Characterization of the Roles of SGT1/RAR1, EDS1/NDR1, NPR1, and NRC/ADR1/NRG1 in Sw-5b-Mediated Resistance to Tomato Spotted Wilt Virus
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plasmid Construction
2.2. Host Plant and Virus
2.3. VIGS Assay
2.4. Total RNA Isolation and Quantitative RT-PCR Analysis
2.5. Western Blot Assays
3. Results
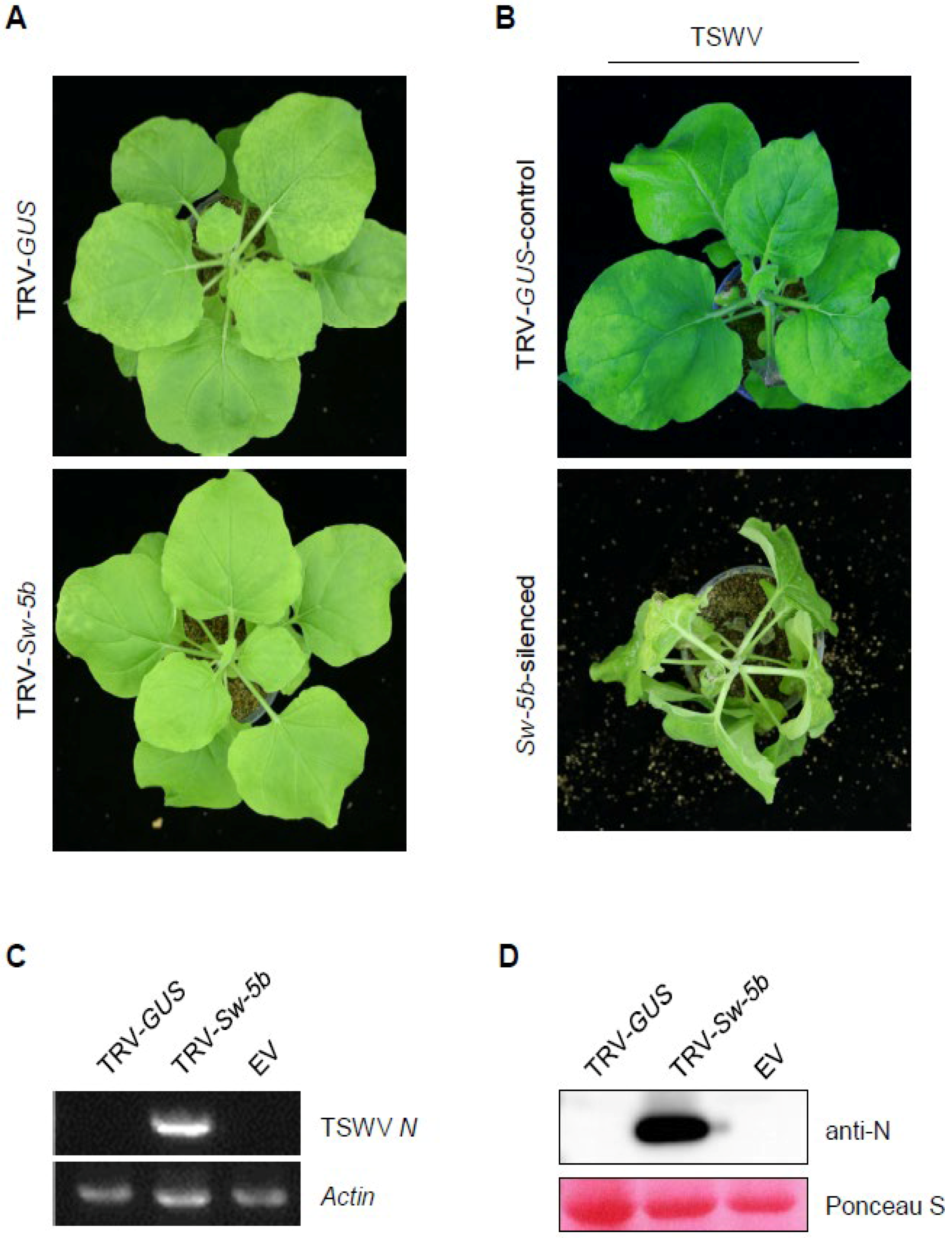
3.1. Silencing Sw-5b by TRV-Based VIGS Abolished the Resistance of Sw-5b-Transgenic N. benthamiana to TSWV Infection
3.2. SGT1, But Not Rar1, Was Required for Sw-5b-Mediated Resistance to TSWV
3.3. Suppression of EDS1 and NDR1 Did Not Disrupt Sw-5b-Triggered Immunity
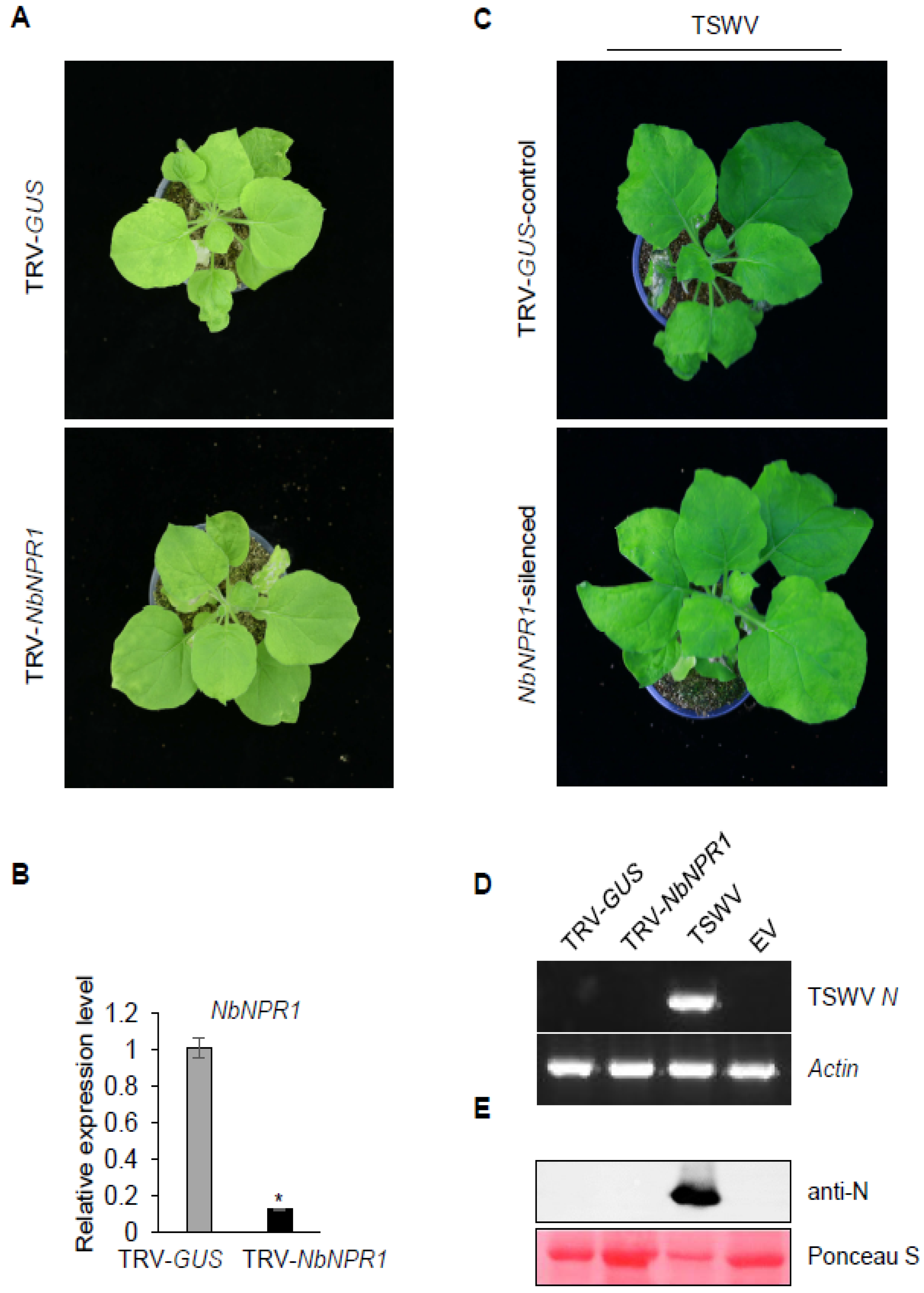
3.4. Silencing Expression of NPR1 Did Not Result in TSWV Systemic Infection in Sw-5b-Transgenic N. benthamiana Plants
3.5. The Helper NLRs NRC2/3/4 Were Required for Sw-5b-Mediated Systemic Resistance to TSWV Infection
3.6. Silencing Expression of the Helper NLRs NRG1 and ADR1 Did Not Affect Sw-5b-Mediated Systemic Resistance to TSWV
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jones, J.D.; Dangl, J.L. The plant immune system. Nature 2006, 444, 323–329. [Google Scholar] [CrossRef] [Green Version]
- Kourelis, J.; van der Hoorn, R.A.L. Defended to the nines: 25 years of resistance gene cloning identifies nine mechanisms for R protein function. Plant Cell 2018, 30, 285–299. [Google Scholar] [CrossRef] [Green Version]
- Van der Biezen, E.A.; Jones, J.D. The NB-ARC domain: A novel signalling motif shared by plant resistance gene products and regulators of cell death in animals. Curr. Biol. 1998, 8, R226–R227. [Google Scholar] [CrossRef] [Green Version]
- Jones, J.D.; Vance, R.E.; Dangl, J.L. Intracellular innate immune surveillance devices in plants and animals. Science 2016, 354. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bonardi, V.; Cherkis, K.; Nishimura, M.T.; Dangl, J.L. A new eye on NLR proteins: Focused on clarity or diffused by complexity? Curr. Opin. Immunol. 2012, 24, 41–50. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dangl, J.L.; Horvath, D.M.; Staskawicz, B.J. Pivoting the plant immune system from dissection to deployment. Science 2013, 341, 746–751. [Google Scholar] [CrossRef] [Green Version]
- Parrella, G.; Gognalons, P.; Gebre-Selassie, K.; Vovlas, C.; Marchoux, G. An update of the host range of tomato spotted wilt virus. J. Plant. Pathol. 2003, 85, 227–264. [Google Scholar]
