The Input of Structural Vaccinology in the Search for Vaccines against Bunyaviruses
Abstract
1. The Concept of Structural Vaccinology
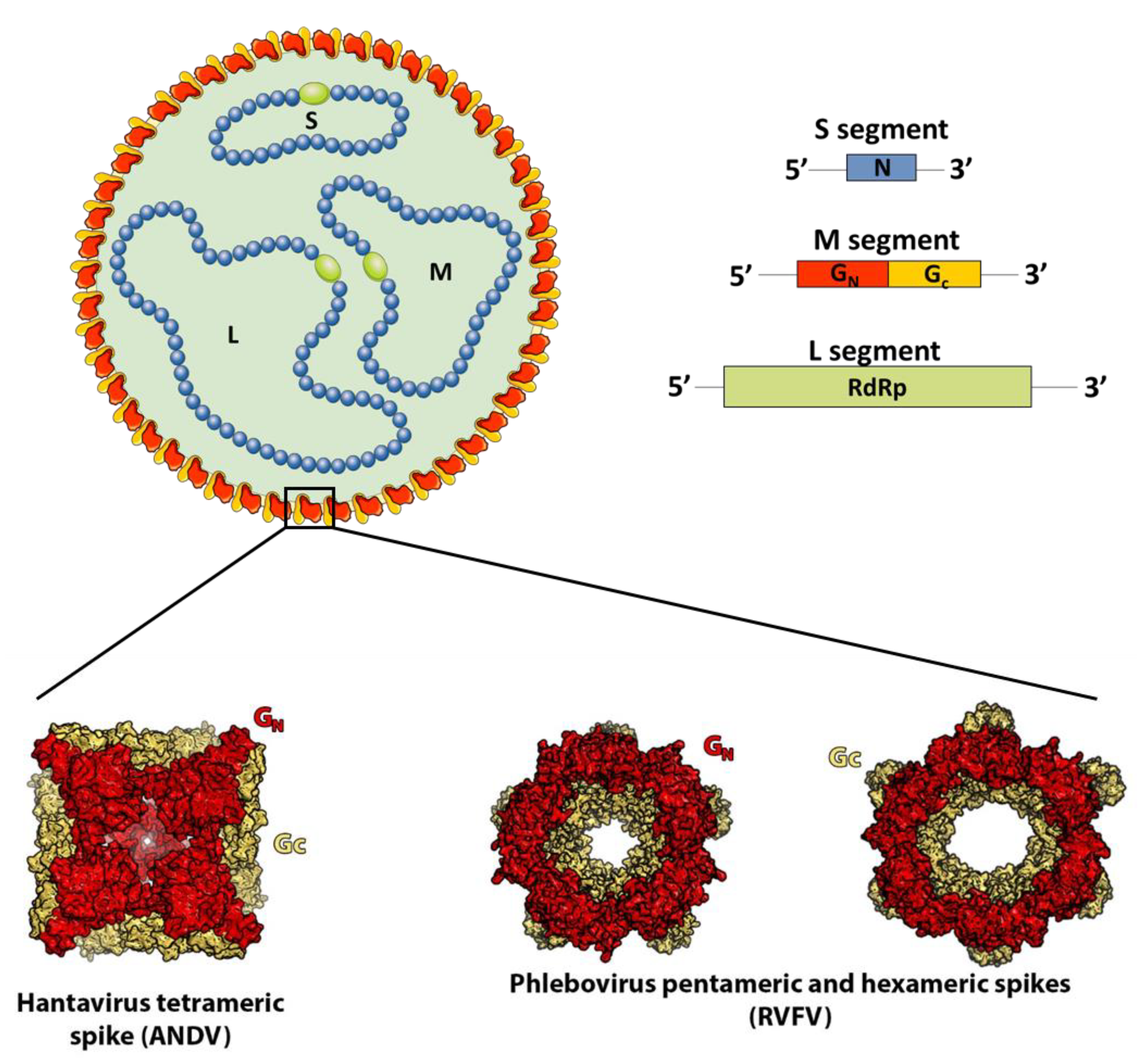
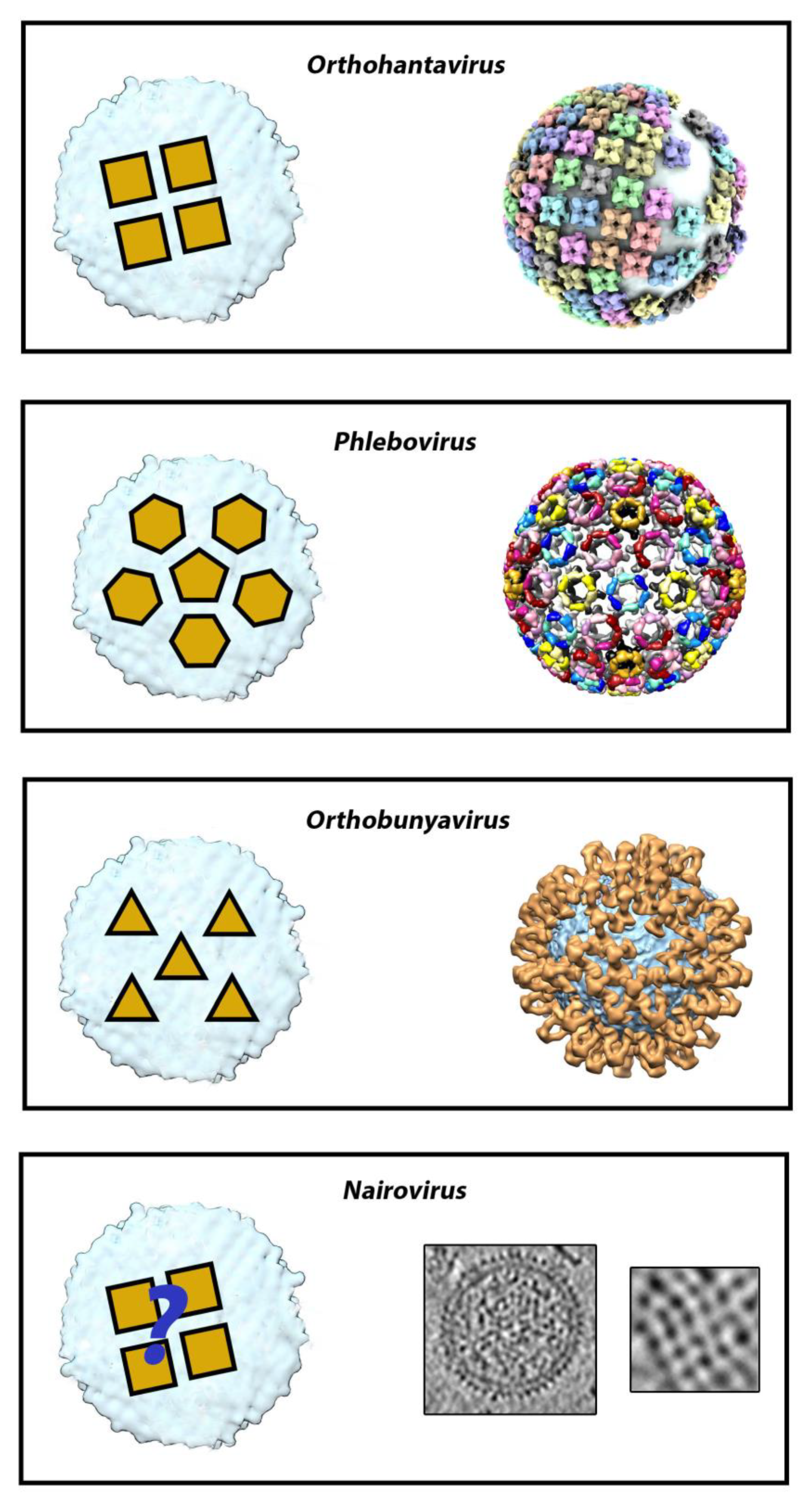
2. The Surface Glycoprotein Lattice of Bunyavirus Particles Constitutes a Challenge for Vaccine Design
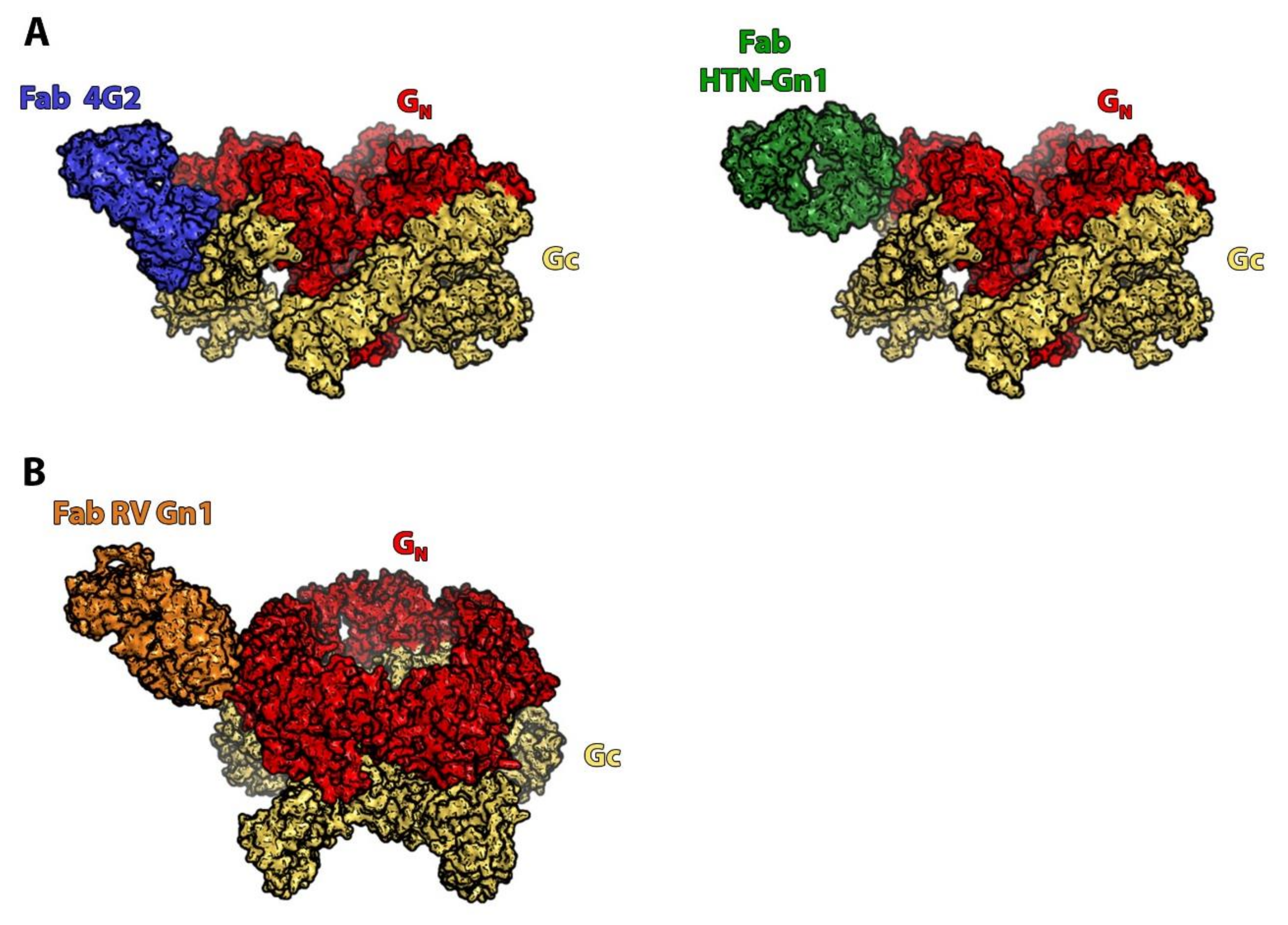
3. Orthohantaviruses (Hantaviridae Family): An Example of the Importance of the Quaternary Spike Structure and the Global Lattice Organization
4. Phleboviruses (Family Phenuiviridae): Identifying the Precise Target of Neutralizing mAbs to Pave the Way for the Design of a Pan-Phlebovirus Vaccine
5. Nairoviruses (Family Nairoviridae): Protection Is Not Always Mediated Only by Neutralization
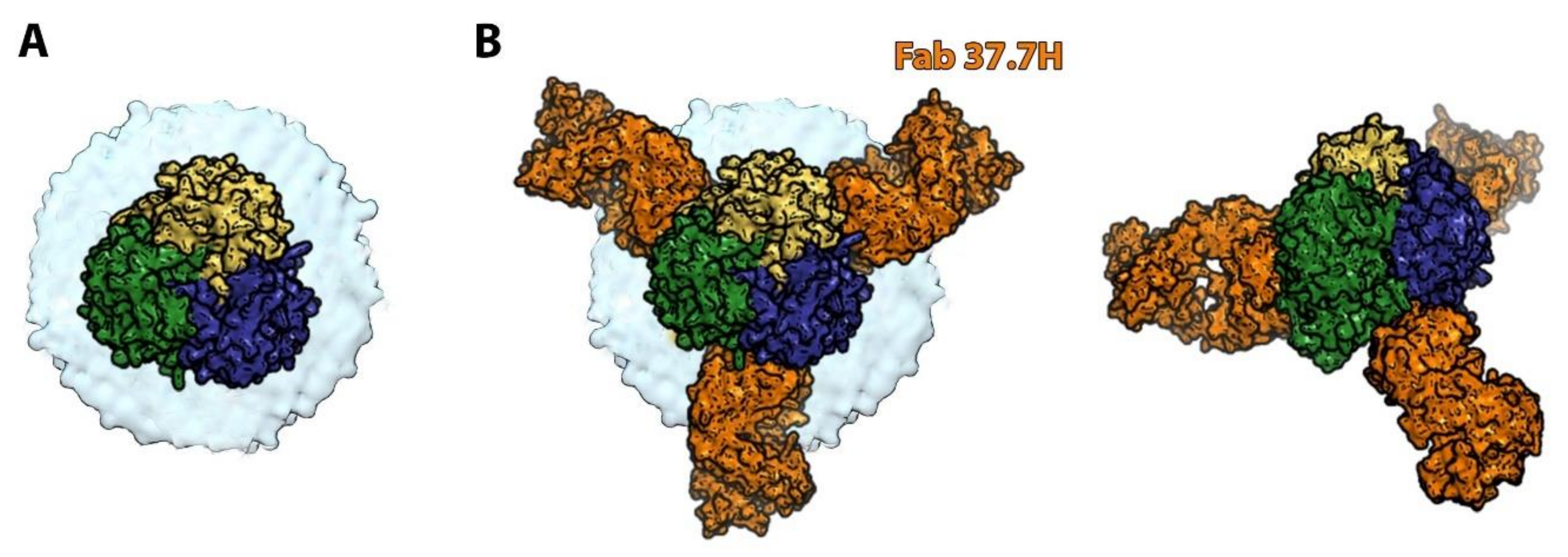
6. Orthobunyaviruses (Family Peribunyaviridae): A Sub-Domain Can Sometimes Be Sufficient to Elicit a Protective Immune Response
7. Arenaviruses (Family Arenaviridae): Differences in the Target-Cell Entry Pathway Result in Different Neutralization Mechanisms within the Same Family
8. Conclusions
Funding
Acknowledgments
Conflicts of Interest
References
- Rappuoli, R.; Bottomley, M.J.; D’Oro, U.; Finco, O.; De Gregorio, E. Reverse vaccinology 2.0: Human immunology instructs vaccine antigen design. J. Exp. Med. 2016, 213, 469–481. [Google Scholar] [CrossRef]
- Rey, F.A.; Lok, S.-M. Common Features of Enveloped Viruses and Implications for Immunogen Design for Next-Generation Vaccines. Cell 2018, 172, 1319–1334. [Google Scholar] [CrossRef]
- Gilman, M.S.A.; Castellanos, C.A.; Chen, M.; Ngwuta, J.O.; Goodwin, E.; Moin, S.; Mas, V.; Melero, J.A.; Wright, P.F.; Graham, B.S.; et al. Rapid profiling of RSV antibody repertoires from the memory B cells of naturally infected adult donors. Sci. Immunol. 2016, 1, eaaj1879. [Google Scholar] [CrossRef] [PubMed]
- McLellan, J.; Chen, M.; Leung, S.; Graepel, K.W.; Du, X.; Yang, Y.; Zhou, T.; Baxa, U.; Yasuda, E.; Beaumont, T.; et al. Structure of RSV Fusion Glycoprotein Trimer Bound to a Prefusion-Specific Neutralizing Antibody. Science 2013, 340, 1113–1117. [Google Scholar] [CrossRef] [PubMed]
- Pallesen, J.; Wang, N.; Corbett, K.S.; Wrapp, D.; Kirchdoerfer, R.N.; Turner, H.L.; Cottrell, C.A.; Becker, M.M.; Wang, L.; Shi, W.; et al. Immunogenicity and structures of a rationally designed prefusion MERS-CoV spike antigen. Proc. Natl. Acad. Sci. USA 2017, 114, E7348–E7357. [Google Scholar] [CrossRef] [PubMed]
