Implications of Dengue Virus Maturation on Vaccine Induced Humoral Immunity in Mice
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animal Ethics
2.2. Cell Culture
2.3. Virus Generation
2.4. Virus Propagation and Purification
2.5. Recombaint Monoclonal Antibody Expression and Purification
2.6. Immunisation of AG129 Mice with bDENV-2
2.7. Virus Challenge
2.8. ELISAs
2.9. Plaque Reduction Neutralisation Tests (PRNTs)
2.10. Quantification of Viraemia by Immunoplaque Assay
2.11. NS1 Quantitative Capture ELISA
2.12. Antibody Dependent Enhancment Assay
2.13. Statistical Analysis
3. Results
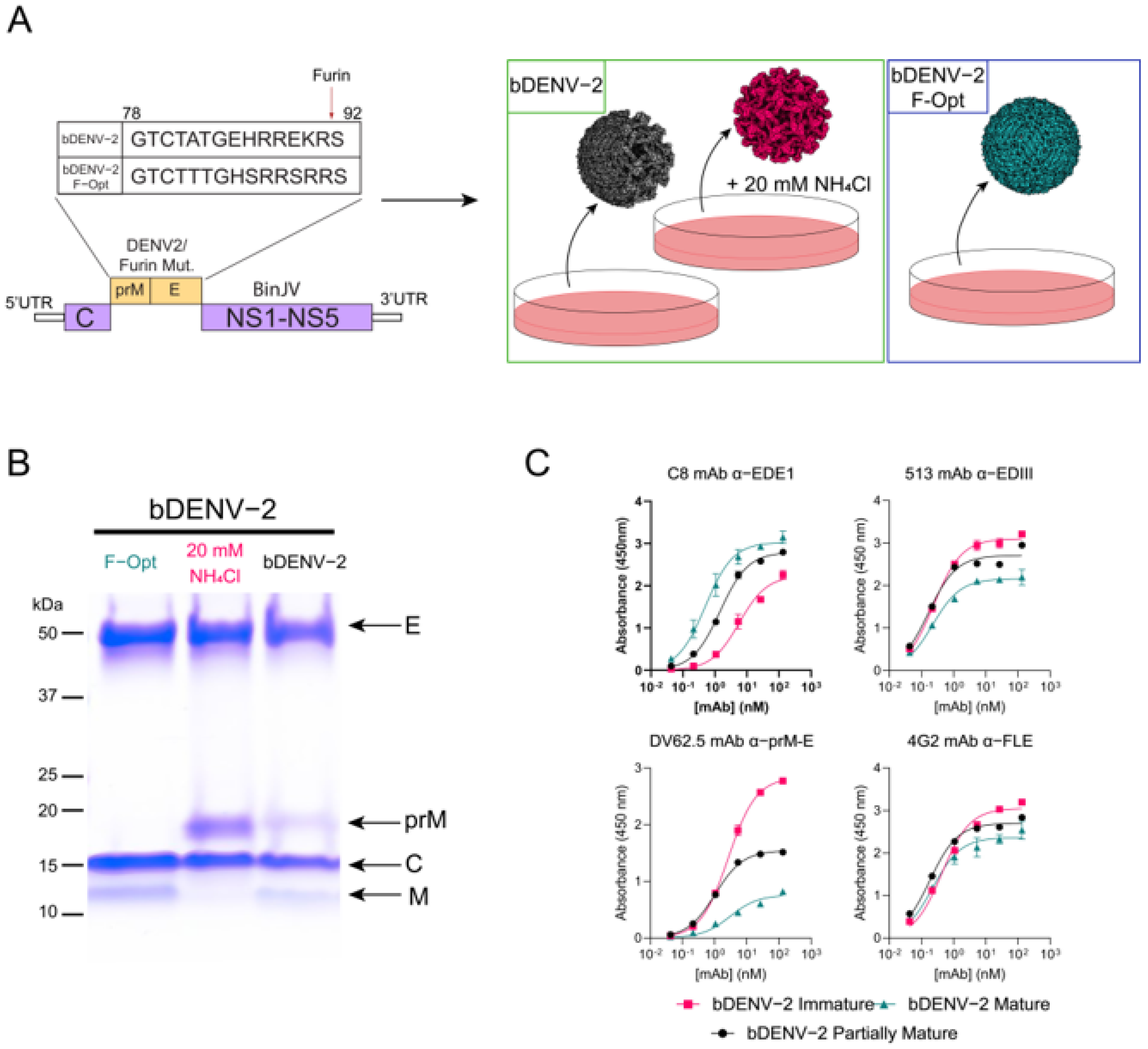
3.1. Generation of Mature, Immature and Partially Mature bDENV-2
3.2. Humoral Immune Responses to Immunisation with bDENV-2
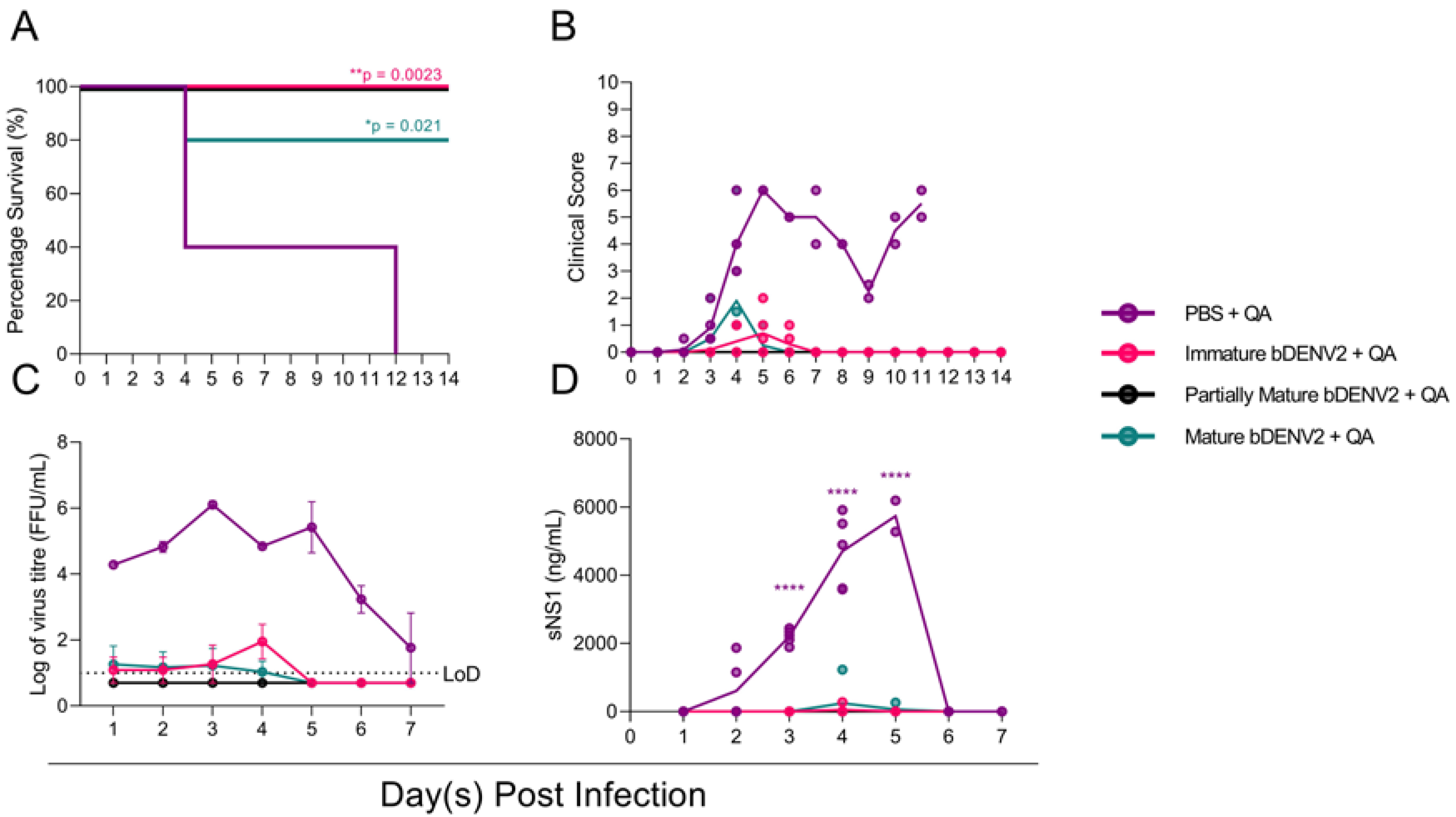
3.3. All Maturities of bDENV-2 Provide Protection in AG129 Mice
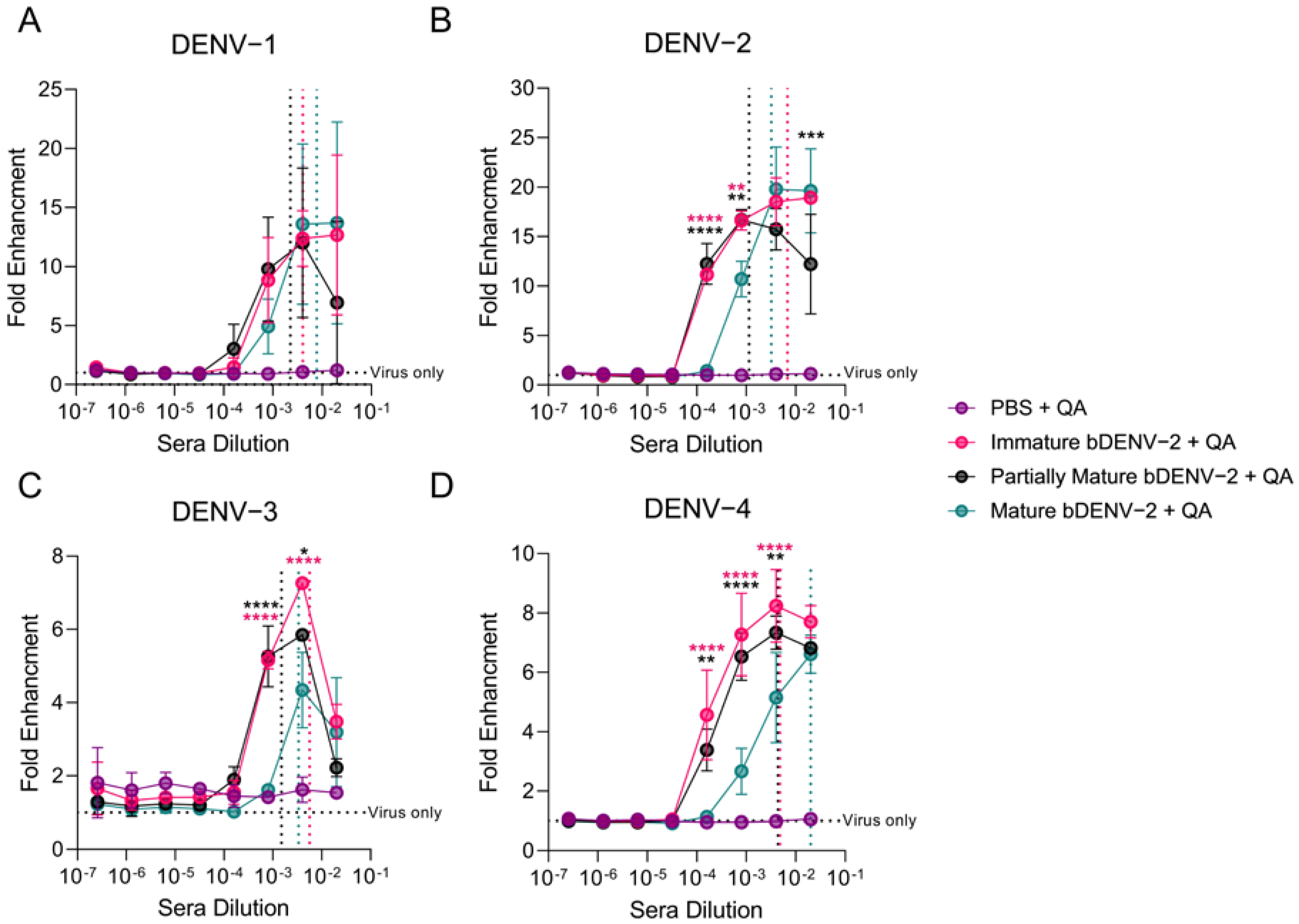
3.4. In Vitro Enhancment of DENV by bDENV-2 Immune Sera
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bhatt, S.; Gething, P.W.; Brady, O.J.; Messina, J.P.; Farlow, A.W.; Moyes, C.L.; Drake, J.M.; Brownstein, J.S.; Hoen, A.G.; Sankoh, O.; et al. The global distribution and burden of dengue. Nature 2013, 496, 504–507. [Google Scholar] [CrossRef]
- Cohen, S.N.; Halstead, S.B. Shock associated with dengue infection. I. Clinical and physiologic manifestations of dengue hemorrhagic fever in Thailand, 1964. J. Pediatrics 1966, 68, 448–456. [Google Scholar] [CrossRef]
- Halstead, S.B. Pathogenesis of Dengue: Dawn of a New Era. F1000Research 2015, 4, 1353. [Google Scholar] [CrossRef] [PubMed]
- Montoya, M.; Gresh, L.; Mercado, J.C.; Williams, K.L.; Vargas, M.J.; Gutierrez, G.; Kuan, G.; Gordon, A.; Balmaseda, A.; Harris, E. Symptomatic Versus Inapparent Outcome in Repeat Dengue Virus Infections Is Influenced by the Time Interval between Infections and Study Year. PLoS Negl. Trop. Dis. 2013, 7, e2357. [Google Scholar] [CrossRef] [Green Version]
- Thein, S.; Aung, M.M.; Shwe, T.N.; Aye, M.; Zaw, A.; Aye, K.; Aye, K.M.; Aaskov, J. Risk Factors in Dengue Shock Syndrome. Am. J. Trop. Med. Hyg. 1997, 56, 566–572. [Google Scholar] [CrossRef] [PubMed]
- Narayan, R.; Tripathi, S. Intrinsic ADE: The Dark Side of Antibody Dependent Enhancement During Dengue Infection. Front. Cell. Infect. Microbiol. 2020, 10, 580096. [Google Scholar] [CrossRef]