- Kormelink, R.; Garcia, M.L.; Goodin, M.; Sasaya, T.; Haenni, A.L. Negative-strand RNA viruses: The plant-infecting counterparts. Virus Res. 2011, 162, 184–202. [Google Scholar] [CrossRef]
- Scholthof, K.B.; Adkins, S.; Czosnek, H.; Palukaitis, P.; Jacquot, E.; Hohn, T.; Hohn, B.; Saunders, K.; Candresse, T.; Ahlquist, P.; et al. Top 10 plant viruses in molecular plant pathology. Mol. Plant Pathol. 2011, 12, 938–954. [Google Scholar] [CrossRef] [PubMed]
- Oliver, J.E.; Whitfield, A.E. The genus tospovirus: Emerging bunyaviruses that threaten food security. Annu. Rev. Virol. 2016, 3, 101–124. [Google Scholar] [CrossRef]
- Prins, M.; Goldbach, R. The emerging problem of tospovirus infection and nonconventional methods of control. Trends MicroBiol. 1998, 6, 31–35. [Google Scholar] [CrossRef]
- Boiteux, L.S.; Giordano, L.D. Genetic-basis of resistance against 2 tospovirus species in tomato (Lycopersicon-Esculentum). Euphytica 1993, 71, 151–154. [Google Scholar] [CrossRef]
- Brommonschenkel, S.H.; Frary, A.; Frary, A.; Tanksley, S.D. The broad-spectrum tospovirus resistance gene Sw-5 of tomato is a homolog of the root-knot nematode resistance gene Mi. Mol. Plant Microbe Interact. 2000, 13, 1130–1138. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Spassova, M.I.; Prins, T.W.; Folkertsma, R.T.; Klein-Lankhorst, R.M.; Hille, J.; Goldbach, R.W.; Prins, M. The tomato gene Sw5 is a member of the coiled coil, nucleotide binding, leucine-rich repeat class of plant resistance genes and confers resistance to TSWV in tobacco. Mol. Breed. 2001, 7, 151–161. [Google Scholar] [CrossRef] [Green Version]
- Bendahmane, A.; Farnham, G.; Moffett, P.; Baulcombe, D.C. Constitutive gain-of-function mutants in a nucleotide binding site-leucine rich repeat protein encoded at the Rx locus of potato. Plant J. 2002, 32, 195–204. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hallwass, M.; de Oliveira, A.S.; de Campos Dianese, E.; Lohuis, D.; Boiteux, L.S.; Inoue-Nagata, A.K.; Resende, R.O.; Kormelink, R. The Tomato spotted wilt virus cell-to-cell movement protein (NSM) triggers a hypersensitive response in Sw-5-containing resistant tomato lines and in Nicotiana benthamiana transformed with the functional Sw-5b resistance gene copy. Mol. Plant Pathol. 2014, 15, 871–880. [Google Scholar] [CrossRef]
