabarre, P.; Dumas, G.; Dupont, T.; Darmon, M.; Azoulay, E.; Zafrani, L. Acute kidney injury in critically ill patients with COVID-19. Intensiv. Care Med. 2020, 46, 1339–1348. [Google Scholar] [CrossRef] [PubMed]
- Ahmadian, E.; Khatibi, S.M.H.; Soofiyani, S.R.; Abediazar, S.; Shoja, M.M.; Ardalan, M.; Vahed, S.Z. COVID-19 and kidney injury: Pathophysiology and molecular mechanisms. Rev. Med. Virol. 2021, 31, e2176. [Google Scholar] [CrossRef] [PubMed]
- Lan, H.Y. Diverse Roles of TGF-β/Smads in Renal Fibrosis and Inflammation. Int. J. Biol. Sci. 2011, 7, 1056–1067. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qin, W.; Chung, A.C.K.; Huang, X.R.; Meng, X.-M.; Hui, D.S.C.; Yu, C.-M.; Sung, J.J.Y.; Lan, H.Y. TGF-β/Smad3 Signaling Promotes Renal Fibrosis by Inhibiting miR-29. J. Am. Soc. Nephrol. 2011, 22, 1462–1474. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhu, X.; Jiang, X.-M.; Guo, J.; Fu, Z.; Zhou, Z.; Yang, P.; Guo, H.; Guo, X.; Liang, G.; et al. Decreased inhibition of exosomal miRNAs on SARS-CoV-2 replication underlies poor outcomes in elderly people and diabetic patients. Signal Transduct. Target Ther. 2021, 6, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Kumar, V.; Singhal, P.C. APOL1 and kidney cell function. Am. J. Physiol.-Ren. Physiol. 2019, 317, F463–F477. [Google Scholar] [CrossRef]
- Friedman, D.; Pollak, M.R. APOL1 and Kidney Disease: From Genetics to Biology. Annu. Rev. Physiol. 2020, 82, 323–342. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hirsch, J.S.; Ng, J.H.; Ross, D.W.; Sharma, P.; Shah, H.H.; Barnett, R.L.; Hazzan, A.D.; Fishbane, S.; Jhaveri, K.D.; Abate, M.; et al. Acute kidney injury in patients hospitalized with COVID-19. Kidney Int. 2020, 98, 209–218. [Google Scholar] [CrossRef] [PubMed]
- Parzibut, G.; Henket, M.; Moermans, C.; Struman, I.; Louis, E.; Malaise, M.; Louis, R.; Misset, B.; Njock, M.-S.; Guiot, J. A Blood Exosomal miRNA Signature in Acute Respiratory Distress Syndrome. Front. Mol. Biosci. 2021, 8. [Google Scholar] [CrossRef] [PubMed]
- Safdar, M.; Khan, M.S.; Karim, A.Y.; Omar, S.A.; Smail, S.W.; Saeed, M.; Zaheer, S.; Ali, M.; Ahmad, B.; Tasleem, M.; et al. SNPs at 3′UTR of APOL1 and miR-6741-3p target sites associated with kidney diseases more susceptible to SARS-CoV-2 infection: In silco and in vitro studies. Mamm. Genome 2021, 32, 389–400. [Google Scholar] [CrossRef] [PubMed]
- Saikumar, J.; Hoffmann, D.; Kim, T.-M.; Gonzalez, V.R.; Zhang, Q.; Goering, P.L.; Brown, R.P.; Bijol, V.; Park, P.J.; Waikar, S.S.; et al. Expression, Circulation, and Excretion Profile of MicroRNA-21, -155, and -18a Following Acute Kidney Injury. Toxicol. Sci. 2012, 129, 256–267. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, R.; Wang, K.; Yu, J.; Howard, D.; French, L.; Chen, Z.; Wen, C.; Xu, Z. The Spatial and Cell-Type Distribution of SARS-CoV-2 Receptor ACE2 in the Human and Mouse Brains. Front. Neurol. 2021, 11, 573095. [Google Scholar] [CrossRef]
