Roles of the Endogenous Lunapark Protein during Flavivirus Replication
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Viruses
2.3. Preparation of Gene Constructs
2.4. Establishment of Cell Line Expressing RNA WNVKUN or LGTV Replicons
2.5. Transfection
2.6. Virus Infections and Cell Harvesting
2.7. Antibodies
2.8. Plaque Assays
2.9. Quantitative Real-Time PCR (qPCR)
2.10. Luciferase Assay
2.11. Protein Electrophoresis and Immunoblotting
2.12. Immunoprecipitations (IP)
2.13. Transmission Electron Microscopy (TEM)
2.14. Immunofluorescence Labeling
2.15. Statistics
3. Results
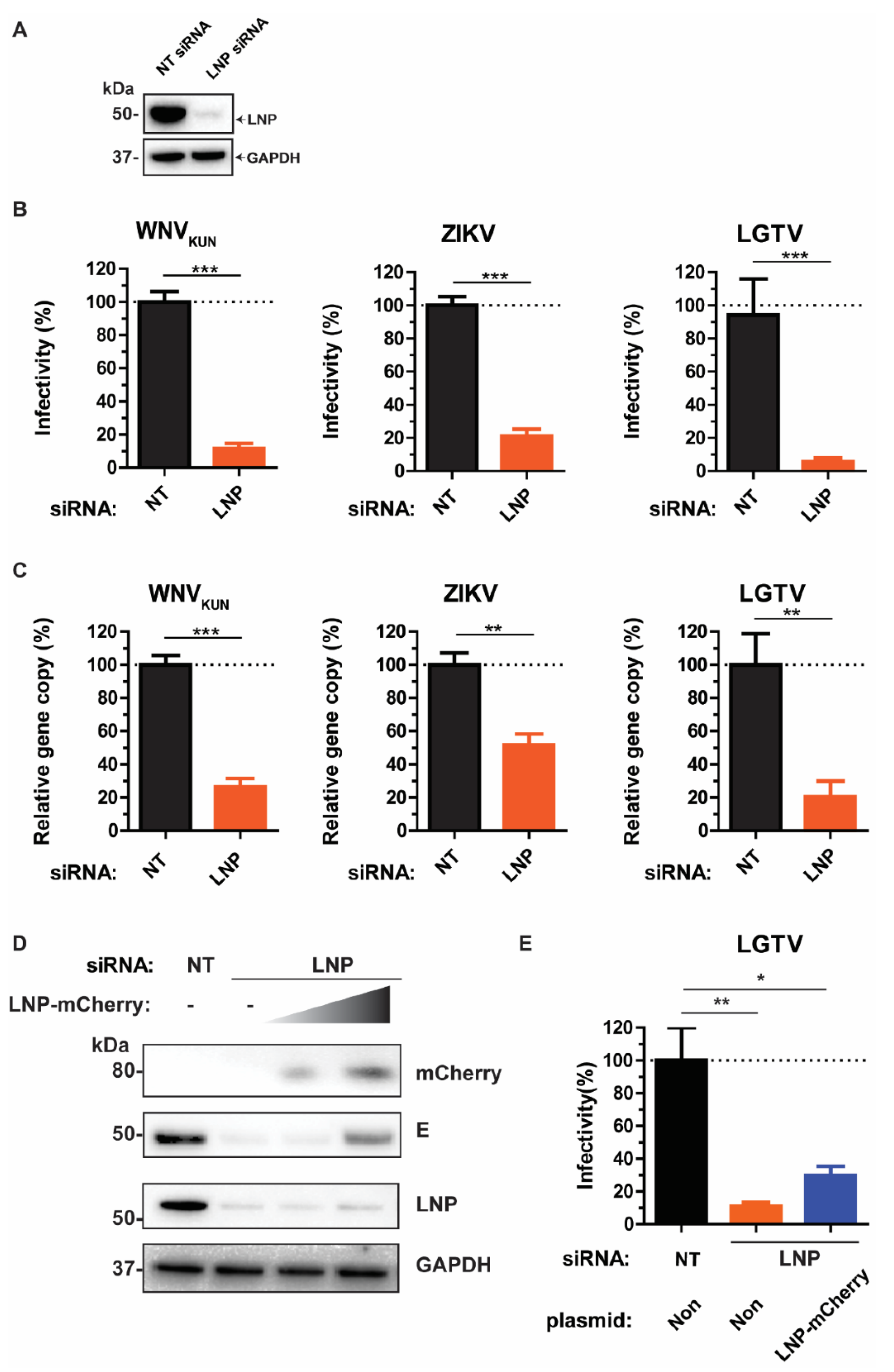
3.1. LNP Is Essential for WNVKUN, ZIKV, and LGTV Production
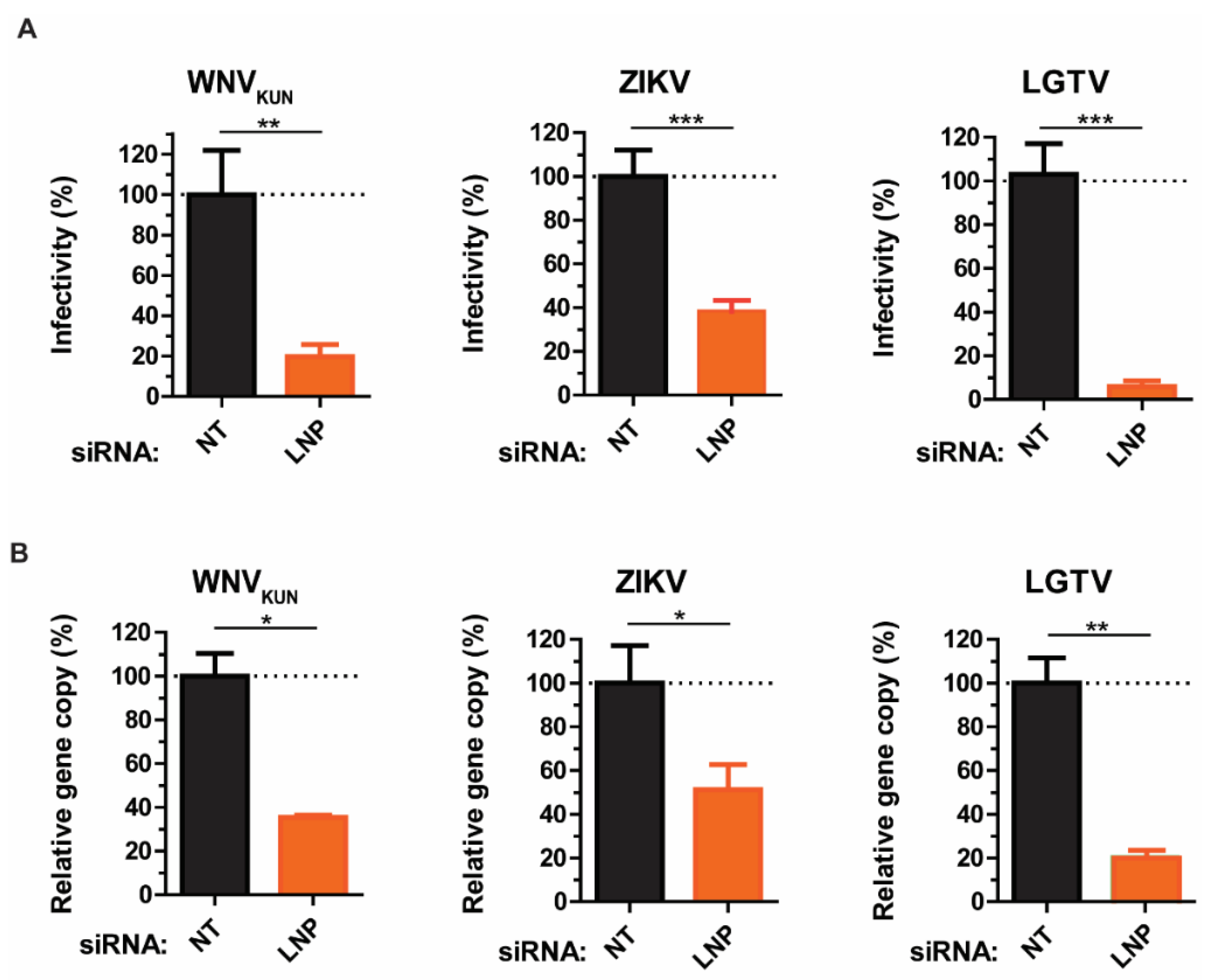
3.2. Requirement of LNP for Intracellular WNVKUN, ZIKV, and LGTV
3.3. LNP Has a Role in WNVKUN and LGTV Replication
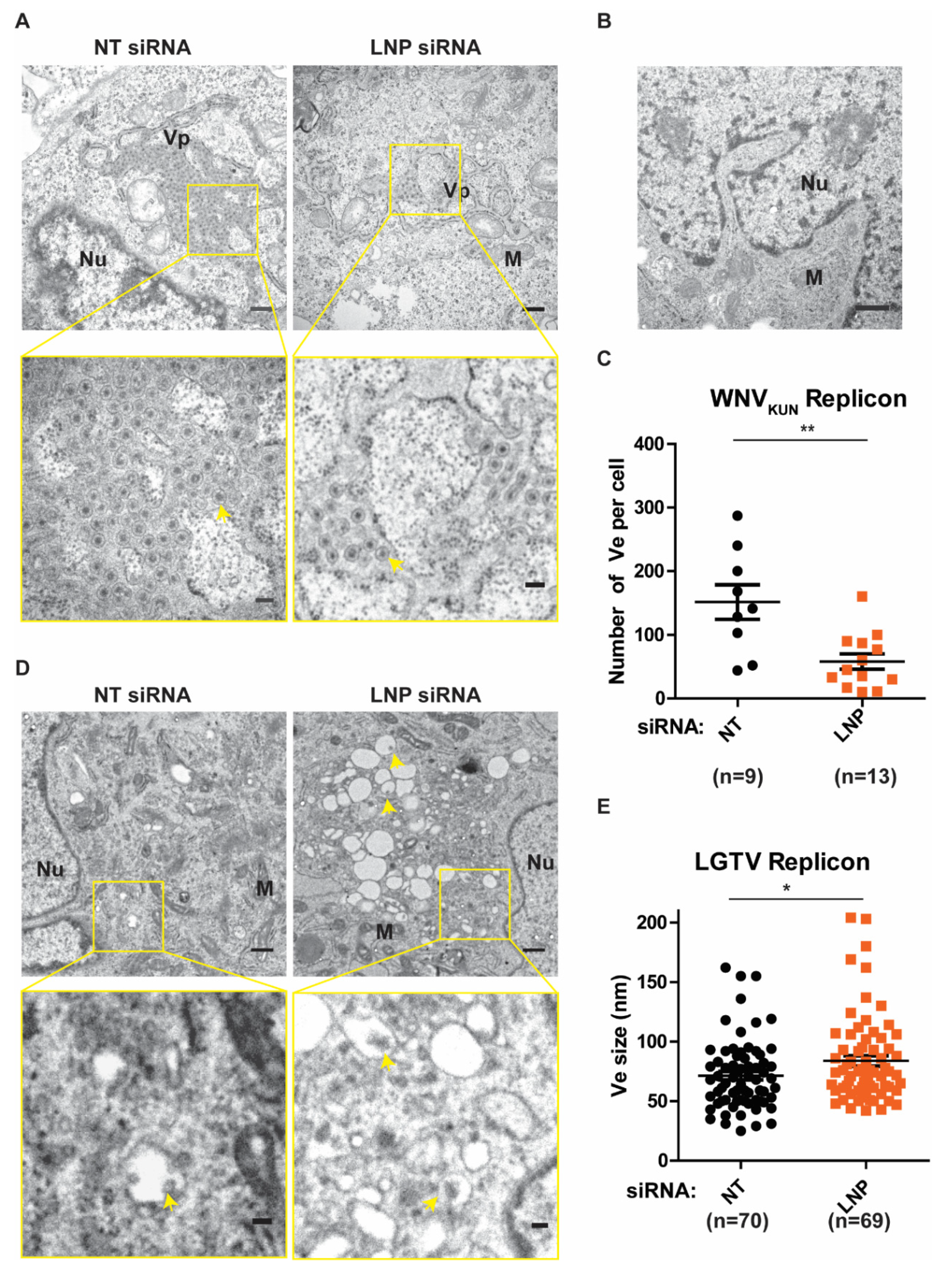
3.4. Function of LNP in Regulating the Number and the Size of Replication Vesicles (Ve)
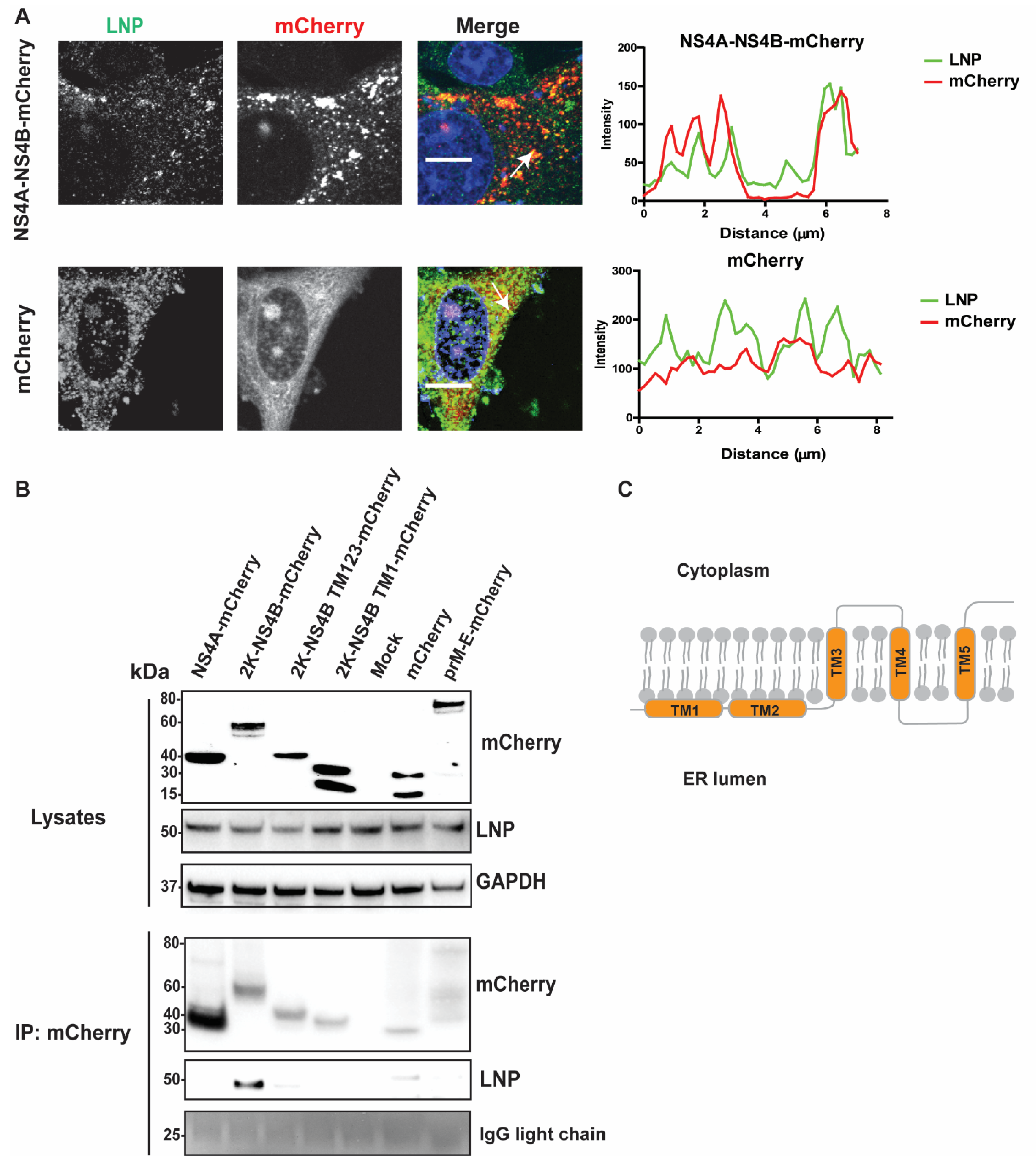
3.5. LNP Interacts with NS4B
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Calisher, C.H.; Karabatsos, N.; Dalrymple, J.M.; Shope, R.E.; Porterfield, J.S.; Westaway, E.G.; Brandt, W.E. Antigenic Relationships between Flaviviruses as Determined by Cross-neutralization Tests with Polyclonal Antisera. J. Gen. Virol. 1989, 70, 37–43. [Google Scholar] [CrossRef]
- Krauer, F.; Riesen, M.; Reveiz, L.; Oladapo, O.T.; Martínez-Vega, R.; Porgo, T.V.; Haefliger, A.; Broutet, N.J.; Low, N.; WHO Zika Causality Working Group. Zika virus infection as a cause of congenital brain abnormalities and Guillain–Barré syndrome: Systematic review. PLoS Med. 2017, 14, e1002203. [Google Scholar] [CrossRef] [Green Version]