- Peiro, A.; Canizares, M.C.; Rubio, L.; Lopez, C.; Moriones, E.; Aramburu, J.; Sanchez-Navarro, J. The movement protein (NSm) of Tomato spotted wilt virus is the avirulence determinant in the tomato Sw-5 gene-based resistance. Mol. Plant Pathol. 2014, 15, 802–813. [Google Scholar] [CrossRef]
- Zhao, W.Y.; Jiang, L.; Feng, Z.K.; Chen, X.J.; Huang, Y.; Xue, F.; Huang, C.J.; Liu, Y.; Li, F.; Liu, Y.T.; et al. Plasmodesmata targeting and intercellular trafficking of Tomato spotted wilt tospovirus movement protein NSm is independent of its function in HR induction. J. Gen. Virol. 2016, 97, 1990–1997. [Google Scholar] [CrossRef]
- Zhu, M.; Jiang, L.; Bai, B.H.; Zhao, W.Y.; Chen, X.J.; Li, J.; Liu, Y.; Chen, Z.Q.; Wang, B.T.; Wang, C.L.; et al. The Intracellular Immune Receptor Sw-5b confers broad-spectrum resistance to tospoviruses through recognition of a conserved 21-amino acid viral effector epitope. Plant Cell 2017, 29, 2214–2232. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Zhu, M.; Jiang, L.; Zhao, W.; Li, J.; Wu, J.; Li, C.; Bai, B.; Lu, G.; Chen, H.; et al. A multilayered regulatory mechanism for the autoinhibition and activation of a plant CC-NB-LRR resistance protein with an extra N-terminal domain. New Phytol. 2016, 212, 161–175. [Google Scholar] [CrossRef]
- Li, J.; Huang, H.; Zhu, M.; Huang, S.; Zhang, W.; Dinesh-Kumar, S.P.; Tao, X. A plant immune receptor adopts a two-step recognition mechanism to enhance viral effector perception. Mol. Plant 2019, 12, 248–262. [Google Scholar] [CrossRef] [Green Version]
- Azevedo, C.; Sadanandom, A.; Kitagawa, K.; Freialdenhoven, A.; Shirasu, K.; Schulze-Lefert, P. The RAR1 interactor SGT1, an essential component of R gene-triggered disease resistance. Science 2002, 295, 2073–2076. [Google Scholar] [CrossRef]
- Kadota, Y.; Shirasu, K.; Guerois, R. NLR sensors meet at the SGT1-HSP90 crossroad. Trends Biochem. Sci. 2010, 35, 199–207. [Google Scholar] [CrossRef]
- Liu, Z.Q.; Liu, Y.Y.; Shi, L.P.; Yang, S.; Shen, L.; Yu, H.X.; Wang, R.Z.; Wen, J.Y.; Tang, Q.; Hussain, A.; et al. SGT1 is required in PcINF1/SRC2-1 induced pepper defense response by interacting with SRC2-1. Sci. Rep. 2016, 6, 21651. [Google Scholar] [CrossRef]