- McLellan, J.S.; Chen, M.; Joyce, M.G.; Sastry, M.; Stewart-Jones, G.B.E.; Yang, Y.; Zhang, B.; Chen, L.; Srivatsan, S.; Zheng, A.; et al. Structure-based design of a fusion glycoprotein vaccine for respiratory syncytial virus. Science 2013, 342, 592–598. [Google Scholar] [CrossRef] [PubMed]
- Crank, M.C.; Ruckwardt, T.J.; Chen, M.; Morabito, K.M.; Phung, E.; Costner, P.J.; Holman, L.A.; Hickman, S.P.; Berkowitz, N.M.; Gordon, I.J.; et al. A proof of concept for structure-based vaccine design targeting RSV in humans. Science 2019, 365, 505–509. [Google Scholar] [CrossRef]
- Ruckwardt, T.J.; Morabito, K.M.; Phung, E.; Crank, M.C.; Costner, P.J.; Holman, L.A.; Chang, L.A.; Hickman, S.P.; Berkowitz, N.M.; Gordon, I.J.; et al. Safety, tolerability, and immunogenicity of the respiratory syncytial virus prefusion F subunit vaccine DS-Cav1: A phase 1, randomised, open-label, dose-escalation clinical trial. Lancet Respir. Med. 2021. [Google Scholar] [CrossRef]
- Corbett, K.S.; Edwards, D.K.; Leist, S.R.; Abiona, O.M.; Boyoglu-Barnum, S.; Gillespie, R.A.; Himansu, S.; Schäfer, A.; Ziwawo, C.T.; DiPi-azza, A.T.; et al. SARS-CoV-2 mRNA vaccine design enabled by prototype pathogen preparedness. Nature 2020, 586, 567–571. [Google Scholar] [CrossRef]
- Abudurexiti, A.; Adkins, S.; Alioto, D.; Alkhovsky, S.V.; Avšič-Županc, T.; Ballinger, M.J.; Bente, D.A.; Beer, M.; Bergeron, É.; Blair, C.D.; et al. Taxonomy of the order Bunyavirales: Update 2019. Arch. Virol. 2019, 164, 1949–1965. [Google Scholar] [CrossRef] [PubMed]
- Guardado-Calvo, P.; Rey, F.A. The Envelope Proteins of the Bunyavirales. Adv. Virus Res. 2017, 98, 83–118. [Google Scholar] [CrossRef]
- Bowden, T.A.; Bitto, D.; McLees, A.; Yeromonahos, C.; Elliott, R.M.; Huiskonen, J.T. Orthobunyavirus Ultrastructure and the Curious Tripodal Glycoprotein Spike. PLOS Pathog. 2013, 9, e1003374. [Google Scholar] [CrossRef]
- Li, S.; Rissanen, I.; Zeltina, A.; Hepojoki, J.; Raghwani, J.; Harlos, K.; Pybus, O.G.; Huiskonen, J.T.; Bowden, T.A. A Molecular-Level Account of the Antigenic Hantaviral Surface. Cell Rep. 2016, 16, 278. [Google Scholar] [CrossRef]
- Serris, A.; Stass, R.; Bignon, E.A.; Muena, N.A.; Manuguerra, J.-C.; Jangra, R.K.; Li, S.; Chandran, K.; Tischler, N.D.; Huiskonen, J.T.; et al. The Hantavirus Surface Glycoprotein Lattice and Its Fusion Control Mechanism. Cell 2020, 183, 442–456.e16. [Google Scholar] [CrossRef]
