HDV-Like Viruses
Abstract
:1. Introduction
2. Current Hypotheses of HDV Origin
3. Discovery of HDV-Like Viruses
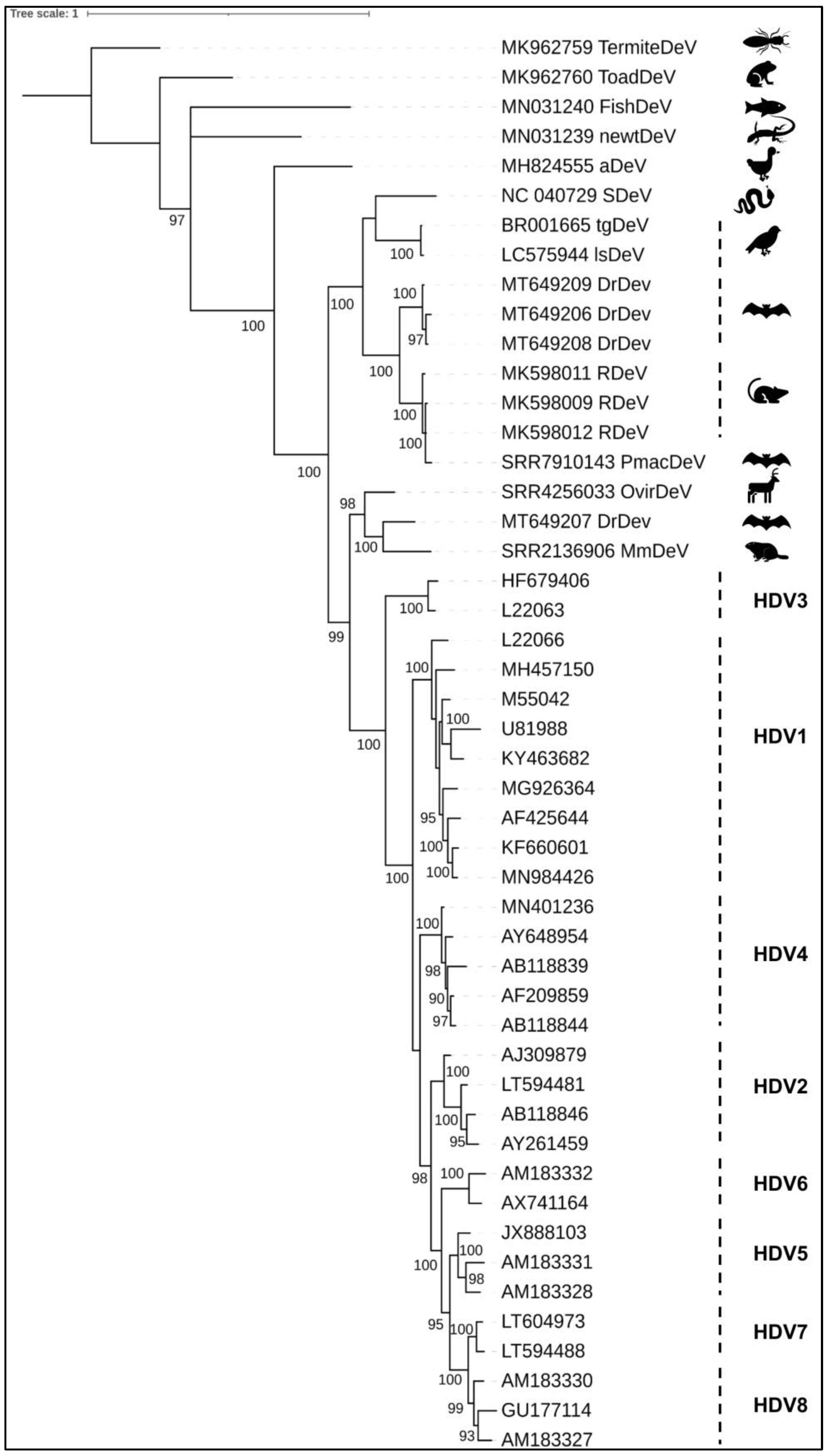
| Deltavirus | Host | CXXQ Motif | Putative Co-Infecting Viruses | Tested Helper Viruses (In Vitro) | HBsAg Usage (In Vitro) | Refs |
|---|---|---|---|---|---|---|
| HDV | Human | Yes | HBV | a HBV,b VSV, c HCV, d DENV, WNV, e LCMV, f HMPV | Yes | [21,30,31,32] |
| aDeV | ducks | no | Influenza A virus | nd | nd | [22] |
| RDeV | rodent P. semispinosus | no | Hepacivirus | nd | nd | [27] |
| SDeV | Boa constrictor | no | Reptarenavirus, hartmanivirus | g UHV-2, UGV-1 h HISV-1 e LCMV, JUNV f PUUV | no | [23,33] |
| tgDeV | Zebra finch Taeniopygia guttata | no | none | nd | no | [25] |
| mmDeV | Eastern woodchuck Marmota monax | no | WHV, herpesvirus, flavivirus, retrovirus | nd | no | [25,26] |
| DrDeV | Vampire bats D. rotundus | no | Herpesvirus, flavivirus, retrovirus | nd | nd | [26] |
| OvirDeV | White-tailed deer Odocoileus virginianus | no | Herpesvirus, flavivirus, retrovirus | nd | nd | [25,26] |
| PmacDeV | Lesser dog-like bat Peropteryx macrotis | no | Herpesvirus, flavivirus, retrovirus | nd | nd | [26] |
4. Phylogeny
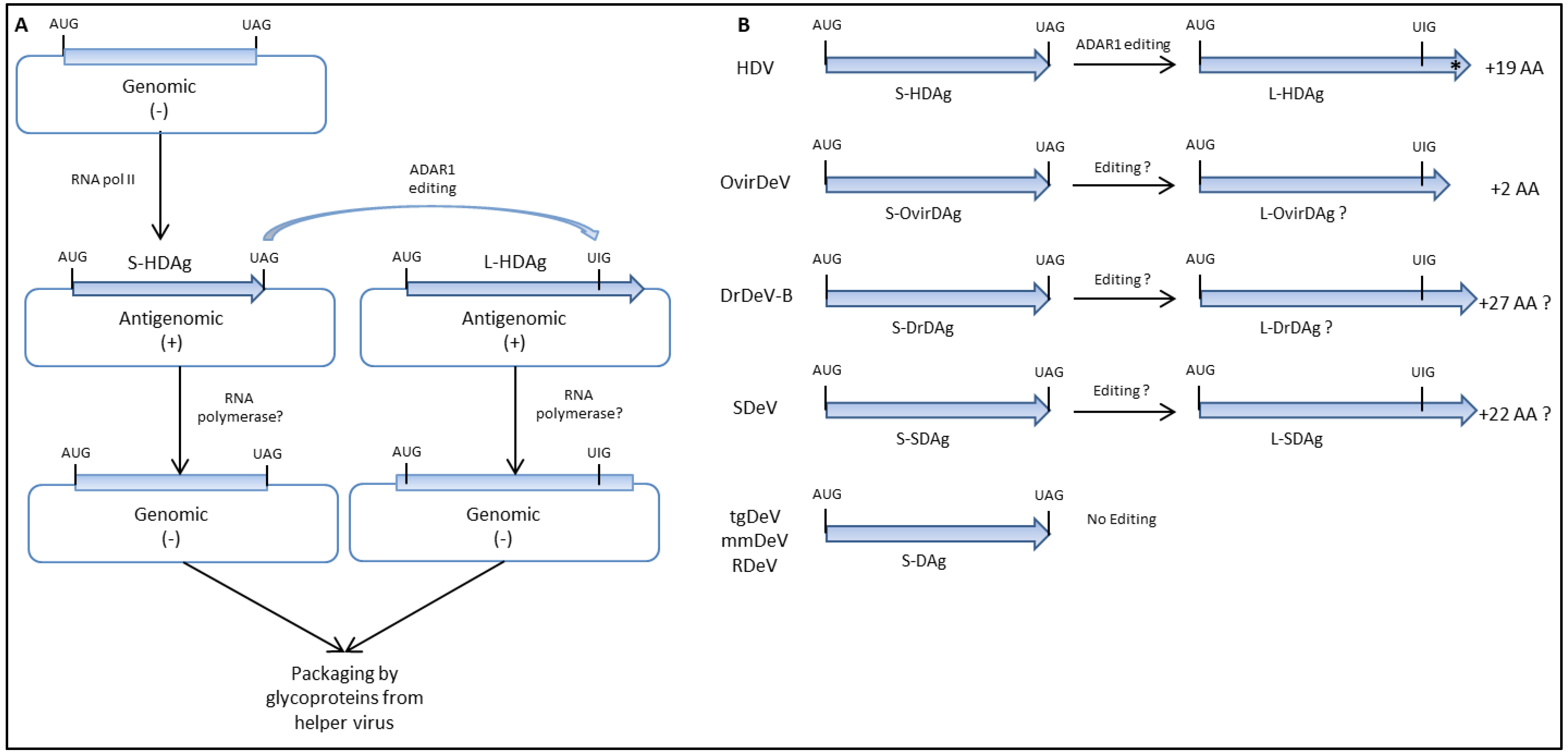
5. Virus Replication, Genome Editing, and Viral Proteins Expression
6. Helper Virus’s Functions
7. Candidate Helper Viruses
8. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Rizzetto, M.; Hoyer, B.; Canese, M.G.; Shih, J.W.; Purcell, R.H.; Gerin, J.L. Delta Agent: Association of delta antigen with hepatitis B surface antigen and RNA in serum of delta-infected chimpanzees. Proc. Natl. Acad. Sci. USA 1980, 77, 6124–6128. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, B.C.; Defenbaugh, D.A.; Casey, J.L. Multimerization of Hepatitis Delta Antigen Is a Critical Determinant of RNA Binding Specificity. J. Virol. 2010, 84, 1406–1413. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rizzetto, M.; Canese, M.G.; Arico, S.; Crivelli, O.; Trepo, C.; Bonino, F.; Verme, G. Immunofluorescence Detection of New Antigen-Antibody System (Delta/Anti-Delta) Associated to Hepatitis-B Virus in Liver and in Serum of Hbsag Carriers. Gut 1977, 18, 997–1003. [Google Scholar] [CrossRef] [Green Version]