- Su, H.; Chen, G.; Yang, L.; Zhang, Y.; Wang, Y.; Fang, Z.; Lv, H. Proteomic variations after short-term heat shock treatment reveal differentially expressed proteins involved in early microspore embryogenesis in cabbage (Brassica oleracea). PeerJ 2020, 8, e8897. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chapman, A.V.E.; Hunt, M.; Surana, P.; Velasquez-Zapata, V.; Xu, W.; Fuerst, G.; Wise, R.P. Disruption of barley immunity to powdery mildew by an in-frame Lys-Leu deletion in the essential protein SGT1. Genetics 2021, 217, iyaa026. [Google Scholar] [CrossRef]
- Azevedo, C.; Betsuyaku, S.; Peart, J.; Takahashi, A.; Noel, L.; Sadanandom, A.; Casais, C.; Parker, J.; Shirasu, K. Role of SGT1 in resistance protein accumulation in plant immunity. EMBO J. 2006, 25, 2007–2016. [Google Scholar] [CrossRef] [Green Version]
- Hoser, R.; Zurczak, M.; Lichocka, M.; Zuzga, S.; Dadlez, M.; Samuel, M.A.; Ellis, B.E.; Stuttmann, J.; Parker, J.E.; Hennig, J.; et al. Nucleocytoplasmic partitioning of tobacco N receptor is modulated by SGT1. New Phytol. 2013, 200, 158–171. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.H.; Abd-El-Haliem, A.; Bozkurt, T.O.; Belhaj, K.; Terauchi, R.; Vossen, J.H.; Kamoun, S. NLR network mediates immunity to diverse plant pathogens. Proc. Natl. Acad. Sci. USA 2017, 114, 8113–8118. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chandra-Shekara, A.C.; Navarre, D.; Kachroo, A.; Kang, H.G.; Klessig, D.; Kachroo, P. Signaling requirements and role of salicylic acid in HRT- and rrt-mediated resistance to turnip crinkle virus in Arabidopsis. Plant J. 2004, 40, 647–659. [Google Scholar] [CrossRef] [PubMed]
- Xiao, S.; Calis, O.; Patrick, E.; Zhang, G.; Charoenwattana, P.; Muskett, P.; Parker, J.E.; Turner, J.G. The atypical resistance gene, RPW8, recruits components of basal defence for powdery mildew resistance in Arabidopsis. Plant J. 2005, 42, 95–110. [Google Scholar] [CrossRef] [PubMed]
- Day, B.; Dahlbeck, D.; Staskawicz, B.J. NDR1 interaction with RIN4 mediates the differential activation of multiple disease resistance pathways in Arabidopsis. Plant Cell 2006, 18, 2782–2791. [Google Scholar] [CrossRef] [Green Version]
- Cao, H.; Glazebrook, J.; Clarke, J.D.; Volko, S.; Dong, X.N. The Arabidopsis NPR1 gene that controls systemic acquired resistance encodes a novel protein containing ankyrin repeats. Cell 1997, 88, 57–63. [Google Scholar] [CrossRef] [Green Version]