- Li, Y.; Bai, W.; Hashikawa, T. The neuroinvasive potential of SARS-CoV2 may play a role in the respiratory failure of COVID-19 patients. J. Med. Virol. 2020, 92, 552–555. [Google Scholar] [CrossRef]
- Desforges, M.; Le Coupanec, A.; Dubeau, P.; Bourgouin, A.; Lajoie, L.; Dube, M.; Talbot, P.J. Human Coronaviruses and Other Respiratory Viruses: Underestimated Opportunistic Pathogens of the Central Nervous System? Viruses 2019, 12, 14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moriguchi, T.; Harii, N.; Goto, J.; Harada, D.; Sugawara, H.; Takamino, J.; Ueno, M.; Sakata, H.; Kondo, K.; Myose, N.; et al. A first case of meningitis/encephalitis associated with SARS-Coronavirus-2. Int. J. Infect. Dis. 2020, 94, 55–58. [Google Scholar] [CrossRef]
- Ye, M.; Ren, Y.; Lv, T. Encephalitis as a clinical manifestation of COVID-19. Brain Behav. Immun. 2020, 88, 945–946. [Google Scholar] [CrossRef] [PubMed]
- Vohora, D.; Jain, S.; Tripathi, M.; Potschka, H. COVID-19 and seizures: Is there a link? Epilepsia 2020, 61, 1840–1853. [Google Scholar] [CrossRef] [PubMed]
- Prasad, K.; AlOmar, S.Y.; Alqahtani, S.A.M.; Malik, Z.; Kumar, V. Brain Disease Network Analysis to Elucidate the Neurological Manifestations of COVID-19. Mol. Neurobiol. 2021, 58, 1875–1893. [Google Scholar] [CrossRef]
- Nannoni, S.; de Groot, R.; Bell, S.; Markus, H.S. Stroke in COVID-19: A systematic review and meta-analysis. Int. J. Stroke 2020, 16, 137–149. [Google Scholar] [CrossRef]
- Kim, T.; Mehta, S.L.; Morris-Blanco, K.C.; Chokkalla, A.K.; Chelluboina, B.; Lopez, M.; Sullivan, R.; Kim, H.T.; Cook, T.D.; Kim, J.Y.; et al. The microRNA miR-7a-5p ameliorates ischemic brain damage by repressing α-synuclein. Sci. Signal. 2018, 11, eaat4285. [Google Scholar] [CrossRef] [Green Version]
- Mone, P.; Gambardella, J.; Wang, X.; Jankauskas, S.S.; Matarese, A.; Santulli, G. miR-24 targets SARS-CoV-2 co-factor Neuropilin-1 in human brain microvascular endothelial cells: In-sights for COVID-19 neurological manifestations. Res. Sq. 2021, rs-3. [Google Scholar] [CrossRef]
- Davies, J.; Randeva, H.S.; Chatha, K.; Hall, M.; Spandidos, D.A.; Karteris, E.; Kyrou, I. Neuropilin-1 as a new potential SARS-CoV-2 infection mediator implicated in the neurologic features and central nervous system involvement of COVID-19. Mol. Med. Rep. 2020, 22, 4221–4226. [Google Scholar] [CrossRef]
- Daly, J.L.; Simonetti, B.; Klein, K.; Chen, K.-E.; Williamson, M.K.; Antón-Plágaro, C.; Shoemark, D.K.; Simón-Gracia, L.; Bauer, M.; Hollandi, R.; et al. Neuropilin-1 is a host factor for SARS-CoV-2 infection. Science 2020, 370, 861–865. [Google Scholar] [CrossRef] [PubMed]
- Keikha, R.; Hashemi-Shahri, S.M.; Jebali, A. The miRNA neuroinflammatory biomarkers in COVID-19 patients with different severity of illness. Neurologia 2021. [Google Scholar] [CrossRef]