- Yu, I.M.; Zhang, W.; Holdaway, H.A.; Li, L.; Kostyuchenko, V.A.; Chipman, P.R.; Kuhn, R.J.; Rossmann, M.G.; Chen, J. Structure of the Immature Dengue Virus at Low pH Primes Proteolytic Maturation. Science 2008, 319, 1834. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Lok, S.-M.; Yu, I.M.; Zhang, Y.; Kuhn, R.J.; Chen, J.; Rossmann, M.G. The Flavivirus Precursor Membrane-Envelope Protein Complex: Structure and Maturation. Science 2008, 319, 1830. [Google Scholar] [CrossRef] [Green Version]
- Newton, N.D.; Hardy, J.M.; Modhiran, N.; Hugo, L.E.; Amarilla, A.A.; Bibby, S.; Venugopal, H.; Harrison, J.J.; Traves, R.J.; Hall, R.A.; et al. The structure of an infectious immature flavivirus redefines viral architecture and maturation. Sci. Adv. 2021, 7, eabe4507. [Google Scholar] [CrossRef] [PubMed]
- Junjhon, J.; Lausumpao, M.; Supasa, S.; Noisakran, S.; Songjaeng, A.; Saraithong, P.; Chaichoun, K.; Utaipat, U.; Keelapang, P.; Kanjanahaluethai, A.; et al. Differential modulation of prM cleavage, extracellular particle distribution, and virus infectivity by conserved residues at nonfurin consensus positions of the dengue virus pr-M junction. J. Virol. 2008, 82, 10776–10791. [Google Scholar] [CrossRef] [Green Version]
- Junjhon, J.; Edwards, T.J.; Utaipat, U.; Bowman, V.D.; Holdaway, H.A.; Zhang, W.; Keelapang, P.; Puttikhunt, C.; Perera, R.; Chipman, P.R.; et al. Influence of pr-M cleavage on the heterogeneity of extracellular dengue virus particles. J. Virol. 2010, 84, 8353–8358. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tse, L.V.; Meganck, R.M.; Dong, S.; Adams, L.E.; White, L.J.; de Silva, A.M.; Baric, R.S. Genetically Engineered DENV Produces Antigenically Distinct Mature Particles. bioRxiv 2021. [Google Scholar] [CrossRef]
- Mukherjee, S.; Sirohi, D.; Dowd, K.A.; Chen, Z.; Diamond, M.S.; Kuhn, R.J.; Pierson, T.C. Enhancing dengue virus maturation using a stable furin over-expressing cell line. Virology 2016, 497, 33–40. [Google Scholar] [CrossRef] [PubMed]
- Wirawan, M.; Fibriansah, G.; Marzinek, J.K.; Lim, X.X.; Ng, T.-S.; Sim, A.Y.L.; Zhang, Q.; Kostyuchenko, V.A.; Shi, J.; Smith, S.A.; et al. Mechanism of Enhanced Immature Dengue Virus Attachment to Endosomal Membrane Induced by prM Antibody. Structure 2019, 27, 253–267.e258. [Google Scholar] [CrossRef] [Green Version]
- Colpitts, T.M.; Rodenhuis-Zybert, I.; Moesker, B.; Wang, P.; Fikrig, E.; Smit, J.M. prM-antibody renders immature West Nile virus infectious in vivo. J. Gen. Virol. 2011, 92, 2281–2285. [Google Scholar] [CrossRef]