- Ryals, J.; Weymann, K.; Lawton, K.; Friedrich, L.; Ellis, D.; Steiner, H.Y.; Johnson, J.; Delaney, T.P.; Jesse, T.; Vos, P.; et al. The Arabidopsis NIM1 protein shows homology to the mammalian transcription factor inhibitor I kappa B. Plant Cell 1997, 9, 425–439. [Google Scholar] [CrossRef] [Green Version]
- Fu, Z.Q.; Yan, S.; Saleh, A.; Wang, W.; Ruble, J.; Oka, N.; Mohan, R.; Spoel, S.H.; Tada, Y.; Zheng, N.; et al. NPR3 and NPR4 are receptors for the immune signal salicylic acid in plants. Nature 2012, 486, 228–232. [Google Scholar] [CrossRef] [Green Version]
- Cao, H.; Bowling, S.A.; Gordon, A.S.; Dong, X.N. Characterization of an arabidopsis mutant that is nonresponsive to inducers of systemic acquired-resistance. Plant Cell 1994, 6, 1583–1592. [Google Scholar] [CrossRef]
- Liu, Y.; Schiff, M.; Marathe, R.; Dinesh-Kumar, S.P. Tobacco Rar1, EDS1 and NPR1/NIM1 like genes are required for N-mediated resistance to tobacco mosaic virus. Plant J. 2002, 30, 415–429. [Google Scholar] [CrossRef]
- Sun, Y.; Detchemendy, T.W.; Pajerowska-Mukhtar, K.M.; Mukhtar, M.S. NPR1 in JazzSet with pathogen effectors. Trends Plant Sci. 2018, 23, 469–472. [Google Scholar] [CrossRef] [PubMed]
- Janda, M.; Ruelland, E. Magical mystery tour: Salicylic acid signalling. Environ. Exp. Bot. 2015, 114, 117–128. [Google Scholar] [CrossRef]
- Kachroo, P.; Yoshioka, K.; Shah, J.; Dooner, H.K.; Klessig, D.F. Resistance to turnip crinkle virus in Arabidopsis is regulated by two host genes and is salicylic acid dependent but NPR1, ethylene, and jasmonate independent. Plant Cell 2000, 12, 677–690. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, C.H.; Belhaj, K.; Bozkurt, T.O.; Birk, M.S.; Kamoun, S. Helper NLR proteins NRC2a/b and NRC3 but not NRC1 are required for Pto-mediated cell death and resistance in Nicotiana benthamiana. New Phytol. 2016, 209, 1344–1352. [Google Scholar] [CrossRef] [Green Version]
- Jubic, L.M.; Saile, S.; Furzer, O.J.; El Kasmi, F.; Dangl, J.L. Help wanted: Helper NLRs and plant immune responses. Curr. Opin. Plant Biol. 2019, 50, 82–94. [Google Scholar] [CrossRef]
- Van Wersch, S.; Tian, L.; Hoy, R.; Li, X. Plant NLRs: The Whistleblowers of Plant Immunity. Plant Commun. 2020, 1, 100016. [Google Scholar]
