Development and Validation of an ELISA for the Detection of Bluetongue Virus Serotype 4-Specific Antibodies
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sera Tested
2.2. Cells, Plasmids and Viruses
2.3. VP2 Expression and Purification
2.4. VP2 Labelling with HRP
2.5. VP2 BTV-4 ELISA
3. Results
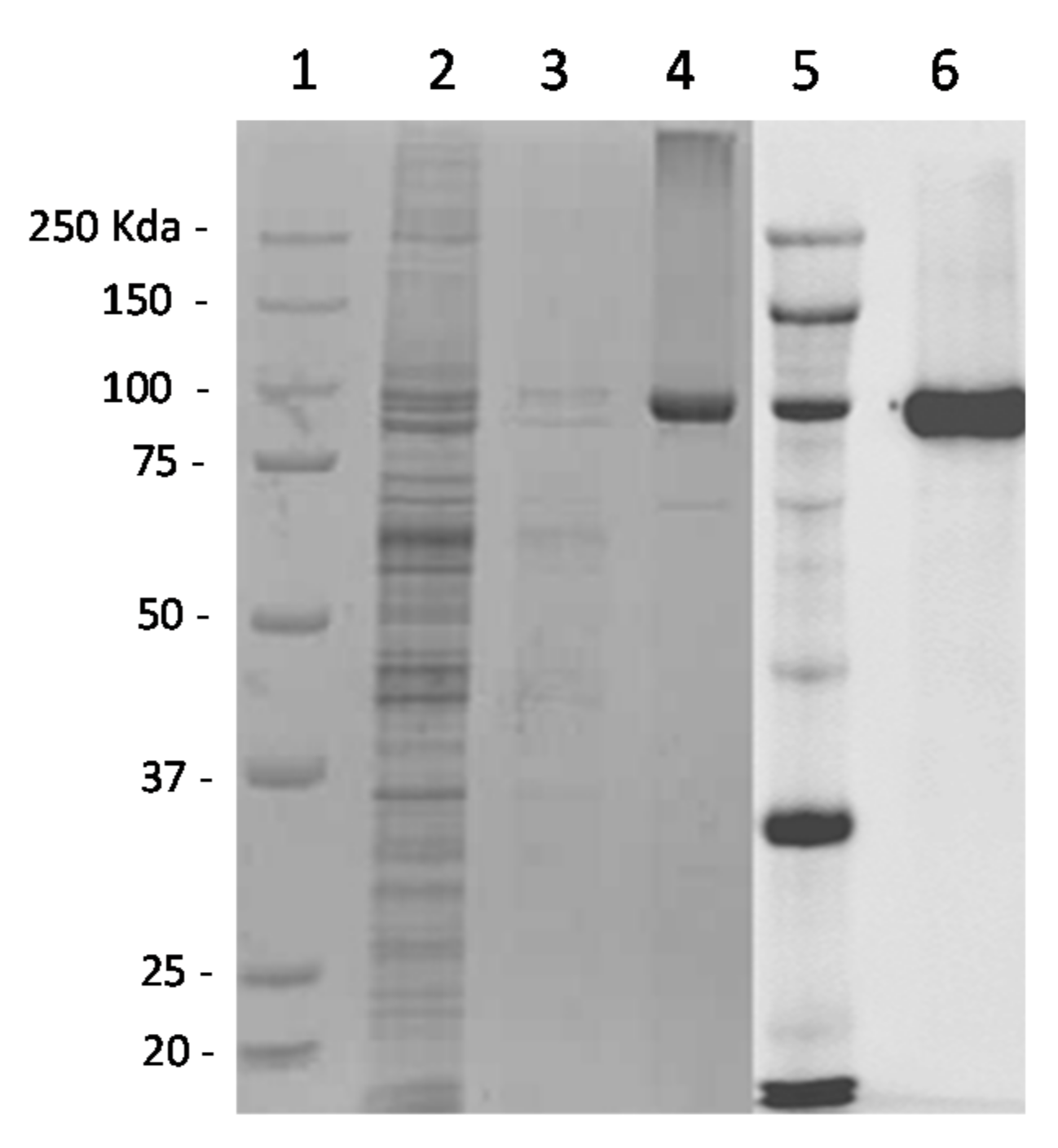
3.1. VP2 Expression and Purification
3.2. ELISA Results on Naïve or BTV-1, 2, 8, 9 or 16 Infected or Vaccinated Animals
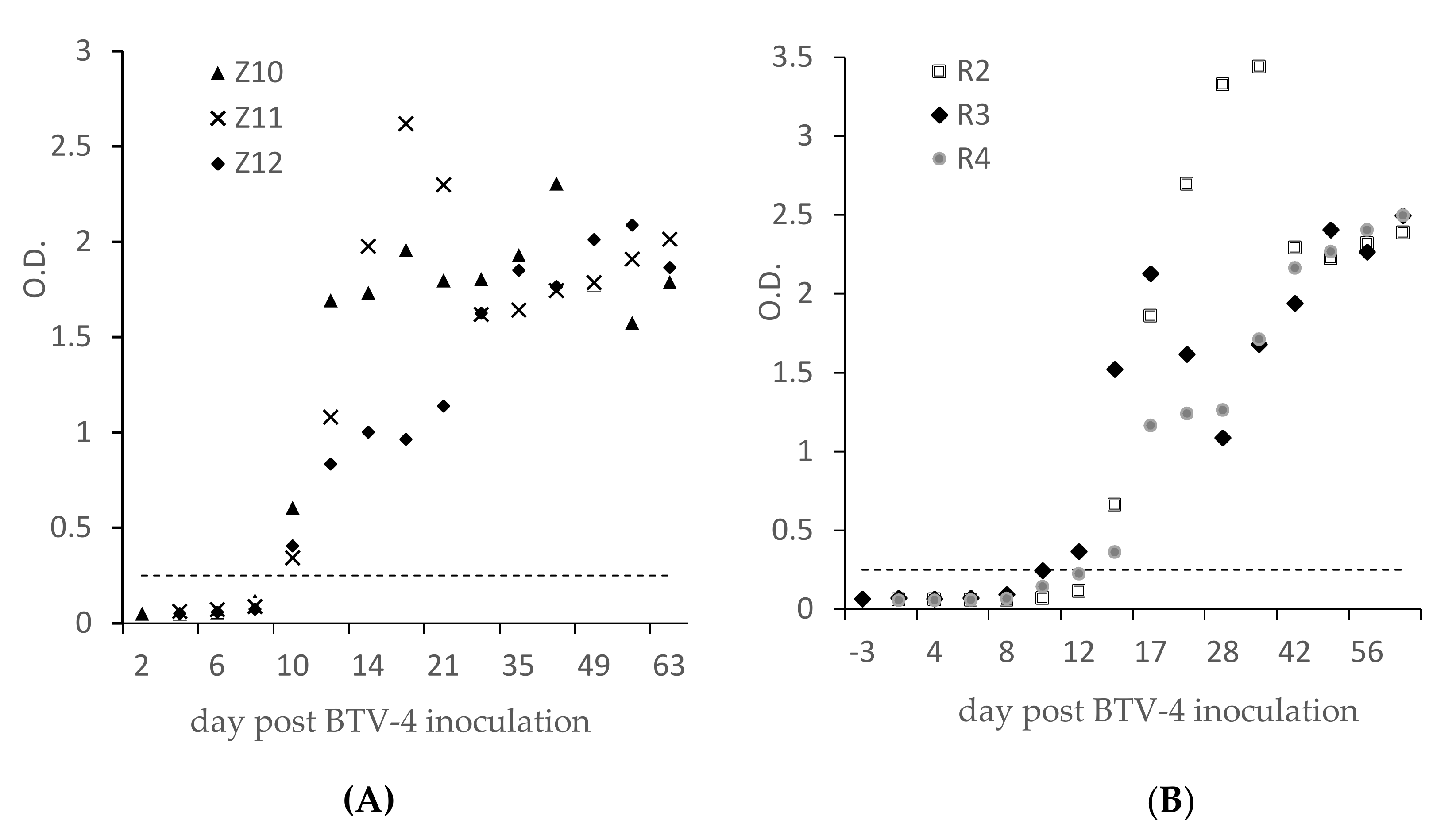
3.3. ELISA Results on Experimental or Field BTV-4 Infected Ruminants
3.4. Detection of BTV-4-Specific Antibodies in Vaccinated Animals
3.5. ELISA Analytical Sensitivity
3.6. ELISA Results with BTV Reference Sera
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mertens, P.P.; Brown, F.; Sangar, D.V. Assignment of the genome segments of bluetongue virus type 1 to the proteins which they encode. Virology 1984, 135, 207–217. [Google Scholar] [CrossRef]
- Batten, C.; Darpel, K.; Henstock, M.; Fay, P.; Veronesi, E.; Gubbins, S.; Graves, S.; Frost, L.; Oura, C. Evidence for transmission of bluetongue virus serotype 26 through direct contact. PLoS ONE 2014, 9, e96049. [Google Scholar] [CrossRef] [Green Version]
- Toussaint, J.F.; Sailleau, C.; Mast, J.; Houdart, P.; Czaplicki, G.; Demeestere, L.; VandenBussche, F.; van Dessel, W.; Goris, N.; Breard, E.; et al. Bluetongue in Belgium, 2006. Emerg. Infect. Dis. 2007, 13, 614–616. [Google Scholar] [CrossRef]
- Kundlacz, C.; Caignard, G.; Sailleau, C.; Viarouge, C.; Postic, L.; Vitour, D.; Zientara, S.; Breard, E. Bluetongue Virus in France: An Illustration of the European and Mediterranean Context since the 2000s. Viruses 2019, 11, 672. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sailleau, C.; Breard, E.; Viarouge, C.; Gorlier, A.; Leroux, A.; Hirchaud, E.; Lucas, P.; Blanchard, Y.; Vitour, D.; Grandcollot-Chabot, M.; et al. Emergence of bluetongue virus serotype 4 in mainland France in November 2017. Transbound Emerg. Dis. 2018, 65, 1158–1162. [Google Scholar] [CrossRef]
- Belbis, G.; Zientara, S.; Breard, E.; Sailleau, C.; Caignard, G.; Vitour, D.; Attoui, H. Bluetongue Virus: From BTV-1 to BTV-27. Adv. Virus Res. 2017, 99, 161–197. [Google Scholar] [CrossRef]