- Dejnirattisai, W.; Jumnainsong, A.; Onsirisakul, N.; Fitton, P.; Vasanawathana, S.; Limpitikul, W.; Puttikhunt, C.; Edwards, C.; Duangchinda, T.; Supasa, S.; et al. Cross-Reacting Antibodies Enhance Dengue Virus Infection in Humans. Science 2010, 328, 745. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.; Ge, P.; Yu, X.; Brannan, J.M.; Bi, G.; Zhang, Q.; Schein, S.; Zhou, Z.H. Cryo-EM structure of the mature dengue virus at 3.5-Å resolution. Nat. Struct. Mol. Biol. 2013, 20, 105–110. [Google Scholar] [CrossRef] [Green Version]
- Cherrier, M.V.; Kaufmann, B.; Nybakken, G.E.; Lok, S.M.; Warren, J.T.; Chen, B.R.; Nelson, C.A.; Kostyuchenko, V.A.; Holdaway, H.A.; Chipman, P.R.; et al. Structural basis for the preferential recognition of immature flaviviruses by a fusion-loop antibody. EMBO J. 2009, 28, 3269–3276. [Google Scholar] [CrossRef] [Green Version]
- Dowd, K.A.; Pierson, T.C. The Many Faces of a Dynamic Virion: Implications of Viral Breathing on Flavivirus Biology and Immunogenicity. Annu. Rev. Virol. 2018, 5, 185–207. [Google Scholar] [CrossRef]
- Fibriansah, G.; Ng, T.-S.; Kostyuchenko, V.A.; Lee, J.; Lee, S.; Wang, J.; Lok, S.-M. Structural changes in dengue virus when exposed to a temperature of 37 °C. J. Virol. 2013, 87, 7585–7592. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Medits, I.; Vaney, M.-C.; Rouvinski, A.; Rey, M.; Chamot-Rooke, J.; Rey, F.A.; Heinz, F.X.; Stiasny, K. Extensive flavivirus E trimer breathing accompanies stem zippering of the post-fusion hairpin. EMBO Rep. 2020, 21, e50069. [Google Scholar] [CrossRef]
- Beltramello, M.; Williams, K.L.; Simmons, C.P.; Macagno, A.; Simonelli, L.; Quyen, N.T.; Sukupolvi-Petty, S.; Navarro-Sanchez, E.; Young, P.R.; de Silva, A.M.; et al. The human immune response to Dengue virus is dominated by highly cross-reactive antibodies endowed with neutralizing and enhancing activity. Cell Host Microbe 2010, 8, 271–283. [Google Scholar] [CrossRef] [Green Version]
- Lai, C.Y.; Tsai, W.Y.; Lin, S.R.; Kao, C.L.; Hu, H.P.; King, C.C.; Wu, H.C.; Chang, G.J.; Wang, W.K. Antibodies to envelope glycoprotein of dengue virus during the natural course of infection are predominantly cross-reactive and recognize epitopes containing highly conserved residues at the fusion loop of domain II. J. Virol. 2008, 82, 6631–6643. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dejnirattisai, W.; Wongwiwat, W.; Supasa, S.; Zhang, X.; Dai, X.; Rouvinski, A.; Jumnainsong, A.; Edwards, C.; Quyen, N.T.H.; Duangchinda, T.; et al. A new class of highly potent, broadly neutralizing antibodies isolated from viremic patients infected with dengue virus. Nat. Immunol. 