- Qin, Z.; Wang, P.-Y.; Su, D.-F.; Liu, X. miRNA-124 in Immune System and Immune Disorders. Front. Immunol. 2016, 7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brennan, G.P.; Dey, D.; Chen, Y.; Patterson, K.P.; Magnetta, E.J.; Hall, A.M.; Dube, C.M.; Mei, Y.-T.; Baram, T.Z. Dual and Opposing Roles of MicroRNA-124 in Epilepsy Are Mediated through Inflammatory and NRSF-Dependent Gene Networks. Cell Rep. 2016, 14, 2402–2412. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Terrinoni, A.; Calabrese, C.; Basso, D.; Aita, A.; Caporali, S.; Plebani, M.; Bernardini, S. The circulating miRNAs as diagnostic and prognostic markers. Clin. Chem. Lab. Med. 2018, 57, 932–953. [Google Scholar] [CrossRef] [Green Version]
- Jafarinejad-Farsangi, S.; Jazi, M.M.; Rostamzadeh, F.; Hadizadeh, M. High affinity of host human microRNAs to SARS-CoV-2 genome: An in silico analysis. Non-Coding RNA Res. 2020, 5, 222–231. [Google Scholar] [CrossRef]
- Rossato, M.; Affandi, A.J.; Thordardottir, S.; Wichers, C.G.K.; Cossu, M.; Broen, J.C.A.; Moret, F.M.; Bossini-Castillo, L.; Chouri, E.; Van Bon, L.; et al. Association of MicroRNA-618 Expression with Altered Frequency and Activation of Plasmacytoid Dendritic Cells in Patients with Systemic Sclerosis. Arthritis Rheumatol. 2017, 69, 1891–1902. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Song, Y.; Pang, Y.; Yu, Z.; Hua, W.; Gu, Y.; Qi, J.; Wu, H. miR-183-5p alleviates early injury after intracerebral hemorrhage by inhibiting heme oxygenase-1 expression. Aging 2020, 12, 12869–12895. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.J.; Kim, I.S.; Lee, S.G.; Kim, Y.J.; Silwal, P.; Kim, J.Y.; Kim, J.K.; Seo, W.; Chung, C.; Cho, H.K.; et al. MiR-144-3p is associated with pathological inflammation in patients infected with Mycobacteroides abscessus. Exp. Mol. Med. 2021, 53, 136–149. [Google Scholar] [CrossRef]
- Zhong, G.; Cheng, X.; Long, H.; He, L.; Qi, W.; Xiang, T.; Zhao, Z.; Zhu, B. Dynamically expressed microRNA-15b modulates the activities of CD8+ T lymphocytes in mice with Lewis lung carcinoma. J. Transl. Med. 2013, 11, 71. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gagnon, J.D.; Kageyama, R.; Shehata, H.M.; Fassett, M.S.; Mar, D.J.; Wigton, E.L.; Johansson, K.; Litterman, A.J.; Odorizzi, P.; Simeonov, D.; et al. miR-15/16 Restrain Memory T Cell Differentiation, Cell Cycle, and Survival. Cell Rep. 2019, 28, 2169–2181.e4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luo, Q.; Zhu, J.; Zhang, Q.; Xie, J.; Yi, C.; Li, T. MicroRNA-486-5p Promotes Acute Lung Injury via Inducing Inflammation and Apoptosis by Targeting OTUD7B. Inflammation 2020, 43, 975–984. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.; Wang, H.; Xiao, Y.; Jin, J.; Chang, J.H.; Zou, Q.; Xiq, X.; Cheng, X.; Sun, S.-C. Otud7b facilitates T cell activation and inflammatory re-sponses by regulating Zap70 ubiquitination. J. Exp. Med. 2016, 213, 399–414. [Google Scholar] [CrossRef] [Green Version]