- Gritsun, T.S.; Lashkevich, V.A.; Gould, E.A. Tick-borne encephalitis. Antivir. Res. 2003, 57, 129–146. [Google Scholar] [CrossRef]
- Mandl, C.W. Steps of the tick-borne encephalitis virus replication cycle that affect neuropathogenesis. Virus Res. 2005, 111, 161–174. [Google Scholar] [CrossRef]
- Apte-Sengupta, S.; Sirohi, D.; Kuhn, R.J. Coupling of replication and assembly in flaviviruses. Curr. Opin. Virol. 2014, 9, 134–142. [Google Scholar] [CrossRef] [Green Version]
- Mackenzie, J.M.; Jones, M.K.; Westaway, E.G. Markers for trans-Golgi membranes and the intermediate compartment localize to induced membranes with distinct replication functions in flavivirus-infected cells. J. Virol. 1999, 73, 9555–9567. [Google Scholar] [CrossRef] [Green Version]
- Westaway, E.G.; Mackenzie, J.M.; Kenney, M.T.; Jones, M.K.; Khromykh, A.A. Ultrastructure of Kunjin virus-infected cells: Colocalization of NS1 and NS3 with double-stranded RNA, and of NS2B with NS3, in virus-induced membrane structures. J. Virol. 1997, 71, 6650–6661. [Google Scholar] [CrossRef] [Green Version]
- Roosendaal, J.; Westaway, E.G.; Khromykh, A.; Mackenzie, J.M. Regulated cleavages at the West Nile virus NS4A-2K-NS4B junctions play a major role in rearranging cytoplasmic membranes and Golgi trafficking of the NS4A protein. J. Virol. 2006, 80, 4623–4632. [Google Scholar] [CrossRef] [Green Version]
- Kaufusi, P.H.; Kelley, J.F.; Yanagihara, R.; Nerurkar, V.R. Induction of endoplasmic reticulum-derived replication-competent membrane structures by West Nile virus non-structural protein 4B. PLoS ONE 2014, 9, e84040. [Google Scholar] [CrossRef] [Green Version]