- Freiberg, A.N.; Sherman, M.B.; Morais, M.C.; Holbrook, M.R.; Watowich, S.J. Three-Dimensional Organization of Rift Valley Fever Virus Revealed by Cryoelectron Tomography. J. Virol. 2008, 82, 10341–10348. [Google Scholar] [CrossRef]
- Hellert, J.; Aebischer, A.; Wernike, K.; Haouz, A.; Brocchi, E.; Reiche, S.; Guardado-Calvo, P.; Beer, M.; Rey, F.A. Orthobunyavirus spike architecture and recognition by neutralizing antibodies. Nat. Commun. 2019, 10, 879. [Google Scholar] [CrossRef]
- Punch, E.K.; Hover, S.; Blest, H.T.W.; Fuller, J.; Hewson, R.; Fontana, J.; Mankouri, J.; Barr, J.N. Potassium is a trigger for conformational change in the fusion spike of an enveloped RNA virus. J. Biol. Chem. 2018, 293, 9937–9944. [Google Scholar] [CrossRef] [PubMed]
- Wright, D.; Allen, E.R.; Clark, M.H.; Gitonga, J.N.; Karanja, H.K.; Hulswit, R.J.; Taylor, I.; Biswas, S.; Marshall, J.; Mwololo, D.; et al. Naturally Acquired Rift Valley Fever Virus Neutralizing Antibodies Predominantly Target the Gn Glycoprotein. iScience 2020, 23, 101669. [Google Scholar] [CrossRef] [PubMed]
- Bertolotti-Ciarlet, A.; Smith, J.; Strecker, K.; Paragas, J.; Altamura, L.A.; McFalls, J.M.; Frias-Stäheli, N.; García-Sastre, A.; Schmaljohn, C.S.; Doms, R.W. Cellular localization and antigenic characterization of Crimean-Congo hemorrhagic fever virus glycoproteins. J. Virol. 2005, 79, 6152–6161. [Google Scholar] [CrossRef] [PubMed]
- Kingsford, L.; Ishizawa, L.D.; Hill, D.W. Biological activities of monoclonal antibodies reactive with antigenic sites mapped on the G1 glycoprotein of La Crosse virus. Virology 1983, 129, 443–455. [Google Scholar] [CrossRef]
- Engdahl, T.B.; Crowe, J.E. Humoral Immunity to Hantavirus Infection. mSphere 2020, 5, e00482-20. [Google Scholar] [CrossRef] [PubMed]
- Duehr, J.; McMahon, M.; Williamson, B.; Amanat, F.; Durbin, A.; Hawman, D.W.; Noack, D.; Uhl, S.; Tan, G.S.; Feldmann, H.; et al. Neutralizing Monoclonal Antibodies against the Gn and the Gc of the Andes Virus Glycoprotein Spike Complex Protect from Virus Challenge in a Preclinical Hamster Model. mBio 2020, 11, e00028-20. [Google Scholar] [CrossRef] [PubMed]
- Jonsson, C.B.; Figueiredo, L.T.M.; Vapalahti, O. A Global Perspective on Hantavirus Ecology, Epidemiology, and Disease. Clin. Microbiol. Rev. 2010, 23, 412–441. [Google Scholar] [CrossRef] [PubMed]