2015, 16, 170–177. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsai, W.-Y.; Chen, H.-L.; Tsai, J.-J.; Dejnirattisai, W.; Jumnainsong, A.; Mongkolsapaya, J.; Screaton, G.; Crowe, J.E.; Wang, W.-K. Potent Neutralizing Human Monoclonal Antibodies Preferentially Target Mature Dengue Virus Particles: Implication for Novel Strategy for Dengue Vaccine. J. Virol. 2018, 92, e00556-18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Capeding, M.R.; Tran, N.H.; Hadinegoro, S.R.S.; Ismail, H.I.H.J.M.; Chotpitayasunondh, T.; Chua, M.N.; Luong, C.Q.; Rusmil, K.; Wirawan, D.N.; Nallusamy, R.; et al. Clinical efficacy and safety of a novel tetravalent dengue vaccine in healthy children in Asia: A phase 3, randomised, observer-masked, placebo-controlled trial. Lancet 2014, 384, 1358–1365. [Google Scholar] [CrossRef]
- Pitisuttithum, P.; Bouckenooghe, A. The first licensed dengue vaccine: An important tool for integrated preventive strategies against dengue virus infection. Expert Rev. Vaccines 2016, 15, 795–798. [Google Scholar] [CrossRef]
- Precioso, A.R.; Palacios, R.; Thomé, B.; Mondini, G.; Braga, P.; Kalil, J. Clinical evaluation strategies for a live attenuated tetravalent dengue vaccine. Vaccine 2015, 33, 7121–7125. [Google Scholar] [CrossRef]
- Biswal, S.; Borja-Tabora, C.; Martinez Vargas, L.; Velásquez, H.; Theresa Alera, M.; Sierra, V.; Johana Rodriguez-Arenales, E.; Yu, D.; Wickramasinghe, V.P.; Duarte Moreira, E., Jr.; et al. Efficacy of a tetravalent dengue vaccine in healthy children aged 4-16 years: A randomised, placebo-controlled, phase 3 trial. Lancet 2020, 395, 1423–1433. [Google Scholar] [CrossRef]
- Danko, J.R.; Kochel, T.; Teneza-Mora, N.; Luke, T.C.; Raviprakash, K.; Sun, P.; Simmons, M.; Moon, J.E.; De La Barrera, R.; Martinez, L.J.; et al. Safety and Immunogenicity of a Tetravalent Dengue DNA Vaccine Administered with a Cationic Lipid-Based Adjuvant in a Phase 1 Clinical Trial. Am. J. Trop. Med. Hyg. 2018, 98, 849–856. [Google Scholar] [CrossRef]
- Manoff, S.B.; Sausser, M.; Falk Russell, A.; Martin, J.; Radley, D.; Hyatt, D.; Roberts, C.C.; Lickliter, J.; Krishnarajah, J.; Bett, A.; et al. Immunogenicity and safety of an investigational tetravalent recombinant subunit vaccine for dengue: Results of a Phase I randomized clinical trial in flavivirus-naïve adults. Hum. Vaccines Immunother. 2019, 15, 2195–2204. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, A.C.; Lin, L.; Martinez, L.J.; Ruck, R.C.; Eckels, K.H.; Collard, A.; De La Barrera, R.; Paolino, K.M.; Toussaint, J.F.; Lepine, E.; et al. Phase 1 Randomized Study of a Tetravalent Dengue Purified Inactivated Vaccine in Healthy Adults in the United States. Am. J. Trop. Med. Hyg. 2017, 96, 1325–1337. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guy, B.; Guirakhoo, F.; Barban, V.; Higgs, S.; Monath, T.P.; Lang, J. Preclinical and clinical development of YFV 17D-based chimeric vaccines against dengue, West Nile and Japanese encephalitis viruses. Vaccine 2010, 28, 632–649. [Google Scholar] [CrossRef] [PubMed]
- Raut, R.; Corbett, K.S.; Tennekoon, R.N.; Premawansa, S.; Wijewickrama, A.; Premawansa, G.; Mieczkowski, P.; Rückert, C.; Ebel, G.D.; De Silva, A.D.; et al. Dengue type 1 viruses circulating in humans are highly infectious and poorly neutralized by human antibodies. Proc. Natl. Acad. Sci. USA 2019, 116, 227. [Google Scholar] [CrossRef] [Green Version]
- Choo, J.J.Y.; Vet, L.J.; McMillan, C.L.D.; Harrison, J.J.; Scott, C.A.P.; Depelsenaire, A.C.I.; Fernando, G.J.P.; Watterson, D.; Hall, R.A.; Young, P.R.; et al. A chimeric dengue virus vaccine candidate delivered by high density microarray patches protects against infection in mice. NPJ Vaccines 2021, 6, 66. [Google Scholar] [CrossRef]
- Harrison Jessica, J.; Hobson-Peters, J.; Colmant Agathe, M.G.; Koh, J.; Newton Natalee, D.; Warrilow, D.; Bielefeldt-Ohmann, H.; Piyasena Thisun, B.H.; O’Brien Caitlin, A.; Vet Laura, J.; et al. Antigenic Characterization of New Lineage II Insect-Specific Flaviviruses in Australian Mosquitoes and Identification of Host Restriction Factors. mSphere 2020, 5, e00095-20. [Google Scholar] [CrossRef]
- Hobson-Peters, J.; Harrison, J.J.; Watterson, D.; Hazlewood, J.E.; Vet, L.J.; Newton, N.D.; Warrilow, D.; Colmant, A.M.G.; Taylor, C.; Huang, B.; et al. A recombinant platform for flavivirus vaccines and diagnostics using chimeras of a new insect-specific virus. Sci. Transl. Med. 2019, 11, eaax7888. [Google Scholar] [CrossRef]
- Piyasena, T.B.H.; Setoh, Y.X.; Hobson-Peters, J.; Newton, N.D.; Bielefeldt-Ohmann, H.; McLean, B.J.; Vet, L.J.; Khromykh, A.A.; Hall, R.A. Infectious DNAs derived from insect-specific flavivirus genomes enable identification of pre- and post-entry host restrictions in vertebrate cells. Sci. Rep. 2017, 7, 2940. [Google Scholar] [CrossRef] [PubMed]