- Miller, S.; Kastner, S.; Krijnse-Locker, J.; Bühler, S.; Bartenschlager, R. The non-structural protein 4A of dengue virus is an integral membrane protein inducing membrane alterations in a 2K-regulated manner. J. Biol. Chem. 2007, 282, 8873–8882. [Google Scholar] [CrossRef] [Green Version]
- English, A.R.; Voeltz, G.K. Endoplasmic reticulum structure and interconnections with other organelles. Cold Spring Harb. Perspect. Biol. 2013, 5, a013227. [Google Scholar] [CrossRef] [Green Version]
- Shibata, Y.; Voss, C.; Rist, J.M.; Hu, J.; Rapoport, T.A.; Prinz, W.A.; Voeltz, G.K. The reticulon and DP1/Yop1p proteins form immobile oligomers in the tubular endoplasmic reticulum. J. Biol. Chem. 2008, 283, 18892–18904. [Google Scholar] [CrossRef] [Green Version]
- Hu, J.; Shibata, Y.; Voss, C.; Shemesh, T.; Li, Z.; Coughlin, M.; Kozlov, M.M.; Rapoport, T.A.; Prinz, W.A. Membrane Proteins of the Endoplasmic Reticulum Induce High-Curvature Tubules. Science 2008, 319, 1247. [Google Scholar] [CrossRef]
- Hu, J.; Shibata, Y.; Zhu, P.-P.; Voss, C.; Rismanchi, N.; Prinz, W.A.; Rapoport, T.A.; Blackstone, C. A Class of Dynamin-like GTPases Involved in the Generation of the Tubular ER Network. Cell 2009, 138, 549–561. [Google Scholar] [CrossRef] [Green Version]
- Anwar, K.; Klemm, R.W.; Condon, A.; Severin, K.N.; Zhang, M.; Ghirlando, R.; Hu, J.; Rapoport, T.A.; Prinz, W.A. The dynamin-like GTPase Sey1p mediates homotypic ER fusion in S. cerevisiae. J. Cell Biol. 2012, 197, 209–217. [Google Scholar] [CrossRef] [Green Version]
- Chen, S.; Novick, P.; Ferro-Novick, S. ER network formation requires a balance of the dynamin-like GTPase Sey1p and the Lunapark family member Lnp1p. Nat. Cell Biol. 2012, 14, 707–716. [Google Scholar] [CrossRef]