- Rizzetto, M.; Canese, M.G.; Gerin, J.L.; London, W.T.; Sly, D.L.; Purcell, R.H. Transmission of the Hepatitis B Virus-Associated Delta Antigen to Chimpanzees. J. Infect. Dis. 1980, 141, 590–602. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.S.; Choo, Q.L.; Weiner, A.J.; Ou, J.H.; Najarian, R.C.; Thayer, R.M.; Mullenbach, G.T.; Denniston, K.J.; Gerin, J.L.; Houghton, M. Structure, Sequence and Expression of the Hepatitis Delta (Delta) Viral Genome. Nature 1986, 323, 508–514. [Google Scholar] [CrossRef]
- Lai, M.M.C. RNA Replication without RNA-Dependent RNA Polymerase: Surprises from Hepatitis Delta Virus. J. Virol. 2005, 79, 7951–7958. [Google Scholar] [CrossRef] [Green Version]
- Riccitelli, N.; Lupták, A. HDV Family of Self-Cleaving Ribozymes. Prog. Mol. Biol. Transl. Sci. 2013, 120, 123–171. [Google Scholar] [CrossRef] [Green Version]
- Memczak, S.; Jens, M.; Elefsinioti, A.; Torti, F.; Krueger, J.; Rybak-Wolf, A.; Maier, L.; Mackowiak, S.; Gregersen, L.H.; Munschauer, M.; et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013, 495, 333–338. [Google Scholar] [CrossRef]
- Casey, J.L.; Gerin, J.L. Hepatitis-D Virus-RNA Editing—Specific Modification of Adenosine in the Anti-genomic RNA. J. Virol. 1995, 69, 7593–7600. [Google Scholar] [CrossRef] [Green Version]
- Weiner, A.J.; Choo, Q.L.; Wang, K.S.; Govindarajan, S.; Redeker, A.G.; Gerin, J.L.; Houghton, M. A Single Antigenomic Open Reading Frame of the Hepatitis Delta Virus Encodes the Epitope(S) of Both Hepatitis Delta Antigen Polypeptides P24-Delta and P27-Delta. J. Virol. 1988, 62, 594–599. [Google Scholar] [CrossRef] [Green Version]
- Kuo, M.Y.P.; Chao, M.; Taylor, J. Initiation of Replication of the Human Hepatitis-Delta Virus Genome from Cloned DNA—Role of Delta-Antigen. J. Virol. 1989, 63, 1945–1950. [Google Scholar] [CrossRef] [Green Version]
- Modahl, L.E.; Lai, M.M.C. The Large Delta Antigen of Hepatitis Delta Virus Potently Inhibits Genomic but Not Antigenomic RNA Synthesis: A Mechanism Enabling Initiation of Viral Replication. J. Virol. 2000, 74, 7375–7380. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chao, M.; Hsieh, S.Y.; Taylor, J. Role of 2 Forms of Hepatitis Delta Virus-Antigen—Evidence for a Mechanism of Self-Limiting Genome Replication. J. Virol. 1990, 64, 5066–5069. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chang, F.L.; Chen, P.J.; Tu, S.J.; Wang, C.J.; Chen, D.-S. The large form of hepatitis delta antigen is crucial for assembly of hepatitis delta virus. Proc. Natl. Acad. Sci. USA 1991, 88, 8490–8494. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brazas, R.; Ganem, D. A Cellular Homolog of Hepatitis Delta Antigen: Implications for Viral Replication and Evolution. Science 1996, 274, 90–94. [Google Scholar] [CrossRef]