- Hardy, J.M.; Newton, N.D.; Modhiran, N.; Scott, C.A.P.; Venugopal, H.; Vet, L.J.; Young, P.R.; Hall, R.A.; Hobson-Peters, J.; Coulibaly, F.; et al. A unified route for flavivirus structures uncovers essential pocket factors conserved across pathogenic viruses. Nat. Commun. 2021, 12, 3266. [Google Scholar] [CrossRef]
- Warrilow, D.; Watterson, D.; Hall, R.A.; Davis, S.S.; Weir, R.; Kurucz, N.; Whelan, P.; Allcock, R.; Hall-Mendelin, S.; O’Brien, C.A.; et al. A New Species of Mesonivirus from the Northern Territory, Australia. PLoS ONE 2014, 9, e91103. [Google Scholar] [CrossRef] [PubMed]
- Jones, M.L.; Seldon, T.; Smede, M.; Linville, A.; Chin, D.Y.; Barnard, R.; Mahler, S.M.; Munster, D.; Hart, D.; Gray, P.P.; et al. A method for rapid, ligation-independent reformatting of recombinant monoclonal antibodies. J. Immunol. Methods 2010, 354, 85–90. [Google Scholar] [CrossRef]
- Gentry, M.K.; Henchal, E.A.; McCown, J.M.; Brandt, W.E.; Dalrymple, J.M. Identification of Distinct Antigenic Determinants on Dengue-2 Virus Using Monoclonal Antibodies. Am. J. Trop. Med. Hyg. 1982, 31, 548–555. [Google Scholar] [CrossRef]
- Robinson, L.N.; Tharakaraman, K.; Rowley, K.J.; Costa, V.V.; Chan, K.R.; Wong, Y.H.; Ong, L.C.; Tan, H.C.; Koch, T.; Cain, D.; et al. Structure-Guided Design of an Anti-dengue Antibody Directed to a Non-immunodominant Epitope. Cell 2015, 162, 493–504. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Orozco, S.; Schmid, M.A.; Parameswaran, P.; Lachica, R.; Henn, M.R.; Beatty, R.; Harris, E. Characterization of a model of lethal dengue virus 2 infection in C57BL/6 mice deficient in the alpha/beta interferon receptor. J. Gen. Virol. 2012, 93, 2152–2157. [Google Scholar] [CrossRef] [PubMed]
- Modhiran, N.; Song, H.; Liu, L.; Bletchly, C.; Brillault, L.; Amarilla, A.A.; Xu, X.; Qi, J.; Chai, Y.; Cheung, S.T.M.; et al. A broadly protective antibody that targets the flavivirus NS1 protein. Science 2021, 371, 190. [Google Scholar] [CrossRef]
- Li, J.; Watterson, D.; Chang, C.W.; Che, X.Y.; Li, X.Q.; Ericsson, D.J.; Qiu, L.W.; Cai, J.P.; Chen, J.; Fry, S.R.; et al. Structural and Functional Characterization of a Cross-Reactive Dengue Virus Neutralizing Antibody that Recognizes a Cryptic Epitope. Structure 2018, 26, 51–59.e54. [Google Scholar] [CrossRef] [Green Version]
- Yamanaka, A.; Miyazaki, K.; Shimizu, J.; Senju, S. Dengue virus susceptibility in novel immortalized myeloid cells. Heliyon 2020, 6, e05407. [Google Scholar] [CrossRef] [PubMed]