- Breard, E.; Sailleau, C.; Nomikou, K.; Hamblin, C.; Mertens, P.P.; Mellor, P.S.; El Harrak, M.; Zientara, S. Molecular epidemiology of bluetongue virus serotype 4 isolated in the Mediterranean Basin between 1979 and 2004. Virus Res. 2007, 125, 191–197. [Google Scholar] [CrossRef]
- Ries, C.; Sharav, T.; Tseren-Ochir, E.O.; Beer, M.; Hoffmann, B. Putative Novel Serotypes ‘33’ and ‘35’ in Clinically Healthy Small Ruminants in Mongolia Expand the Group of Atypical BTV. Viruses 2020, 13, 42. [Google Scholar] [CrossRef]
- Schulz, C.; Breard, E.; Sailleau, C.; Jenckel, M.; Viarouge, C.; Vitour, D.; Palmarini, M.; Gallois, M.; Hoper, D.; Hoffmann, B.; et al. Bluetongue virus serotype 27: Detection and characterization of two novel variants in Corsica, France. J. Gen. Virol. 2016, 97, 2073–2083. [Google Scholar] [CrossRef]
- Ratinier, M.; Caporale, M.; Golder, M.; Franzoni, G.; Allan, K.; Nunes, S.F.; Armezzani, A.; Bayoumy, A.; Rixon, F.; Shaw, A.; et al. Identification and characterization of a novel non-structural protein of bluetongue virus. PLoS Pathog. 2011, 7, e1002477. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shaw, A.E.; Ratinier, M.; Nunes, S.F.; Nomikou, K.; Caporale, M.; Golder, M.; Allan, K.; Hamers, C.; Hudelet, P.; Zientara, S.; et al. Reassortment between two serologically unrelated bluetongue virus strains is flexible and can involve any genome segment. J. Virol. 2013, 87, 543–557. [Google Scholar] [CrossRef] [Green Version]
- Mecham, J.O.; Wilson, W.C. Antigen capture competitive enzyme-linked immunosorbent assays using baculovirus-expressed antigens for diagnosis of bluetongue virus and epizootic hemorrhagic disease virus. J. Clin. Microbiol. 2004, 42, 518–523. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hund, A.; Gollnick, N.; Sauter-Louis, C.; Neubauer-Juric, A.; Lahm, H.; Buttner, M. A two year BTV-8 vaccination follow up: Molecular diagnostics and assessment of humoral and cellular immune reactions. Vet. Microbiol. 2012, 154, 247–256. [Google Scholar] [CrossRef]
- Martinelle, L.; Dal Pozzo, F.; Thys, C.; De Leeuw, I.; Van Campe, W.; De Clercq, K.; Thiry, E.; Saegerman, C. Assessment of cross-protection induced by a bluetongue virus (BTV) serotype 8 vaccine towards other BTV serotypes in experimental conditions. Vet. Res. 2018, 49, 63. [Google Scholar] [CrossRef] [Green Version]