- Chen, S.; Desai, T.; McNew, J.A.; Gerard, P.; Novick, P.J.; Ferro-Novick, S. Lunapark stabilizes nascent three-way junctions in the endoplasmic reticulum. Proc. Natl. Acad. Sci. USA 2015, 112, 418–423. [Google Scholar] [CrossRef] [Green Version]
- Aktepe, T.E.; Liebscher, S.; Prier, J.E.; Simmons, C.P.; Mackenzie, J.M. The Host Protein Reticulon 3.1A Is Utilized by Flaviviruses to Facilitate Membrane Remodelling. Cell Rep. 2017, 21, 1639–1654. [Google Scholar] [CrossRef] [Green Version]
- Neufeldt, C.J.; Cortese, M.; Scaturro, P.; Cerikan, B.; Wideman, J.G.; Tabata, K.; Moraes, T.; Oleksiuk, O.; Pichlmair, A.; Bartenschlager, R. ER-shaping atlastin proteins act as central hubs to promote flavivirus replication and virion assembly. Nat. Microbiol. 2019, 4, 2416–2429. [Google Scholar] [CrossRef]
- Monel, B.; Rajah, M.M.; Hafirassou, M.L.; Sid Ahmed, S.; Burlaud-Gaillard, J.; Zhu, P.P.; Nevers, Q.; Buchrieser, J.; Porrot, F.; Meunier, C.; et al. Atlastin Endoplasmic Reticulum-Shaping Proteins Facilitate Zika Virus Replication. J. Virol. 2019, 93. [Google Scholar] [CrossRef]
- Smith, C.E. A virus resembling Russian spring-summer encephalitis virus from an ixodid tick in Malaya. Nature 1956, 178, 581–582. [Google Scholar] [CrossRef]
- Tran, P.T.; Asghar, N.; Höglund, U.; Larsson, O.; Haag, L.; Mirazimi, A.; Johansson, M.; Melik, W. Development of a Multivalent Kunjin Virus Reporter Virus-Like Particle System Inducing Seroconversion for Ebola and West Nile Virus Proteins in Mice. Microorganisms 2020, 8, 1890. [Google Scholar] [CrossRef]