- Lian, H.; Liu, C.; Li, M.; Hu, Y.; Shi, N.; Yu, H.; Liu, H. miR-486-5p attenuates tumor growth and lymphangiogenesis by targeting neuropilin-2 in colorectal carcinoma. OncoTargets Ther. 2016, 9, 2865–2871. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mucka, P.; Levonyak, N.; Geretti, E.; Zwaans, B.M.M.; Li, X.; Adini, I.; Klagsbrun, M.; Adam, R.M.; Bielenberg, D.R. Inflammation and Lymphedema Are Exacerbated and Prolonged by Neuropilin 2 Deficiency. Am. J. Pathol. 2016, 186, 2803–2812. [Google Scholar] [CrossRef] [Green Version]
- Gabryšová, L.; Alvarez-Martinez, M.; Luisier, R.; Cox, L.S.; Sodenkamp, J.; Hosking, C.; Pérez-Mazliah, D.; Whicher, C.; Kannan, Y.; Potempa, K.; et al. c-Maf controls immune responses by regulating disease-specific gene networks and repressing IL-2 in CD4+ T cells. Nat. Immunol. 2018, 19, 497–507. [Google Scholar] [CrossRef]
- Galicia, J.; Naqvi, A.R.; Ko, C.-C.; Nares, S.; Khan, A.A. MiRNA-181a regulates Toll-like receptor agonist-induced inflammatory response in human fibroblasts. Genes Immun. 2014, 15, 333–337. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hutchison, E.R.; Kawamoto, E.M.; Taub, D.D.; Lal, A.; Abdelmohsen, K.; Zhang, Y.; Wood, W.H.; Lehrmann, E.; Camandola, S.; Becker, K.G.; et al. Evidence for miR-181 involvement in neuroinflammatory responses of astrocytes. Glia 2013, 61, 1018–1028. [Google Scholar] [CrossRef]
- Lin, Y.; Deng, W.; Pang, J.; Kemper, T.; Hu, J.; Yin, J.; Zhang, J.; Lu, M. The microRNA-99 family modulates hepatitis B virus replication by promoting IGF-1R/PI3K/Akt/mTOR/ULK1 signaling-induced autophagy. Cell. Microbiol. 2017, 19, e12709. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Erlandsson, M.C.; Silfverswärd, S.T.; Nadali, M.; Turkkila, M.; Svensson, M.N.D.; Jonsson, I.-M.; Andersson, K.M.E.; Bokarewa, M.I. IGF-1R signalling contributes to IL-6 production and T cell dependent inflammation in rheumatoid arthritis. Biochim. Biophys. Acta Mol. Basis Dis. 2017, 1863, 2158–2170. [Google Scholar] [CrossRef] [PubMed]
- Farr, R.L.; Rootes, C.L.; Rowntree, L.C.; Nguyen, T.H.O.; Hensen, L.; Kedzierski, L.; Cheng, A.C.; Kedzierska, K.; Au, G.G.; Marsh, G.; et al. Altered microRNA expression in COVID-19 patients enables identification of SARS-CoV-2 infection. PLoS Pathog. 2021, 17, e1009759. [Google Scholar] [CrossRef] [PubMed]
- De Gonzalo-Calvo, D.; Benítez, I.D.; Pinilla, L.; Carratalá, A.; Moncusí-Moix, A.; Gort-Paniello, C.; Molinero, M.; González, J.; Torres, G.; Bernal, M.; et al. Circulating microRNA profiles predict the severity of COVID-19 in hospitalized patients. Transl. Res. 2021, 236, 147–159. [Google Scholar] [CrossRef]