- Diener, T. The viroid: Biological oddity or evolutionary fossil? Adv. Virus Res. 2001, 57, 137–184. [Google Scholar] [CrossRef]
- Dinter-Gottlieb, G. The Unique Hepatitis Delta Virus; Springer: Berlin/Heidelberg, Germany, 1995. [Google Scholar]
- Taylor, J.M. Host RNA circles and the origin of hepatitis delta virus. World J. Gastroenterol. 2014, 20, 2971–2978. [Google Scholar] [CrossRef]
- Polo, J.M.; Jeng, K.S.; Lim, B.; Govindarajan, S.; Hofman, F.; Sangiorgi, F.; Lai, M.M.C. Transgenic Mice Support Replication of Hepatitis-Delta Virus-RNA in Multiple Tissues, Particularly in Skeletal-Muscle. J. Virol. 1995, 69, 4880–4887. [Google Scholar] [CrossRef] [Green Version]
- Giersch, K.; Helbig, M.; Volz, T.; Allweiss, L.; Mancke, L.V.; Lohse, A.W.; Polywka, S.; Pollok, J.M.; Petersen, J.; Taylor, J.; et al. Persistent hepatitis D virus mono-infection in humanized mice is efficiently converted by hepatitis B virus to a productive co-infection. J. Hepatol. 2014, 60, 538–544. [Google Scholar] [CrossRef]
- Perez-Vargas, J.; Amirache, F.; Boson, B.; Mialon, C.; Freitas, N.; Sureau, C.; Fusil, F.; Cosset, F.-L. Enveloped viruses distinct from HBV induce dissemination of hepatitis D virus in vivo. Nat. Commun. 2019, 10, 2098. [Google Scholar] [CrossRef] [Green Version]
- Wille, M.; Netter, H.J.; Littlejohn, M.; Yuen, L.; Shi, M.; Eden, J.-S.; Klaassen, M.; Holmes, E.C.; Hurt, A.C. A Divergent Hepatitis D-Like Agent in Birds. Viruses 2018, 10, 720. [Google Scholar] [CrossRef] [Green Version]
- Hetzel, U.; Szirovicza, L.; Smura, T.; Prähauser, B.; Vapalahti, O.; Kipar, A.; Hepojoki, J. Identification of a Novel Deltavirus in Boa Constrictors. mBio 2019, 10, e00014-19. [Google Scholar] [CrossRef] [Green Version]
- Chang, W.S.; Pettersson, J.H.O.; Le Lay, C.; Shi, M.; Lo, N.; Wille, M.; Eden, J.S.; Holmes, E.C. Novel hepatitis D-like agents in vertebrates and invertebrates. Virus Evol. 2019, 5, vez021. [Google Scholar] [CrossRef] [PubMed]
- Iwamoto, M.; Shibata, Y.; Kawasaki, J.; Kojima, S.; Li, Y.-T.; Iwami, S.; Muramatsu, M.; Wu, H.-L.; Wada, K.; Tomonaga, K.; et al. Identification of novel avian and mammalian deltaviruses provides new insights into deltavirus evolution. Virus Evol. 2021, 7, veab003. [Google Scholar] [CrossRef]