- Gabriels, S.H.; Takken, F.L.; Vossen, J.H.; de Jong, C.F.; Liu, Q.; Turk, S.C.; Wachowski, L.K.; Peters, J.; Witsenboer, H.M.; de Wit, P.J.; et al. CDNA-AFLP combined with functional analysis reveals novel genes involved in the hypersensitive response. Mol. Plant Microbe Interact. 2006, 19, 567–576. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bonardi, V.; Tang, S.; Stallmann, A.; Roberts, M.; Cherkis, K.; Dangl, J.L. Expanded functions for a family of plant intracellular immune receptors beyond specific recognition of pathogen effectors. Proc. Natl. Acad. Sci. USA 2011, 108, 16463–16468. [Google Scholar] [CrossRef] [Green Version]
- Collier, S.M.; Hamel, L.P.; Moffett, P. Cell death mediated by the N-terminal domains of a unique and highly conserved class of NB-LRR protein. Mol. Plant Microbe Interact. 2011, 24, 918–931. [Google Scholar] [CrossRef] [Green Version]
- Wu, Z.; Li, M.; Dong, O.X.; Xia, S.; Liang, W.; Bao, Y.; Wasteneys, G.; Li, X. Differential regulation of TNL-mediated immune signaling by redundant helper CNLs. New Phytol. 2019, 222, 938–953. [Google Scholar] [CrossRef]
- Castel, B.; Ngou, P.M.; Cevik, V.; Redkar, A.; Kim, D.S.; Yang, Y.; Ding, P.; Jones, J.D.G. Diverse NLR immune receptors activate defence via the RPW8-NLR NRG1. New Phytol. 2019, 222, 966–980. [Google Scholar] [CrossRef] [PubMed]
- Peart, J.R.; Mestre, P.; Lu, R.; Malcuit, I.; Baulcombe, D.C. NRG1, a CC-NB-LRR protein, together with N, a TIR-NB-LRR protein, mediates resistance against tobacco mosaic virus. Curr. Biol. 2005, 15, 968–973. [Google Scholar] [CrossRef] [Green Version]
- Qi, T.; Seong, K.; Thomazella, D.P.T.; Kim, J.R.; Pham, J.; Seo, E.; Cho, M.J.; Schultink, A.; Staskawicz, B.J. NRG1 functions downstream of EDS1 to regulate TIR-NLR-mediated plant immunity in Nicotiana benthamiana. Proc. Natl. Acad. Sci. USA 2018, 115, E10979–E10987. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grech-Baran, M.; Witek, K.; Szajko, K.; Witek, A.I.; Morgiewicz, K.; Wasilewicz-Flis, I.; Jakuczun, H.; Marczewski, W.; Jones, J.D.G.; Hennig, J. Extreme resistance to Potato virus Y in potato carrying the Rysto gene is mediated by a TIR-NLR immune receptor. Plant Biotechnol. J. 2020, 18, 655–667. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qian, X.; Xiang, Q.; Yang, T.Q.; Ma, H.Y.; Ding, X.S.; Tao, X.R. Molecular co-chaperone SGT1 is critical for cell-to-cell movement and systemic infection of tomato spotted wilt virus in Nicotiana benthamiana. Viruses 2018, 10, 647. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, Z.Z.; Feng, Z.K.; Zhang, Z.J.; Liu, Y.B.; Tao, X.R. Complete genome sequence of a tomato spotted wilt virus isolate from China and comparison to other TSWV isolates of different geographic origin. Arch. Virol. 2011, 156, 1905–1908. [Google Scholar] [CrossRef]