- Asghar, N.; Lee, Y.P.; Nilsson, E.; Lindqvist, R.; Melik, W.; Kröger, A.; Överby, A.K.; Johansson, M. The role of the poly(A) tract in the replication and virulence of tick-borne encephalitis virus. Sci. Rep. 2016, 6, 39265. [Google Scholar] [CrossRef] [Green Version]
- Wang, S.; Tukachinsky, H.; Romano, F.B.; Rapoport, T.A. Cooperation of the ER-shaping proteins atlastin, lunapark, and reticulons to generate a tubular membrane network. Elife 2016, 5. [Google Scholar] [CrossRef] [PubMed]
- Bílý, T.; Palus, M.; Eyer, L.; Elsterová, J.; Vancová, M.; Růžek, D. Electron Tomography Analysis of Tick-Borne Encephalitis Virus Infection in Human Neurons. Sci. Rep. 2015, 5, 10745. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Offerdahl, D.K.; Dorward, D.W.; Hansen, B.T.; Bloom, M.E. A Three-Dimensional Comparison of Tick-Borne Flavivirus Infection in Mammalian and Tick Cell Lines. PLoS ONE 2012, 7, e47912. [Google Scholar] [CrossRef] [PubMed]
- Miorin, L.; Romero-Brey, I.; Maiuri, P.; Hoppe, S.; Krijnse-Locker, J.; Bartenschlager, R.; Marcello, A. Three-Dimensional Architecture of Tick-Borne Encephalitis Virus Replication Sites and Trafficking of the Replicated RNA. J. Virol. 2013, 87, 6469–6481. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yau, W.-L.; Nguyen-Dinh, V.; Larsson, E.; Lindqvist, R.; Överby, A.K.; Lundmark, R. Model System for the Formation of Tick-Borne Encephalitis Virus Replication Compartments without Viral RNA Replication. J. Virol. 2019, 93, e00292-19. [Google Scholar] [CrossRef] [Green Version]