- Friedrich, M.; Pracht, K.; Mashreghi, M.-F.; Jäck, H.-M.; Radbruch, A.; Seliger, B. The role of the miR-148/-152 family in physiology and disease. Eur. J. Immunol. 2017, 47, 2026–2038. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, P.; Zhao, Y.; Li, J.; Liu, C.; Zhu, L.; Zhang, J.; Yu, Y.; Wang, W.-J.; Lei, G.; Yan, J.; et al. Downregulated miR-451a as a feature of the plasma cfRNA landscape reveals regulatory networks of IL-6/IL-6R-associated cytokine storms in COVID-19 patients. Cell. Mol. Immunol. 2021, 18, 1064–1066. [Google Scholar] [CrossRef]
- Ren, F.-J.; Yao, Y.; Cai, X.-Y.; Fang, G.-Y. Emerging Role of MiR-192-5p in Human Diseases. Front. Pharmacol. 2021, 12. [Google Scholar] [CrossRef]
- Ge, L.; Habiel, D.M.; Hansbro, P.M.; Kim, R.Y.; Gharib, S.A.; Edelman, J.D.; Koenigshoff, M.; Parimon, T.; Brauer, R.; Huang, Y.; et al. miR-323a-3p regulates lung fibrosis by targeting multiple profibrotic pathways. JCI Insight 2016, 1, e90301. [Google Scholar] [CrossRef]
- Fu, Z.; Wang, J.; Wang, Z.; Sun, Y.; Wu, J.; Zhang, Y.; Liu, X.; Zhou, Z.; Zhou, L.; Zhang, C.-Y.; et al. A virus-derived microRNA-like small RNA serves as a serum biomarker to prioritize the COVID-19 patients at high risk of developing severe disease. Cell Discov. 2021, 7, 48. [Google Scholar] [CrossRef]
- Donyavi, T.; Bokharaei-Salim, F.; Baghi, H.B.; Khanaliha, K.; Janat-Makan, M.A.; Karimi, B.; Nahand, J.S.; Mirzaei, H.; Khatami, A.; Garshasbi, S.; et al. Acute and post-acute phase of COVID-19: Analyzing expression patterns of miRNA-29a-3p, 146a-3p, 155–5p, and let-7b-3p in PBMC. Int. Immunopharmacol. 2021, 97, 107641. [Google Scholar] [CrossRef]
- Zheng, H.-Y.; Xu, M.; Yang, C.-X.; Tian, R.-R.; Zhang, M.; Li, J.-J.; Wang, X.-C.; Ding, Z.-L.; Li, G.-M.; Li, X.-L.; et al. Longitudinal transcriptome analyses show robust T cell immunity during recovery from COVID-19. Signal Transduct. Target. Ther. 2020, 5, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Sabbatinelli, J.; Giuliani, A.; Matacchione, G.; Latini, S.; Laprovitera, N.; Pomponio, G.; Ferrarini, A.; Baroni, S.S.; Pavani, M.; Moretti, M.; et al. Decreased serum levels of the inflammaging marker miR-146a are associated with clinical non-response to tocilizumab in COVID-19 patients. Mech. Ageing Dev. 2021, 193, 111413. [Google Scholar] [CrossRef] [PubMed]
- Mensà, E.; Giuliani, A.; Matacchione, G.; Gurău, F.; Bonfigli, A.R.; Romagnoli, F.; De Luca, M.; Sabbatinelli, J.; Olivieri, F. Circulating miR-146a in healthy aging and type 2 diabetes: Age- and gender-specific trajectories. Mech. Ageing Dev. 2019, 180, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Tian, Y.; Xu, J.; Li, Y.; Zhao, R.; Du, S.; Lv, C.; Wu, W.; Liu, R.; Sheng, X.; Song, Y.; et al. MicroRNA-31 Reduces Inflammatory Signaling and Promotes Regeneration in Colon Epithelium, and Delivery of Mimics in Microspheres Reduces Colitis in Mice. Gastroenterology 2019, 156, 2281–2296.e6. [Google Scholar] [CrossRef] [PubMed]