- Shirasu, K. The HSP90-SGT1 chaperone complex for NLR immune sensors. Annu. Rev. Plant Biol. 2009, 60, 139–164. [Google Scholar] [CrossRef] [Green Version]
- Zhang, M.; Kadota, Y.; Prodromou, C.; Shirasu, K.; Pearl, L.H. Structural basis for assembly of Hsp90-Sgt1-CHORD protein complexes: Implications for chaperoning of NLR innate immunity receptors. Mol. Cell 2010, 39, 269–281. [Google Scholar] [CrossRef]
- Cui, H.; Tsuda, K.; Parker, J.E. Effector-triggered immunity: From pathogen perception to robust defense. Annu. Rev. Plant Biol. 2015, 66, 487–511. [Google Scholar] [CrossRef] [PubMed]
- Horsefield, S.; Burdett, H.; Zhang, X.; Manik, M.K.; Shi, Y.; Chen, J.; Qi, T.; Gilley, J.; Lai, J.S.; Rank, M.X.; et al. NAD(+) cleavage activity by animal and plant TIR domains in cell death pathways. Science 2019, 365, 793–799. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wan, L.; Essuman, K.; Anderson, R.G.; Sasaki, Y.; Monteiro, F.; Chung, E.H.; Osborne Nishimura, E.; DiAntonio, A.; Milbrandt, J.; Dangl, J.L.; et al. TIR domains of plant immune receptors are NAD(+)-cleaving enzymes that promote cell death. Science 2019, 365, 799–803. [Google Scholar] [CrossRef] [PubMed]
- Century, K.S.; Holub, E.B.; Staskawicz, B.J. Ndr1, a locus of Arabidopsis thaliana that is required for disease resistance to both a bacterial and a fungal pathogen. Proc. Natl. Acad. Sci. USA 1995, 92, 6597–6601. [Google Scholar] [CrossRef] [Green Version]
- Century, K.S.; Shapiro, A.D.; Repetti, P.P.; Dahlbeck, D.; Holub, E.; Staskawicz, B.J. NDR1, a pathogen-induced component required for Arabidopsis disease resistance. Science 1997, 278, 1963–1965. [Google Scholar] [CrossRef]
- Selote, D.; Shine, M.B.; Robin, G.P.; Kachroo, A. Soybean NDR1-like proteins bind pathogen effectors and regulate resistance signaling. New Phytol. 2014, 202, 485–498. [Google Scholar] [CrossRef]
- Saile, S.C.; Jacob, P.; Castel, B.; Jubic, L.M.; Salas-Gonzales, I.; Backer, M.; Jones, J.D.G.; Dangl, J.L.; El Kasmi, F. Two unequally redundant “helper” immune receptor families mediate Arabidopsis thaliana intracellular “sensor” immune receptor functions. PLoS Biol. 2020, 18, e3000783. [Google Scholar] [CrossRef]