- Breard, E.; Garnier, A.; Despres, P.; Blaise Boisseau, S.; Comtet, L.; Viarouge, C.; Bakkali-Kassimi, L.; Pourquier, P.; Hudelet, P.; Vitour, D.; et al. Development of a Double-Antigen Microsphere Immunoassay for Simultaneous Group and Serotype Detection of Bluetongue Virus Antibodies. Transbound Emerg. Dis. 2017, 64, 1837–1847. [Google Scholar] [CrossRef]
- Moss, B. Genetically engineered poxviruses for recombinant gene expression, vaccination, and safety. Proc. Natl. Acad. Sci. USA 1996, 93, 11341–11348. [Google Scholar] [CrossRef] [Green Version]
- Pradeau-Aubreton, K.; Ruff, M.; Garnier, J.M.; Schultz, P.; Drillien, R. Vectors for recombinational cloning and gene expression in mammalian cells using modified vaccinia virus Ankara. Anal. Biochem. 2010, 404, 103–105. [Google Scholar] [CrossRef]
- Breard, E.; Belbis, G.; Viarouge, C.; Nomikou, K.; Haegeman, A.; De Clercq, K.; Hudelet, P.; Hamers, C.; Moreau, F.; Lilin, T.; et al. Evaluation of adaptive immune responses and heterologous protection induced by inactivated bluetongue virus vaccines. Vaccine 2015, 33, 512–518. [Google Scholar] [CrossRef]
- Schulz, C.; Sailleau, C.; Breard, E.; Flannery, J.; Viarouge, C.; Zientara, S.; Beer, M.; Batten, C.; Hoffmann, B. Experimental infection of sheep, goats and cattle with a bluetongue virus serotype 4 field strain from Bulgaria, 2014. Transbound Emerg. Dis. 2018, 65, e243–e250. [Google Scholar] [CrossRef] [PubMed]
- Sailleau, C.; Breard, E.; Viarouge, C.; Belbis, G.; Lilin, T.; Vitour, D.; Zientara, S. Experimental infection of calves with seven serotypes of Epizootic Hemorrhagic Disease virus: Production and characterization of reference sera. Vet. Ital. 2019, 55, 339–346. [Google Scholar] [CrossRef] [PubMed]
- Stewart, M.; Bhatia, Y.; Athmaran, T.N.; Noad, R.; Gastaldi, C.; Dubois, E.; Russo, P.; Thiery, R.; Sailleau, C.; Breard, E.; et al. Validation of a novel approach for the rapid production of immunogenic virus-like particles for bluetongue virus. Vaccine 2010, 28, 3047–3054. [Google Scholar] [CrossRef] [PubMed]