- Martínez, V.P.; Di Paola, N.; Alonso, D.O.; Pérez-Sautu, U.; Bellomo, C.M.; Iglesias, A.A.; Coelho, R.M.; López, B.; Periolo, N.; Larson, P.A.; et al. “Super-Spreaders” and Person-to-Person Transmission of Andes Virus in Argentina. N. Engl. J. Med. 2020, 383, 2230–2241. [Google Scholar] [CrossRef]
- Ferrés, M.; Martínez-Valdebenito, C.; Angulo, J.; Henríquez, C.; Vera-Otárola, J.; Vergara, M.J.; Pérez, J.; Fernández, J.; Sotomayor, V.; Valdés, M.F.; et al. Mother-to-Child Transmission of Andes Virus through Breast Milk, Chile. Emerg. Infect. Dis. 2020, 26, 1885–1888. [Google Scholar] [CrossRef]
- Song, J.Y.; Woo, H.J.; Cheong, H.J.; Noh, J.Y.; Baek, L.J.; Kim, W.J. Long-term immunogenicity and safety of inactivated Hantaan virus vaccine (Hantavax™) in healthy adults. Vaccine 2016, 34, 1289–1295. [Google Scholar] [CrossRef]
- Mushtaq, A.; El-Azizi, M.; Khardori, N. Category C Potential Bioterrorism Agents and Emerging Pathogens. Infect. Dis. Clin. N. Am. 2006, 20, 423–441. [Google Scholar] [CrossRef]
- Pettersson, L.; Thunberg, T.; Rocklöv, J.; Klingström, J.; Evander, M.; Ahlm, C. Viral load and humoral immune response in association with disease severity in Puumala hantavirus-infected patients—Implications for treatment. Clin. Microbiol. Infect. 2014, 20, 235–241. [Google Scholar] [CrossRef]
- Garrido, J.L.; Prescott, J.; Calvo, M.; Bravo, F.; Alvarez, R.; Salas, A.; Riquelme, R.; Rioseco, M.L.; Williamson, B.N.; Haddock, E.; et al. Two recombinant human monoclonal antibodies that protect against lethal Andes hantavirus infection in vivo. Sci. Transl. Med. 2018, 10, eaat6420. [Google Scholar] [CrossRef]
- Hooper, J.; Paolino, K.M.; Mills, K.; Kwilas, S.; Josleyn, M.; Cohen, M.; Somerville, B.; Wisniewski, M.; Norris, S.; Hill, B.; et al. A Phase 2a Randomized, Double-Blind, Dose-Optimizing Study to Evaluate the Immunogenicity and Safety of a Bivalent DNA Vaccine for Hemorrhagic Fever with Renal Syndrome Delivered by Intramuscular Electroporation. Vaccines 2020, 8, 377. [Google Scholar] [CrossRef]
- Engdahl, T.B.; Kuzmina, N.A.; Ronk, A.J.; Mire, C.E.; Hyde, M.A.; Kose, N.; Josleyn, M.D.; Sutton, R.E.; Mehta, A.; Wolters, R.M.; et al. Broad and potently neutralizing monoclonal antibodies isolated from human survivors of New World hantavirus infection. Cell Rep. 2021, 35, 109086. [Google Scholar] [CrossRef] [PubMed]