- Bergner, L.M.; Orton, R.J.; Broos, A.; Tello, C.; Becker, D.J.; Carrera, J.E.; Patel, A.H.; Biek, R.; Streicker, D.G. Diversification of mammalian deltaviruses by host shifting. Proc. Natl. Acad. Sci. USA 2021, 118, e2019907118. [Google Scholar] [CrossRef]
- Paraskevopoulou, S.; Pirzer, F.; Goldmann, N.; Schmid, J.; Corman, V.M.; Gottula, L.T.; Schroeder, S.; Rasche, A.; Muth, D.; Drexler, J.F.; et al. Mammalian deltavirus without hepadnavirus coinfection in the neotropical rodent Proechimys semispinosus. Proc. Natl. Acad. Sci. USA 2020, 117, 17977–17983. [Google Scholar] [CrossRef]
- Gudima, S.; Wu, S.-Y.; Chiang, C.-M.; Moraleda, G.; Taylor, J. Origin of Hepatitis Delta Virus mRNA. J. Virol. 2000, 74, 7204–7210. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De la Peña, M.; Ceprián, R.; Casey, J.L.; Cervera, A. Hepatitis delta virus-like circular RNAs from diverse metazoans encode conserved hammerhead ribozymes. Virus Evol. 2021, 7, veab016. [Google Scholar] [CrossRef]
- Barrera, A.; Guerra, B.; Lee, H.; Lanford, R.E. Analysis of Host Range Phenotypes of Primate Hepadnaviruses by In Vitro Infections of Hepatitis D Virus Pseudotypes. J. Virol. 2004, 78, 5233–5243. [Google Scholar] [CrossRef] [Green Version]
- Drexler, J.F.; Geipel, A.; König, A.; Corman, V.M.; van Riel, D.; Leijten, L.M.; Bremer, C.M.; Rasche, A.; Cottontail, V.M.; Maganga, G.D.; et al. Bats carry pathogenic hepadnaviruses antigenically related to hepatitis B virus and capable of infecting human hepatocytes. Proc. Natl. Acad. Sci. USA 2013, 110, 16151–16156. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gudima, S.; He, Y.; Chai, N.; Bruss, V.; Urban, S.; Mason, W.; Taylor, J. Primary Human Hepatocytes Are Susceptible to Infection by Hepatitis Delta Virus Assembled with Envelope Proteins of Woodchuck Hepatitis Virus. J. Virol. 2008, 82, 7276–7283. [Google Scholar] [CrossRef] [Green Version]
- Szirovicza, L.; Hetzel, U.; Kipar, A.; Martinez-Sobrido, L.; Vapalahti, O.; Hepojoki, J. Snake Deltavirus Utilizes Envelope Proteins of Different Viruses to Generate Infectious Particles. mBio 2020, 11, e03250-19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, H.-Y.; Shen, D.-T.; Ji, D.-Z.; Han, P.-C.; Zhang, W.-M.; Ma, J.-F.; Chen, W.-S.; Goyal, H.; Pan, S.; Xu, H.-G. Prevalence and burden of hepatitis D virus infection in the global population: A systematic review and meta-analysis. Gut 2019, 68, 512–521. [Google Scholar] [CrossRef]