- Matsuo, E.; Celma, C.C.; Boyce, M.; Viarouge, C.; Sailleau, C.; Dubois, E.; Breard, E.; Thiery, R.; Zientara, S.; Roy, P. Generation of replication-defective virus-based vaccines that confer full protection in sheep against virulent bluetongue virus challenge. J. Virol. 2011, 85, 10213–10221. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hebben, M.; Brants, J.; Birck, C.; Samama, J.P.; Wasylyk, B.; Spehner, D.; Pradeau, K.; Domi, A.; Moss, B.; Schultz, P.; et al. High level protein expression in mammalian cells using a safe viral vector: Modified vaccinia virus Ankara. Protein Expr. Purif. 2007, 56, 269–278. [Google Scholar] [CrossRef] [PubMed]
- White, S.D.; Conwell, K.; Langland, J.O.; Jacobs, B.L. Use of a negative selectable marker for rapid selection of recombinant vaccinia virus. Biotechniques 2011, 50, 303–309. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lemasson, M.; Caignard, G.; Unterfinger, Y.; Attoui, H.; Bell-Sakyi, L.; Hirchaud, E.; Moutailler, S.; Johnson, N.; Vitour, D.; Richardson, J.; et al. Exploration of binary protein-protein interactions between tick-borne flaviviruses and Ixodes ricinus. Parasit Vectors 2021, 14, 144. [Google Scholar] [CrossRef]
- Falkner, F.G.; Moss, B. Escherichia coli gpt gene provides dominant selection for vaccinia virus open reading frame expression vectors. J. Virol. 1988, 62, 1849–1854. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Celma, C.C.; Boyce, M.; van Rijn, P.A.; Eschbaumer, M.; Wernike, K.; Hoffmann, B.; Beer, M.; Haegeman, A.; De Clercq, K.; Roy, P. Rapid generation of replication-deficient monovalent and multivalent vaccines for bluetongue virus: Protection against virulent virus challenge in cattle and sheep. J. Virol. 2013, 87, 9856–9864. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Golender, N.; Bumbarov, V.; Eldar, A.; Lorusso, A.; Kenigswald, G.; Varsano, J.S.; David, D.; Schainin, S.; Dagoni, I.; Gur, I.; et al. Bluetongue Serotype 3 in Israel 2013-2018: Clinical Manifestations of the Disease and Molecular Characterization of Israeli Strains. Front. Vet. Sci. 2020, 7, 112. [Google Scholar] [CrossRef]
- Lorusso, A.; Guercio, A.; Purpari, G.; Camma, C.; Calistri, P.; D’Alterio, N.; Hammami, S.; Sghaier, S.; Savini, G. Bluetongue virus serotype 3 in Western Sicily, November 2017. Vet. Ital. 2017, 53, 273–275. [Google Scholar] [CrossRef]