- Muller, D.A.; Depelsenaire, A.C.I.; Shannon, A.E.; Watterson, D.; Corrie, S.R.; Owens, N.S.; Agyei-Yeboah, C.; Cheung, S.T.M.; Zhang, J.; Fernando, G.J.P.; et al. Efficient Delivery of Dengue Virus Subunit Vaccines to the Skin by Microprojection Arrays. Vaccines 2019, 7, 189. [Google Scholar] [CrossRef] [Green Version]
- Sridhar, S.; Luedtke, A.; Langevin, E.; Zhu, M.; Bonaparte, M.; Machabert, T.; Savarino, S.; Zambrano, B.; Moureau, A.; Khromava, A.; et al. Effect of Dengue Serostatus on Dengue Vaccine Safety and Efficacy. N. Engl. J. Med. 2018, 379, 327–340. [Google Scholar] [CrossRef] [PubMed]
- Wahala, W.M.; Kraus, A.A.; Haymore, L.B.; Accavitti-Loper, M.A.; de Silva, A.M. Dengue virus neutralization by human immune sera: Role of envelope protein domain III-reactive antibody. Virology 2009, 392, 103–113. [Google Scholar] [CrossRef] [Green Version]
- Williams, K.L.; Wahala, W.M.P.B.; Orozco, S.; de Silva, A.M.; Harris, E. Antibodies targeting dengue virus envelope domain III are not required for serotype-specific protection or prevention of enhancement in vivo. Virology 2012, 429, 12–20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nelson, S.; Jost, C.A.; Xu, Q.; Ess, J.; Martin, J.E.; Oliphant, T.; Whitehead, S.S.; Durbin, A.P.; Graham, B.S.; Diamond, M.S.; et al. Maturation of West Nile virus modulates sensitivity to antibody-mediated neutralization. PLoS Pathog. 2008, 4, e1000060. [Google Scholar] [CrossRef] [Green Version]
- Guy, B.; Jackson, N. Dengue vaccine: Hypotheses to understand CYD-TDV-induced protection. Nat. Rev. Microbiol. 2016, 14, 45–54. [Google Scholar] [CrossRef] [PubMed]
- Shukla, R.; Beesetti, H.; Brown, J.A.; Ahuja, R.; Ramasamy, V.; Shanmugam, R.K.; Poddar, A.; Batra, G.; Krammer, F.; Lim, J.K.; et al. Dengue and Zika virus infections are enhanced by live attenuated dengue vaccine but not by recombinant DSV4 vaccine candidate in mouse models. EBioMedicine 2020, 60, 102991. [Google Scholar] [CrossRef]
- Hueston, L.; Ramirez, R.; Mahalingam, S. Enhancement of Zika Infection by Dengue Virus–Specific Antibody Is Associated With Low Levels of Antiviral Factors. J. Infect. Dis. 2017, 216, 612–614. [Google Scholar] [CrossRef] [PubMed]
- Boonnak, K.; Dambach, K.M.; Donofrio, G.C.; Tassaneetrithep, B.; Marovich, M.A. Cell type specificity and host genetic polymorphisms influence antibody-dependent enhancement of dengue virus infection. J. Virol. 2011, 85, 1671–1683. [Google Scholar] [CrossRef] [Green Version]