- Hooper, J.W.; Josleyn, M.; Ballantyne, J.; Brocato, R. A novel Sin Nombre virus DNA vaccine and its inclusion in a candidate pan-hantavirus vaccine against hantavirus pulmonary syndrome (HPS) and hemorrhagic fever with renal syndrome (HFRS). Vaccine 2013, 31, 4314–4321. [Google Scholar] [CrossRef][Green Version]
- Klasse, P.J. Neutralization of Virus Infectivity by Antibodies: Old Problems in New Perspectives. Adv. Biol. 2014, 2014, 1–24. [Google Scholar] [CrossRef] [PubMed]
- Rissanen, I.; Stass, R.; Krumm, S.A.; Seow, J.; Hulswit, R.J.; Paesen, G.C.; Hepojoki, J.; Vapalahti, O.; Lundkvist, Å.; Reynard, O.; et al. Molecular rationale for antibody-mediated targeting of the hantavirus fusion glycoprotein. eLife 2020, 9, e58242. [Google Scholar] [CrossRef]
- Bignon, E.A.; Albornoz, A.; Guardado-Calvo, P.; Rey, F.A.; Tischler, N.D. Molecular organization and dynamics of the fusion protein Gc at the hantavirus surface. eLife 2019, 8, e46028. [Google Scholar] [CrossRef] [PubMed]
- Rissanen, I.; Krumm, S.A.; Stass, R.; Whitaker, A.; Voss, J.E.; Bruce, E.A.; Rothenberger, S.; Kunz, S.; Burton, D.R.; Huiskonen, J.T.; et al. Structural Basis for a Neutralizing Antibody Response Elicited by a Recombinant Hantaan Virus Gn Immunogen. mBio 2021, e0253120. [Google Scholar] [CrossRef]
- Allen, E.; Krumm, S.A.; Raghwani, J.; Halldorsson, S.; Elliott, A.; Graham, V.A.; Koudriakova, E.; Harlos, K.; Wright, D.; Warimwe, G.M.; et al. A Protective Monoclonal Antibody Targets a Site of Vulnerability on the Surface of Rift Valley Fever Virus. Cell Rep. 2018, 25, 3750–3758.e4. [Google Scholar] [CrossRef] [PubMed]
- Ikegami, T.; Makino, S. The Pathogenesis of Rift Valley Fever. Viruses 2011, 3, 493–519. [Google Scholar] [CrossRef] [PubMed]
- Wright, D.; Kortekaas, J.; Bowden, T.A.; Warimwe, G.M. Rift Valley fever: Biology and epidemiology. J. Gen. Virol. 2019, 100, 1187–1199. [Google Scholar] [CrossRef] [PubMed]
- Elliott, R.M.; Brennan, B. Emerging phleboviruses. Curr. Opin. Virol. 2014, 5, 50–57. [Google Scholar] [CrossRef]
- Faburay, B.; LaBeaud, A.D.; McVey, D.S.; Wilson, W.C.; Richt, J.A. Current Status of Rift Valley Fever Vaccine Development. Vaccines 2017, 5, 29. [Google Scholar] [CrossRef] [PubMed]
- Grobbelaar, A.A.; Weyer, J.; Leman, P.A.; Kemp, A.; Paweska, J.T.; Swanepoel, R. Molecular Epidemiology of Rift Valley Fever Virus. Emerg. Infect. Dis. 2011, 17, 2270–2276. [Google Scholar] [CrossRef] [PubMed]