- Dény, P. Hepatitis Delta Virus Genetic Variability: From Genotypes I, II, III to Eight Major Clades? Curr. Top. Microbiol. Immunol. 2006, 307, 151–171. [Google Scholar] [CrossRef]
- Huang, C.-R.; Lo, S.J. Evolution and Diversity of the Human Hepatitis D Virus Genome. Adv. Bioinform. 2010, 2010, 323654. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Löytynoja, A.; Goldman, N. From the Cover: An algorithm for progressive multiple alignment of sequences with insertions. Proc. Natl. Acad. Sci. USA 2005, 102, 10557–10562. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Löytynoja, A.; Goldman, N. Phylogeny-Aware Gap Placement Prevents Errors in Sequence Alignment and Evolutionary Analysis. Science 2008, 320, 1632–1635. [Google Scholar] [CrossRef] [Green Version]
- Lefort, V.; Longueville, J.-E.; Gascuel, O. SMS: Smart Model Selection in PhyML. Mol. Biol. Evol. 2017, 34, 2422–2424. [Google Scholar] [CrossRef] [Green Version]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL): An online tool for phylogenetic tree display and annotation. Bioinformatics 2007, 23, 127–128. [Google Scholar] [CrossRef] [Green Version]
- Griffin, B.L.; Chasovskikh, S.; Dritschilo, A.; Casey, J.L. Hepatitis Delta Antigen Requires a Flexible Quasi-Double-Stranded RNA Structure to Bind and Condense Hepatitis Delta Virus RNA in a Ribonucleoprotein Complex. J. Virol. 2014, 88, 7402–7411. [Google Scholar] [CrossRef] [Green Version]
- Chang, M.F.; Chen, C.H.; Lin, S.L.; Chen, C.J.; Chang, S.C. Functional Domains of Delta Antigens and Viral-RNA Required for RNA Packaging of Hepatitis-Delta-Virus. J. Virol. 1995, 69, 2508–2514. [Google Scholar] [CrossRef] [Green Version]
- Gudima, S.; Chang, J.; Moraleda, G.; Azvolinsky, A.; Taylor, J. Parameters of Human Hepatitis Delta Virus Genome Replication: The Quantity, Quality, and Intracellular Distribution of Viral Proteins and RNA. J. Virol. 2002, 76, 3709–3719. [Google Scholar] [CrossRef] [Green Version]
- Hsieh, S.Y.; Taylor, J. Regulation of Polyadenylation of Hepatitis Delta-Virus Antigenomic RNA. J. Virol. 1991, 65, 6438–6446. [Google Scholar] [CrossRef] [Green Version]
- Chen, P.J.; Kalpana, G.; Goldberg, J.; Mason, W.; Werner, B.; Gerin, J.; Taylor, J. Structure and Replication of the Genome of the Hepatitis Delta-Virus. Proc. Natl. Acad. Sci. USA 1986, 83, 8774–8778. [Google Scholar] [CrossRef] [Green Version]