- Henein, S.; Swanstrom, J.; Byers, A.M.; Moser, J.M.; Shaik, S.F.; Bonaparte, M.; Jackson, N.; Guy, B.; Baric, R.; de Silva, A.M. Dissecting Antibodies Induced by a Chimeric Yellow Fever-Dengue, Live-Attenuated, Tetravalent Dengue Vaccine (CYD-TDV) in Naive and Dengue-Exposed Individuals. J. Infect. Dis. 2017, 215, 351–358. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Putnak, R.; Cassidy, K.; Conforti, N.; Lee, R.; Sollazzo, D.; Truong, T.; Ing, E.; Dubois, D.; Sparkuhl, J.; Gastle, W.; et al. Immunogenic and protective response in mice immunized with a purified, inactivated, Dengue-2 virus vaccine prototype made in fetal rhesus lung cells. Am. J. Trop. Med. Hyg. 1996, 55, 504–510. [Google Scholar] [CrossRef]
- Yan, K.; Vet, L.J.; Tang, B.; Hobson-Peters, J.; Rawle, D.J.; Le, T.T.; Larcher, T.; Hall, R.A.; Suhrbier, A. A Yellow Fever Virus 17D Infection and Disease Mouse Model Used to Evaluate a Chimeric Binjari-Yellow Fever Virus Vaccine. Vaccines 2020, 8, 368. [Google Scholar] [CrossRef] [PubMed]
- Vet, L.J.; Setoh, Y.X.; Amarilla, A.A.; Habarugira, G.; Suen, W.W.; Newton, N.D.; Harrison, J.J.; Hobson-Peters, J.; Hall, R.A.; Bielefeldt-Ohmann, H. Protective Efficacy of a Chimeric Insect-Specific Flavivirus Vaccine against West Nile Virus. Vaccines 2020, 8, 258. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Scott, C.A.P.; Amarilla, A.A.; Bibby, S.; Newton, N.D.; Hall, R.A.; Hobson-Peters, J.; Muller, D.A.; Chappell, K.J.; Young, P.R.; Modhiran, N.; et al. Implications of Dengue Virus Maturation on Vaccine Induced Humoral Immunity in Mice. Viruses 2021, 13, 1843. https://doi.org/10.3390/v13091843
Scott CAP, Amarilla AA, Bibby S, Newton ND, Hall RA, Hobson-Peters J, Muller DA, Chappell KJ, Young PR, Modhiran N, et al. Implications of Dengue Virus Maturation on Vaccine Induced Humoral Immunity in Mice. Viruses. 2021; 13(9):1843. https://doi.org/10.3390/v13091843
Chicago/Turabian StyleScott, Connor A. P., Alberto A. Amarilla, Summa Bibby, Natalee D. Newton, Roy A. Hall, Jody Hobson-Peters, David A. Muller, Keith J. Chappell, Paul R. Young, Naphak Modhiran, and et al. 2021. "Implications of Dengue Virus Maturation on Vaccine Induced Humoral Immunity in Mice" Viruses 13, no. 9: 1843. https://doi.org/10.3390/v13091843
APA StyleScott, C. A. P., Amarilla, A. A., Bibby, S., Newton, N. D., Hall, R. A., Hobson-Peters, J., Muller, D. A., Chappell, K. J., Young, P. R., Modhiran, N., & Watterson, D. (2021). Implications of Dengue Virus Maturation on Vaccine Induced Humoral Immunity in Mice. Viruses, 13(9), 1843. https://doi.org/10.3390/v13091843