- Hao, M.; Zhang, G.; Zhang, S.; Chen, Z.; Chi, X.; Dong, Y.; Fan, P.; Liu, Y.; Chen, Y.; Song, X.; et al. Characterization of Two Neutralizing Antibodies against Rift Valley Fever Virus Gn Protein. Viruses 2020, 12, 259. [Google Scholar] [CrossRef]
- Lagerqvist, N.; Näslund, J.; Lundkvist, Å.; Bouloy, M.; Ahlm, C.; Bucht, G. Characterisation of immune responses and protective efficacy in mice after immunisation with Rift Valley Fever virus cDNA constructs. Virol. J. 2009, 6, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Ma, T.; Wu, Y.; Chen, Z.; Zeng, H.; Tong, Z.; Gao, F.; Qi, J.; Zhao, Z.; Chai, Y.; et al. Neutralization mechanism of human monoclonal antibodies against Rift Valley fever virus. Nat. Microbiol. 2019, 4, 1231–1241. [Google Scholar] [CrossRef]
- Ikegami, T.; Balogh, A.; Nishiyama, S.; Lokugamage, N.; Saito, T.B.; Morrill, J.C.; Shivanna, V.; Indran, S.V.; Zhang, L.; Smith, J.K.; et al. Distinct virulence of Rift Valley fever phlebovirus strains from different genetic lineages in a mouse model. PLoS ONE 2017, 12, e0189250. [Google Scholar] [CrossRef]
- Scarselli, M.; Aricò, B.; Brunelli, B.; Savino, S.; Di Marcello, F.; Palumbo, E.; Veggi, D.; Ciucchi, L.; Cartocci, E.; Bottomley, M.J.; et al. Rational Design of a Meningococcal Antigen Inducing Broad Protective Immunity. Sci. Transl. Med. 2011, 3, 91ra62. [Google Scholar] [CrossRef] [PubMed]
- Negredo, A.; de la Calle-Prieto, F.; Palencia-Herrejón, E.; Mora-Rillo, M.; Astray-Mochales, J.; Sanchez-Seco, M.P.; Lopez, E.B.; Menárguez, J.; Fernández-Cruz, A.; Sanchez-Artola, B.; et al. Autochthonous Crimean–Congo Hemorrhagic Fever in Spain. New Engl. J. Med. 2017, 377, 154–161. [Google Scholar] [CrossRef]
- Mousavi-Jazi, M.; Karlberg, H.; Papa, A.; Christova, I.; Mirazimi, A. Healthy individuals’ immune response to the Bulgarian Crimean-Congo hemorrhagic fever virus vaccine. Vaccine 2012, 30, 6225–6229. [Google Scholar] [CrossRef]
- Dowall, S.D.; Carroll, M.W.; Hewson, R. Development of vaccines against Crimean-Congo haemorrhagic fever virus. Vaccine 2017, 35, 6015–6023. [Google Scholar] [CrossRef]
- Hawman, D.W.; Ahlén, G.; Appelberg, K.S.; Meade-White, K.; Hanley, P.W.; Scott, D.; Monteil, V.; Devignot, S.; Okumura, A.; Weber, F.; et al. A DNA-based vaccine protects against Crimean-Congo haemorrhagic fever virus disease in a Cynomolgus macaque model. Nat. Microbiol. 2020, 6, 187–195. [Google Scholar] [CrossRef] [PubMed]