- Flores, R.; Grubb, D.; Elleuch, A.; Nohales, M.A.; Delgado, S.; Gago, S. Rolling-circle replication of viroids, viroid-like satellite RNAs and hepatitis delta virus: Variations on a theme. RNA Biol. 2011, 8, 200–206. [Google Scholar] [CrossRef] [Green Version]
- Macnaughton, T.B.; Lai, M.M.C. Genomic but Not Antigenomic Hepatitis Delta Virus RNA Is Preferentially Exported from the Nucleus Immediately after Synthesis and Processing. J. Virol. 2002, 76, 3928–3935. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lazinski, D.W.; Taylor, J.M. Expression of Hepatitis-Delta Virus-RNA Deletions—Cis and Trans Requirements for Self-Cleavage, Ligation, and RNA Packaging. J. Virol. 1994, 68, 2879–2888. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Macnaughton, T.; Gowans, E.; Albert, A.; Burrell, C. Hepatitis delta virus RNA, protein synthesis and associated cytotoxicity in a stably transfected cell line. Virology 1990, 177, 692–698. [Google Scholar] [CrossRef]
- Wang, Y.-H.; Chang, S.C.; Huang, C.; Li, Y.-P.; Lee, C.-H.; Chang, M.-F. Novel Nuclear Export Signal-Interacting Protein, NESI, Critical for the Assembly of Hepatitis Delta Virus. J. Virol. 2005, 79, 8113–8120. [Google Scholar] [CrossRef] [Green Version]
- Glenn, J.S.; Watson, J.A.; Havel, C.M.; White, J.M. Identification of a prenylation site in delta virus large antigen. Science 1992, 256, 1331–1333. [Google Scholar] [CrossRef]
- Komla-Soukha, I.; Sureau, C. A tryptophan-rich motif in the carboxyl-terminus of the small envelope protein of hepatitis B virus is central to the assembly of hepatitis delta virus particles. J. Virol. 2006, 80, 4648–4655. [Google Scholar] [CrossRef] [Green Version]
- O’Malley, B.; Lazinski, D.W. Roles of Carboxyl-Terminal and Farnesylated Residues in the Functions of the Large Hepatitis Delta Antigen. J. Virol. 2005, 79, 1142–1153. [Google Scholar] [CrossRef] [Green Version]
- Zuccola, H.J.; Rozzelle, J.E.; Lemon, S.M.; Erickson, B.W.; Hogle, J.M. Structural basis of the oligomerization of hepatitis delta antigen. Structure 1998, 6, 821–830. [Google Scholar] [CrossRef] [Green Version]
- Yan, H.; Peng, B.; He, W.; Zhong, G.; Qi, Y.; Ren, B.; Gao, Z.; Jing, Z.; Song, M.; Xu, G.; et al. Molecular Determinants of Hepatitis B and D Virus Entry Restriction in Mouse Sodium Taurocholate Cotransporting Polypeptide. J. Virol. 2013, 87, 7977–7991. [Google Scholar] [CrossRef] [Green Version]