- Zulu, G.B.; Venter, E.H. Evaluation of cross-protection of bluetongue virus serotype 4 with other serotypes in sheep. J. S. Afr. Vet. Assoc. 2014, 85, 1041. [Google Scholar] [CrossRef] [Green Version]



| Origin | Animals | Status | Serum Number | Country | Year of Sampling/Ref | Total |
|---|---|---|---|---|---|---|
| field | cattle | Naïve | 96 | France | 1998–2000 | 319 |
| goats | 110 | 2006 | ||||
| sheep | 64 | 2008–2015 | ||||
| experimental assay | sheep | Naïve | 26 | France | [18] | |
| Field | deer | Naïve | 5 | Guyana | 2020 | |
| experimental assay | cattle, goats | Naïve | 11 | Germany | [19] | |
| cattle | EHD | 7 | France | [20] | ||
| experimental assay | sheep | BTV-2 | 8 | France | 2004 | 306 |
| sheep | BTV-1 | 41 | [21,22] | |||
| experimental assay | sheep | BTV-9 | 21 | France | [18] | |
| sheep | BTV-16 | 5 | ||||
| field | cattle | BTV-8 | 202 | 2015 | ||
| sheep | 10 | |||||
| experimental assay | cattle, goats, rabbit | BTV-8 | 16 | Germany | - | |
| MRI | 3 | France | - | |||
| experimental assay | cattle | BTV-4 | 27 | Germany | [19] | 141 |
| goats | 35 | |||||
| field | cattle | BTV-4 | 55 * | France | 2018–2021 | |
| sheep | 21 * | 2017 | ||||
| experimental assay | sheep | BTV-4 | 3 | Germany | - | |
| field | goats | vaccinated BTV-4 | 180 | France | 2019 | 363 |
| experimental assay | sheep | 38 | [18] | |||
| zoo | see Table 3 | 145 | 2018–2021 | |||
| reference sera | 23 | BTV-1-6, 8-24 | 23 | UK (Pirbright) | - | 57 |
| 23 | BTV-1, 2-6, 8-24, 26 | 23 | Australia | - | ||
| 3 | BTV-27 | 3 | France | - | ||
| 8 | BTV-25, 30, 33, 35 | 8 | Germany | - |
| SNT Results | ||||
|---|---|---|---|---|
| Positive | Negative | Total | ||
| BTV-4 ELISA results | positive | 140 | 28 | 168 |
| negative | 1 | 11 | 12 | |
| total | 141 | 39 | 180 | |
| Days after BTV-4 Vaccination (Number of BTV-4 ELISA Positive/Total Tested) | ||||
|---|---|---|---|---|
| 5 | 21 | 28 | 42 | |
| Vac 2 and 4 (2x) | (0/4)a | (5/5)b | (5/5)b | (4/4)b |
| Vac 9, 2 and 4 * | (0/8) | (7/10) | - | - |
| ELISA BTV-4 Results | |||||
|---|---|---|---|---|---|
| Zoo Species | Number of Animals | Negative | Positive | % Positive | |