- Zivcec, M.; Scholte, F.; Spiropoulou, C.F.; Spengler, J.R.; Bergeron, E. Molecular Insights into Crimean-Congo Hemorrhagic Fever Virus. Viruses 2016, 8, 106. [Google Scholar] [CrossRef]
- Golden, J.W.; Shoemaker, C.J.; Lindquist, M.E.; Zeng, X.; Daye, S.P.; Williams, J.A.; Liu, J.; Coffin, K.M.; Olschner, S.; Flusin, O.; et al. GP38-targeting monoclonal antibodies protect adult mice against lethal Crimean-Congo hemorrhagic fever virus infection. Sci. Adv. 2019, 5, eaaw9535. [Google Scholar] [CrossRef] [PubMed]
- Suschak, J.J.; Golden, J.W.; Fitzpatrick, C.J.; Shoemaker, C.J.; Badger, C.V.; Schmaljohn, C.S.; Garrison, A.R. A CCHFV DNA vaccine protects against heterologous challenge and establishes GP38 as immunorelevant in mice. NPJ Vaccines 2021, 6, 1–11. [Google Scholar] [CrossRef]
- Libraty, D.H.; Young, P.R.; Pickering, D.; Endy, T.P.; Kalayanarooj, S.; Green, S.; Vaughn, D.W.; Nisalak, A.; Ennis, F.A.; Rothman, A.L. High circulating levels of the dengue virus nonstructural protein NS1 early in dengue illness correlate with the development of dengue hemorrhagic fever. J. Infect. Dis. 2002, 186, 1165–1168. [Google Scholar] [CrossRef] [PubMed]
- Chung, K.M.; Nybakken, G.E.; Thompson, B.S.; Engle, M.J.; Marri, A.; Fremont, D.H.; Diamond, M.S. Antibodies against West Nile Virus Nonstructural Protein NS1 Prevent Lethal Infection through Fc γ Receptor-Dependent and -Independent Mechanisms. J. Virol. 2006, 80, 1340–1351. [Google Scholar] [CrossRef]
- Fels, J.M.; Maurer, D.P.; Herbert, A.S.; Wirchnianski, A.S.; Vergnolle, O.; Cross, R.W.; Abelson, D.M.; Moyer, C.L.; Mishra, A.K.; Aguilan, J.T.; et al. Protective neutralizing antibodies from human survivors of Crimean-Congo hemorrhagic fever. Cell 2021. [Google Scholar] [CrossRef]
- Evans, A.B.; Winkler, C.W.; Peterson, K.E. Differences in Neuropathogenesis of Encephalitic California Serogroup Viruses. Emerg. Infect. Dis. 2019, 25, 728–738. [Google Scholar] [CrossRef] [PubMed]
- Travassos da Rosa, J.F.; de Souza, W.M.; de Paula Pinheiro, F.; Figueiredo, M.L.; Cardoso, J.F.; Acrani, G.O.; Nunes, M.R.T. Oropouche Virus: Clinical, Epidemiological, and Molecular Aspects of a Neglected Orthobunyavirus. Am. J. Trop. Med. Hyg. 2017, 96, 1019–1030. [Google Scholar]
- Ngari Virus Is a Bunyamwera Virus Reassortant That Can Be Associated with Large Outbreaks of Hemorrhagic Fever in Africa [Internet]. [Cité 30 Août 2018]. Available online: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC479050/ (accessed on 22 August 2021).