- Luo, Y.; Dong, H.-Y.; Zhang, B.; Feng, Z.; Liu, Y.; Gao, Y.-Q.; Dong, M.-Q.; Li, Z.-C. miR-29a-3p Attenuates Hypoxic Pulmonary Hypertension by Inhibiting Pulmonary Adventitial Fibroblast Activation. Hypertension 2015, 65, 414–420. [Google Scholar] [CrossRef] [Green Version]
- Cao, D.; Mikosz, A.M.; Ringsby, A.J.; Anderson, K.C.; Beatman, E.L.; Koike, K.; Petrache, I. MicroRNA-126-3p Inhibits Angiogenic Function of Human Lung Microvascular Endothelial Cells via LAT1 (L-Type Amino Acid Transporter 1)-Mediated mTOR (Mammalian Target of Rapamycin) Signaling. Arter. Thromb. Vasc. Biol. 2020, 40, 1195–1206. [Google Scholar] [CrossRef] [Green Version]
- Rupaimoole, R.; Han, H.-D.; Lopez-Berestein, G.; Sood, A.K. MicroRNA therapeutics: Principles, expectations, and challenges. Chin. J. Cancer 2011, 30, 368–370. [Google Scholar] [CrossRef]
- Hanna, J.; Hossain, G.S.; Kocerha, J. The Potential for microRNA Therapeutics and Clinical Research. Front. Genet. 2019, 10, 478. [Google Scholar] [CrossRef] [Green Version]
- Schmidt, M.F. Drug target miRNAs: Chances and challenges. Trends Biotechnol. 2014, 32, 578–585. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Li, X.; Liu, J.; Don, L.; Chen, Q.; Liu, J.; Kong, H.; Zhang, Q.; Qi, X.; Hou, D.; et al. Honeysuckle-encoded atypical microRNA2911 directly targets influenza A viruses. Cell Res. 2014, 25, 39–49. [Google Scholar] [CrossRef] [PubMed]
- Woods, P.S.; Doolittle, L.M.; Rosas, L.E.; Nana-Sinkam, S.P.; Tili, E.; Davis, I.C. Increased expression of microRNA-155-5p by alveolar type II cells contributes to development of lethal ARDS in H1N1 influenza A virus-infected mice. Virology 2020, 545, 40–52. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Ma, J.; Woods, P.S.; Chesarino, N.M.; Liu, C.; Lee, L.J.; Nana-Sinkam, S.P.; Davis, I.C. Selective targeting of alveolar type II respiratory epithelial cells by anti-surfactant protein-C antibody-conjugated lipoplexes. J. Control. Release 2015, 203, 140–149. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Desjarlais, M.; Wirth, M.; Lahaie, I.; Ruknudin, P.; Hardy, P.; Rivard, A.; Chemtob, S. Nutraceutical Targeting of Inflammation-Modulating microRNAs in Severe Forms of COVID-19: A Novel Approach to Prevent the Cytokine Storm. Front. Pharmacol. 2020, 11, 602999. [Google Scholar] [CrossRef]
- Zhang, S.; Cheng, Z.; Wang, Y.; Han, T. The Risks of miRNA Therapeutics: In a Drug Target Perspective. Drug Des. Dev. Ther. 2021, 15, 721–733. [Google Scholar] [CrossRef] [PubMed]
- Bajan, S.; Hutvagner, G. RNA-Based Therapeutics: From Antisense Oligonucleotides to miRNAs. Cells 2020, 9, 137. [Google Scholar] [CrossRef] [Green Version]
- Lindow, M.; Kauppinen, S. Discovering the first microRNA-targeted drug. J. Cell Biol. 2012, 199, 407–412. [Google Scholar] [CrossRef] [PubMed]