- Rumyantsev, A.A.; Murphy, B.R.; Pletnev, A.G. A tick-borne Langat virus mutant that is temperature sensitive and host range restricted in neuroblastoma cells and lacks neuroinvasiveness for immunodeficient mice. J. Virol. 2006, 80, 1427–1439. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miller, S.; Sparacio, S.; Bartenschlager, R. Subcellular localization and membrane topology of the Dengue virus type 2 Non-structural protein 4B. J. Biol. Chem. 2006, 281, 8854–8863. [Google Scholar] [CrossRef] [Green Version]
- Markoff, L.; Chang, A.; Falgout, B. Processing of flavivirus structural glycoproteins: Stable membrane insertion of premembrane requires the envelope signal peptide. Virology 1994, 204, 526–540. [Google Scholar] [CrossRef]
- Chen, S.; Wu, Z.; Wang, M.; Cheng, A. Innate Immune Evasion Mediated by Flaviviridae Non-Structural Proteins. Viruses 2017, 9, 291. [Google Scholar] [CrossRef]
- Suthar, M.S.; Aguirre, S.; Fernandez-Sesma, A. Innate immune sensing of flaviviruses. PLoS Pathog. 2013, 9, e1003541. [Google Scholar] [CrossRef] [PubMed]
- Khromykh, A.A.; Westaway, E.G. Subgenomic replicons of the flavivirus Kunjin: Construction and applications. J. Virol. 1997, 71, 1497–1505. [Google Scholar] [CrossRef] [Green Version]
- Zhang, R.; Miner, J.J.; Gorman, M.J.; Rausch, K.; Ramage, H.; White, J.P.; Zuiani, A.; Zhang, P.; Fernandez, E.; Zhang, Q.; et al. A CRISPR screen defines a signal peptide processing pathway required by flaviviruses. Nature 2016, 535, 164–168. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zmurko, J.; Neyts, J.; Dallmeier, K. Flaviviral NS4b, chameleon and jack-in-the-box roles in viral replication and pathogenesis, and a molecular target for antiviral intervention. Rev. Med. Virol. 2015, 25, 205–223. [Google Scholar] [CrossRef] [Green Version]
- Lee, C.; Chen, L.B. Dynamic behavior of endoplasmic reticulum in living cells. Cell 1988, 54, 37–46. [Google Scholar] [CrossRef]