- Scofield, S.R.; Huang, L.; Brandt, A.S.; Gill, B.S. Development of a virus-induced gene-silencing system for hexaploid wheat and its use in functional analysis of the Lr21-mediated leaf rust resistance pathway. Plant Physiol. 2005, 138, 2165–2173. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bhattarai, K.K.; Li, Q.; Liu, Y.; Dinesh-Kumar, S.P.; Kaloshian, I. The MI-1-mediated pest resistance requires Hsp90 and Sgt1. Plant Physiol. 2007, 144, 312–323. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kapos, P.; Devendrakumar, K.T.; Li, X. Plant NLRs: From discovery to application. Plant Sci. 2019, 279, 3–18. [Google Scholar] [PubMed]
- Bittner-Eddy, P.D.; Beynon, J.L. The Arabidopsis downy mildew resistance gene, RPP13-Nd, functions independently of NDR1 and EDS1 and does not require the accumulation of salicylic acid. Mol. Plant Microbe Interact. 2001, 14, 416–421. [Google Scholar] [CrossRef] [Green Version]
- Lewis, J.D.; Wu, R.; Guttman, D.S.; Desveaux, D. Allele-specific virulence attenuation of the Pseudomonas syringae HopZ1a type III effector via the Arabidopsis ZAR1 resistance protein. PLoS Genet. 2010, 6, e1000894. [Google Scholar] [CrossRef] [PubMed] [Green Version]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, Z.; Wu, Q.; Tong, C.; Chen, H.; Miao, D.; Qian, X.; Zhao, X.; Jiang, L.; Tao, X. Characterization of the Roles of SGT1/RAR1, EDS1/NDR1, NPR1, and NRC/ADR1/NRG1 in Sw-5b-Mediated Resistance to Tomato Spotted Wilt Virus. Viruses 2021, 13, 1447. https://doi.org/10.3390/v13081447
Chen Z, Wu Q, Tong C, Chen H, Miao D, Qian X, Zhao X, Jiang L, Tao X. Characterization of the Roles of SGT1/RAR1, EDS1/NDR1, NPR1, and NRC/ADR1/NRG1 in Sw-5b-Mediated Resistance to Tomato Spotted Wilt Virus. Viruses. 2021; 13(8):1447. https://doi.org/10.3390/v13081447
Chicago/Turabian StyleChen, Zhengqiang, Qian Wu, Cong Tong, Hongyu Chen, Dan Miao, Xin Qian, Xiaohui Zhao, Lei Jiang, and Xiaorong Tao. 2021. "Characterization of the Roles of SGT1/RAR1, EDS1/NDR1, NPR1, and NRC/ADR1/NRG1 in Sw-5b-Mediated Resistance to Tomato Spotted Wilt Virus" Viruses 13, no. 8: 1447. https://doi.org/10.3390/v13081447
APA StyleChen, Z., Wu, Q., Tong, C., Chen, H., Miao, D., Qian, X., Zhao, X., Jiang, L., & Tao, X. (2021). Characterization of the Roles of SGT1/RAR1, EDS1/NDR1, NPR1, and NRC/ADR1/NRG1 in Sw-5b-Mediated Resistance to Tomato Spotted Wilt Virus. Viruses, 13(8), 1447. https://doi.org/10.3390/v13081447