- Yang, X.; Cai, W.; Sun, X.; Bi, Y.; Zeng, C.; Zhao, X.; Zhou, Q.; Xu, T.; Xie, Q.; Sun, P.; et al. Defined host factors support HBV infection in non-hepatic 293T cells. J. Cell. Mol. Med. 2020, 24, 2507–2518. [Google Scholar] [CrossRef]
- Weller, M.L.; Gardener, M.R.; Bogus, Z.C.; Smith, M.A.; Astorri, E.; Michael, D.G.; Michael, D.A.; Zheng, C.; Burbelo, P.D.; Lai, Z.; et al. Hepatitis Delta Virus Detected in Salivary Glands of Sjogren’s Syndrome Patients and Induces a Sjogren’s Syndrome Phenotype In Vivo. Scand. J. Immunol. 2015, 81, 427. [Google Scholar]
- Seeger, C.; Mason, W.S. Molecular biology of hepatitis B virus infection. Virology 2015, 479, 672–686. [Google Scholar] [CrossRef] [Green Version]
- Freitas, N.; Cunha, C.; Menne, S.; Gudima, S.O. Envelope Proteins Derived from Naturally Integrated Hepatitis B Virus DNA Support Assembly and Release of Infectious Hepatitis Delta Virus Particles. J. Virol. 2014, 88, 5742–5754. [Google Scholar] [CrossRef] [Green Version]
- Bichko, V.; Netter, H.J.; Taylor, J. Introduction of Hepatitis-Delta Virus into Animal-Cell Lines via Cationic Liposomes. J. Virol. 1994, 68, 5247–5252. [Google Scholar] [CrossRef] [Green Version]
- Magnius, L.; Mason, W.S.; Taylor, J.; Kann, M.; Glebe, D.; Dény, P.; Sureau, C.; Norder, H.; ICTV Report Consortium. ICTV Virus Taxonomy Profile: Hepadnaviridae. J. Gen. Virol. 2020, 101, 571–572. [Google Scholar] [CrossRef]
- Hahn, C.M.; Iwanowicz, L.R.; Cornman, R.S.; Conway, C.M.; Winton, J.R.; Blazer, V.S. Characterization of a Novel Hepadnavirus in the White Sucker (Catostomus commersonii) from the Great Lakes Region of the United States. J. Virol. 2015, 89, 11801–11811. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dill, J.A.; Camus, A.C.; Leary, J.H.; Di Giallonardo, F.; Holmes, E.C.; Ng, T.F.F. Distinct Viral Lineages from Fish and Amphibians Reveal the Complex Evolutionary History of Hepadnaviruses. J. Virol. 2016, 90, 7920–7933. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lauber, C.; Seitz, S.; Mattei, S.; Suh, A.; Beck, J.; Herstein, J.; Börold, J.; Salzburger, W.; Kaderali, L.; Briggs, J.A.; et al. Deciphering the Origin and Evolution of Hepatitis B Viruses by Means of a Family of Non-enveloped Fish Viruses. Cell Host Microbe 2017, 22, 387–399.e6. [Google Scholar] [CrossRef] [Green Version]