- Schuh, T.; Schultz, J.; Moelling, K.; Pavlovic, J. DNA-based vaccine against La Crosse virus: Protective immune response mediated by neutralizing antibodies and CD4+ T cells. Hum. Gene Ther. 1999, 10, 1649–1658. [Google Scholar] [CrossRef]
- Nuccitelli, A.; Cozzi, R.; Gourlay, L.J.; Donnarumma, D.; Necchi, F.; Norais, N.; Telford, J.L.; Rappuoli, R.; Bolognesi, M.; Maione, D.; et al. Structure-based approach to rationally design a chimeric protein for an effective vaccine against Group B Streptococcus infections. Proc. Natl. Acad. Sci. USA 2011, 108, 10278–10283. [Google Scholar] [CrossRef] [PubMed]
- Brisse, M.; Ly, H. Hemorrhagic Fever-Causing Arenaviruses: Lethal Pathogens and Potent Immune Suppressors. Front. Immunol. 2019, 10, 372. [Google Scholar] [CrossRef] [PubMed]
- Oloniniyi, O.K.; Unigwe, U.S.; Okada, S.; Kimura, M.; Koyano, S.; Miyazaki, Y.; Iroezindu, M.O.; Ajayi, N.A.; Chukwubike, C.M.; Chika-Igwenyi, N.M.; et al. Genetic characterization of Lassa virus strains isolated from 2012 to 2016 in southeastern Nigeria. PLOS Negl. Trop. Dis. 2018, 12, e0006971. [Google Scholar] [CrossRef] [PubMed]
- Sarute, N.; Ross, S.R. New World Arenavirus Biology. Annu. Rev. Virol. 2017, 4, 141–158. [Google Scholar] [CrossRef] [PubMed]
- Maiztegui, J.I.; McKee, J.K.T.; Oro, J.G.B.; Harrison, L.H.; Gibbs, P.H.; Feuillade, M.R.; Enria, D.A.; Briggiler, A.M.; Levis, S.C.; Ambrosio, A.M.; et al. Protective Efficacy of a Live Attenuated Vaccine against Argentine Hemorrhagic Fever. J. Infect. Dis. 1998, 177, 277–283. [Google Scholar] [CrossRef] [PubMed]
- Hastie, K.M.; Saphire, E.O. Lassa virus glycoprotein: Stopping a moving target. Curr. Opin. Virol. 2018, 31, 52–58. [Google Scholar] [CrossRef] [PubMed]
- Sommerstein, R.; Flatz, L.; Remy, M.M.; Malinge, P.; Magistrelli, G.; Fischer, N.; Sahin, M.; Bergthaler, A.; Igonet, S.; Ter Meulen, J.; et al. Arenavirus Glycan Shield Promotes Neutralizing Antibody Evasion and Protracted Infection. PLOS Pathog. 2015, 11, e1005276. [Google Scholar] [CrossRef] [PubMed]
- Zeltina, A.; Krumm, S.A.; Sahin, M.; Struwe, W.; Harlos, K.; Nunberg, J.H.; Crispin, M.; Pinschewer, D.D.; Doores, K.J.; Bowden, T.A. Convergent immunological solutions to Argentine hemorrhagic fever virus neutralization. Proc. Natl. Acad. Sci. USA 2017, 114, 7031–7036. [Google Scholar] [CrossRef]
- Robinson, J.E.; Hastie, K.M.; Cross, R.W.; Yenni, R.E.; Elliott, D.H.; Rouelle, J.A.; Kannadka, C.B.; Smira, A.A.; Garry, C.E.; Bradley, B.T.; et al. Most neutralizing human monoclonal antibodies target novel epitopes requiring both Lassa virus glycoprotein subunits. Nat. Commun. 2016, 7, 11544. [Google Scholar] [CrossRef]
- Hastie, K.M.; Zandonatti, M.A.; Kleinfelter, L.M.; Heinrich, M.L.; Rowland, M.M.; Chandran, K.; Branco, L.M.; Robinson, J.E.; Garry, R.F.; Saphire, E.O. Structural basis for antibody-mediated neutralization of Lassa virus. Science 2017, 356, 923–928. [Google Scholar] [CrossRef]
| Bunyavirus Genus | Target of Neutralizing/Protecting mAbs | Further Challenges |
|---|---|---|
| Orthohantavirus | Gn/Gc spike |
|
| Phlebovirus | Gn |
|
| Nairovirus | Gc and GP38 |
|
| Orthobunyavirus | Gc N-terminal half (spike domains) |
|
| Arenavirus | GP1 for NWA Trimeric GPC for OWA |
|
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Serris, A. The Input of Structural Vaccinology in the Search for Vaccines against Bunyaviruses. Viruses 2021, 13, 1766. https://doi.org/10.3390/v13091766
Serris A. The Input of Structural Vaccinology in the Search for Vaccines against Bunyaviruses. Viruses. 2021; 13(9):1766. https://doi.org/10.3390/v13091766
Chicago/Turabian StyleSerris, Alexandra. 2021. "The Input of Structural Vaccinology in the Search for Vaccines against Bunyaviruses" Viruses 13, no. 9: 1766. https://doi.org/10.3390/v13091766
APA StyleSerris, A. (2021). The Input of Structural Vaccinology in the Search for Vaccines against Bunyaviruses. Viruses, 13(9), 1766. https://doi.org/10.3390/v13091766