- Janssen, H.L.A.; Reesink, H.W.; Lawitz, E.J.; Zeuzem, S.; Rodriguez-Torres, M.; Patel, K.; van der Meer, A.J.; Patick, A.K.; Chen, A.; Zhou, Y.; et al. Treatment of HCV Infection by Targeting MicroRNA. N. Engl. J. Med. 2013, 368, 1685–1694. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- van der Ree, M.H.; de Vree, J.M.; Stelma, F.; Willemse, S.; van der Valk, M.; Rietdijk, S.; Molenkamp, R.; Schinkel, J.; van Nuenen, A.C.; Beuers, U.; et al. Safety, tolerability, and antiviral effect of RG-101 in patients with chronic hepatitis C: A phase 1B, double-blind, randomised controlled trial. Lancet 2017, 389, 709–717. [Google Scholar] [CrossRef]
- Baldassarre, A.; Paolini, A.; Bruno, S.P.; Felli, C.; Tozzi, A.E.; Masotti, A. Potential use of noncoding RNAs and innovative therapeutic strategies to target the 5′UTR of SARS-CoV-2. Epigenomics 2020, 12, 1349–1361. [Google Scholar] [CrossRef]
- Alam, T.; Lipovich, L. miRCOVID-19: Potential Targets of Human miRNAs in SARS-CoV-2 for RNA-Based Drug Discovery. Non-Coding RNA 2021, 7, 18. [Google Scholar] [CrossRef] [PubMed]
- Rizkita, L.D.; Astuti, I. The potential of miRNA-based therapeutics in severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection: A review. J. Pharm. Anal. 2021, 11, 265–271. [Google Scholar] [CrossRef] [PubMed]
- Demirci, M.D.S.; Adan, A. Computational analysis of microRNA-mediated interactions in SARS-CoV-2 infection. PeerJ 2020, 8, e9369. [Google Scholar] [CrossRef]
- Sardar, R.; Satish, D.; Birla, S.; Gupta, D. Integrative analyses of SARS-CoV-2 genomes from different geographical locations reveal unique features potentially consequential to host-virus interaction, pathogenesis and clues for novel therapies. Heliyon 2020, 6, e04658. [Google Scholar] [CrossRef] [PubMed]
- Bozgeyik, I. Therapeutic potential of miRNAs targeting SARS-CoV-2 host cell receptor ACE2. Meta Gene 2020, 27, 100831. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Du, J.; Yu, X.; Xu, J.; Huang, F.; Li, X.; Zhang, C.; Li, X.; Chang, J.; Shang, D.; et al. miRNA-200c-3p is crucial in acute respiratory distress syndrome. Cell Discov. 2017, 3, 17021. [Google Scholar] [CrossRef]
- Lee, S.W.L.; Paoletti, C.; Campisi, M.; Osaki, T.; Adriani, G.; Kamm, R.D.; Mattu, C.; Chiono, V. MicroRNA delivery through nanoparticles. J. Control Release 2019, 313, 80–95. [Google Scholar] [CrossRef] [PubMed]
- Yang, N. An overview of viral and nonviral delivery systems for microRNA. Int. J. Pharm. Investig. 2015, 5, 179–181. [Google Scholar] [CrossRef] [PubMed]
- Bai, Z.; Wei, J.; Yu, C.; Han, X.; Qin, X.; Zhang, C.; Liao, W.; Li, L.; Huang, W. Non-viral nanocarriers for intracellular delivery of microRNA therapeutics. J. Mater. Chem. B 2019, 7, 1209–1225. [Google Scholar] [CrossRef] [PubMed]