- Freitas, N.; Salisse, J.; Cunha, C.; Toshkov, I.; Menne, S.; Gudima, S.O. Hepatitis delta virus infects the cells of hepadnavirus-induced hepatocellular carcinoma in woodchucks. Hepatology 2012, 56, 76–85. [Google Scholar] [CrossRef] [Green Version]
- Flores, R.; Ruiz-Ruiz, S.; Serra, P. Viroids and Hepatitis Delta Virus. Semin. Liver Dis. 2012, 32, 201–210. [Google Scholar] [CrossRef]
- Flores, R.; Owens, R.A.; Taylor, J. Pathogenesis by subviral agents: Viroids and hepatitis delta virus. Curr. Opin. Virol. 2016, 17, 87–94. [Google Scholar] [CrossRef]
- Van Bogaert, N.; De Jonghe, K.; Van Damme, E.; Maes, M.; Smagghe, G. Quantitation and localization of pospiviroids in aphids. J. Virol. Methods 2015, 211, 51–54. [Google Scholar] [CrossRef]
- Cappy, P.; Lucas, Q.; Kankarafou, N.; Sureau, C.; Laperche, S. No Evidence of Hepatitis C Virus (HCV)-Assisted Hepatitis D Virus Propagation in a Large Cohort of HCV-Positive Blood Donors. J. Infect. Dis. 2021, 223, 1376–1380. [Google Scholar] [CrossRef]
- Chemin, I.; Pujol, F.H.; Scholtès, C.; Loureiro, C.L.; Amirache, F.; Levrero, M.; Zoulim, F.; Pérez-Vargas, J.; Cosset, F. Preliminary Evidence for Hepatitis Delta Virus Exposure in Patients Who Are Apparently Not Infected with Hepatitis B Virus. Hepatology 2021, 73, 861–864. [Google Scholar] [CrossRef]
- Pfluger, L.S.; Schulze Zur Wiesch, J.; Polywka, S.; Lutgehetmann, M. Hepatitis delta virus propagation enabled by hepatitis C virus-Scientifically intriguing, but is it relevant to clinical practice? J. Viral Hepat. 2021, 28, 213–216. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pérez-Vargas, J.; Pereira de Oliveira, R.; Jacquet, S.; Pontier, D.; Cosset, F.-L.; Freitas, N. HDV-Like Viruses. Viruses 2021, 13, 1207. https://doi.org/10.3390/v13071207
Pérez-Vargas J, Pereira de Oliveira R, Jacquet S, Pontier D, Cosset F-L, Freitas N. HDV-Like Viruses. Viruses. 2021; 13(7):1207. https://doi.org/10.3390/v13071207
Chicago/Turabian StylePérez-Vargas, Jimena, Rémi Pereira de Oliveira, Stéphanie Jacquet, Dominique Pontier, François-Loïc Cosset, and Natalia Freitas. 2021. "HDV-Like Viruses" Viruses 13, no. 7: 1207. https://doi.org/10.3390/v13071207
APA StylePérez-Vargas, J., Pereira de Oliveira, R., Jacquet, S., Pontier, D., Cosset, F.-L., & Freitas, N. (2021). HDV-Like Viruses. Viruses, 13(7), 1207. https://doi.org/10.3390/v13071207