| Antilope cervicapra | Blackbuck | 39 | 6 | 33 | 84.6 |
| Axis axis | Chital deer | 33 | 0 | 33 | 100 |
| Axis porcinus | Indian hog deer | 8 | 5 | 3 | 37.5 |
| Budocas taxicolor | Takin Cattle | 5 | 1 | 4 | 80 |
| Camelus dromedarius | Arabian camel | 2 | 2 | 0 | 0 |
| Connochaetes taurinus | Blue wildebeest | 2 | 2 | 0 | 0 |
| Hippotragus niger | Sable antelope | 10 | 1 | 9 | 90 |
| Madoqua kirkii | Kirk’s dik-dik | 2 | 2 | 0 | 0 |
| Muntiacus reevesi | Chinese muntja | 12 | 5 | 7 | 58.3 |
| Oryx beisa | East African oryx | 3 | 1 | 2 | 66.7 |
| Tragelaphus angasii | Nyala | 3 | 0 | 3 | 100 |
| Tragelaphusus eurycerus | Bongo | 7 | 2 | 5 | 71.4 |
| Tragelaphus imberbis | Lesser kudu | 3 | 0 | 3 | 100 |
| Tragelaphus spekii | Marshbuck | 14 | 3 | 11 | 78.6 |
| Tragulus javanicus | Lesser mouse-deer | 2 | 0 | 2 | 100 |
| total | 145 | 30 * | 115 ** | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bréard, E.; Turpaud, M.; Beaud, G.; Postic, L.; Fablet, A.; Beer, M.; Sailleau, C.; Caignard, G.; Viarouge, C.; Hoffmann, B.; et al. Development and Validation of an ELISA for the Detection of Bluetongue Virus Serotype 4-Specific Antibodies. Viruses 2021, 13, 1741. https://doi.org/10.3390/v13091741
Bréard E, Turpaud M, Beaud G, Postic L, Fablet A, Beer M, Sailleau C, Caignard G, Viarouge C, Hoffmann B, et al. Development and Validation of an ELISA for the Detection of Bluetongue Virus Serotype 4-Specific Antibodies. Viruses. 2021; 13(9):1741. https://doi.org/10.3390/v13091741
Chicago/Turabian StyleBréard, Emmanuel, Mathilde Turpaud, Georges Beaud, Lydie Postic, Aurore Fablet, Martin Beer, Corinne Sailleau, Grégory Caignard, Cyril Viarouge, Bernd Hoffmann, and et al. 2021. "Development and Validation of an ELISA for the Detection of Bluetongue Virus Serotype 4-Specific Antibodies" Viruses 13, no. 9: 1741. https://doi.org/10.3390/v13091741
APA StyleBréard, E., Turpaud, M., Beaud, G., Postic, L., Fablet, A., Beer, M., Sailleau, C., Caignard, G., Viarouge, C., Hoffmann, B., Vitour, D., & Zientara, S. (2021). Development and Validation of an ELISA for the Detection of Bluetongue Virus Serotype 4-Specific Antibodies. Viruses, 13(9), 1741. https://doi.org/10.3390/v13091741