- Boca, S.; Gulei, D.; Zimta, A.A.; Onaciu, A.; Magdo, L.; Tigu, A.B.; Ionescu, C.; Irimie, A.; Buiga, R.; Berindan-Neagoe, I. Nanoscale delivery systems for microRNAs in cancer therapy. Cell. Mol. Life Sci. 2020, 77, 1059–1086. [Google Scholar] [CrossRef] [PubMed]
- Scheideler, M.; Vidakovic, I.; Prassl, R. Lipid nanocarriers for microRNA delivery. Chem. Phys. Lipids 2020, 226, 104837. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Piñeiro, I.; Badiola, I.; Sanchez, A. Nanocarriers for microRNA delivery in cancer medicine. Biotechnol. Adv. 2017, 35, 350–360. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, Z.; Gemeinhart, R.A. Progress in microRNA delivery. J. Control Release 2013, 172, 962–974. [Google Scholar] [CrossRef] [PubMed] [Green Version]

 : upregulated.
: upregulated.  : downregulated. Created with BioRender.com.
: downregulated. Created with BioRender.com.
 : upregulated.
: upregulated.  : downregulated. Created with BioRender.com.
: downregulated. Created with BioRender.com.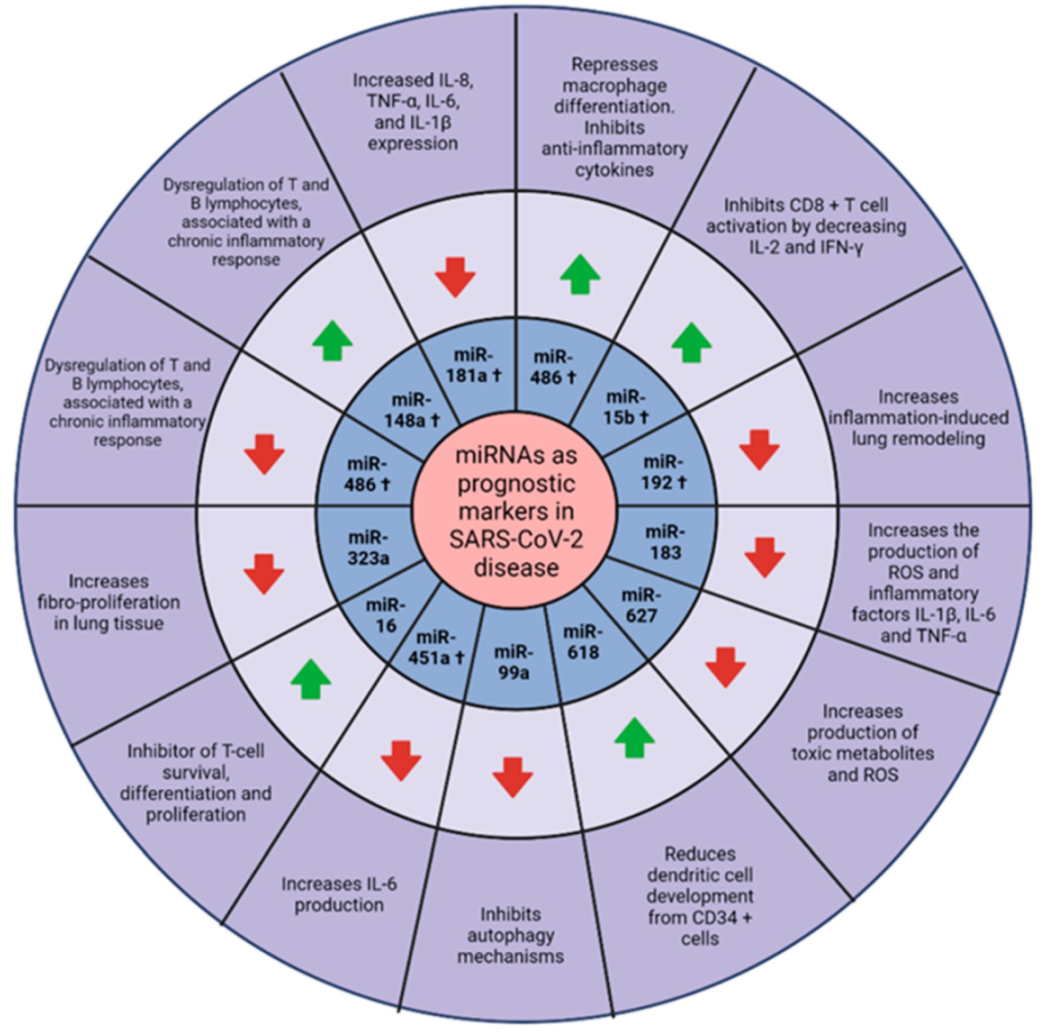
| Rate Ratios Compared to White, Non-Hispanic Population | American Indian or Alaskan Native | Asian | Black or African-American | Hispanic or Latino |
|---|---|---|---|---|
| Cases | 1.7 | 0.7 | 1.1 | 1.9 |
| Hospitalizations | 3.4 | 1.0 | 2.8 | 2.8 |
| Deaths | 2.4 | 1.0 | 2.0 | 2.3 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bautista-Becerril, B.; Pérez-Dimas, G.; Sommerhalder-Nava, P.C.; Hanono, A.; Martínez-Cisneros, J.A.; Zarate-Maldonado, B.; Muñoz-Soria, E.; Aquino-Gálvez, A.; Castillejos-López, M.; Juárez-Cisneros, A.; et al. miRNAs, from Evolutionary Junk to Possible Prognostic Markers and Therapeutic Targets in COVID-19. Viruses 2022, 14, 41. https://doi.org/10.3390/v14010041
Bautista-Becerril B, Pérez-Dimas G, Sommerhalder-Nava PC, Hanono A, Martínez-Cisneros JA, Zarate-Maldonado B, Muñoz-Soria E, Aquino-Gálvez A, Castillejos-López M, Juárez-Cisneros A, et al. miRNAs, from Evolutionary Junk to Possible Prognostic Markers and Therapeutic Targets in COVID-19. Viruses. 2022; 14(1):41. https://doi.org/10.3390/v14010041
Chicago/Turabian StyleBautista-Becerril, Brandon, Guillermo Pérez-Dimas, Paola C. Sommerhalder-Nava, Alejandro Hanono, Julio A. Martínez-Cisneros, Bárbara Zarate-Maldonado, Evangelina Muñoz-Soria, Arnoldo Aquino-Gálvez, Manuel Castillejos-López, Armida Juárez-Cisneros, and et al. 2022. "miRNAs, from Evolutionary Junk to Possible Prognostic Markers and Therapeutic Targets in COVID-19" Viruses 14, no. 1: 41. https://doi.org/10.3390/v14010041
APA StyleBautista-Becerril, B., Pérez-Dimas, G., Sommerhalder-Nava, P. C., Hanono, A., Martínez-Cisneros, J. A., Zarate-Maldonado, B., Muñoz-Soria, E., Aquino-Gálvez, A., Castillejos-López, M., Juárez-Cisneros, A., Lopez-Gonzalez, J. S., & Camarena, A. (2022). miRNAs, from Evolutionary Junk to Possible Prognostic Markers and Therapeutic Targets in COVID-19. Viruses, 14(1), 41. https://doi.org/10.3390/v14010041