- Sun, J.; Movahed, N.; Zheng, H. LUNAPARK Is an E3 Ligase That Mediates Degradation of ROOT HAIR DEFECTIVE3 to Maintain a Tubular ER Network in Arabidopsis. Plant Cell 2020, 32, 2964. [Google Scholar] [CrossRef]
- Yuniati, L.; Lauriola, A.; Gerritsen, M.; Abreu, S.; Ni, E.; Tesoriero, C.; Onireti, J.O.; Low, T.Y.; Heck, A.J.R.; Vettori, A.; et al. Ubiquitylation of the ER-Shaping Protein Lunapark via the CRL3(KLHL12) Ubiquitin Ligase Complex. Cell Rep. 2020, 31, 107664. [Google Scholar] [CrossRef]
- Mackenzie, J.M.; Westaway, E.G. Assembly and maturation of the flavivirus Kunjin virus appear to occur in the rough endoplasmic reticulum and along the secretory pathway, respectively. J. Virol. 2001, 75, 10787–10799. [Google Scholar] [CrossRef] [Green Version]
- Voeltz, G.K.; Prinz, W.A.; Shibata, Y.; Rist, J.M.; Rapoport, T.A. A class of membrane proteins shaping the tubular endoplasmic reticulum. Cell 2006, 124, 573–586. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Craene, J.O.; Coleman, J.; Estrada de Martin, P.; Pypaert, M.; Anderson, S.; Yates, J.R., 3rd; Ferro-Novick, S.; Novick, P. Rtn1p is involved in structuring the cortical endoplasmic reticulum. Mol. Biol. Cell 2006, 17, 3009–3020. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tang, W.F.; Yang, S.Y.; Wu, B.W.; Jheng, J.R.; Chen, Y.L.; Shih, C.H.; Lin, K.H.; Lai, H.C.; Tang, P.; Horng, J.T. Reticulon 3 binds the 2C protein of enterovirus 71 and is required for viral replication. J. Biol. Chem. 2007, 282, 5888–5898. [Google Scholar] [CrossRef] [Green Version]
- Wu, M.-J.; Ke, P.-Y.; Hsu, J.T.A.; Yeh, C.-T.; Horng, J.-T. Reticulon 3 interacts with NS4B of the hepatitis C virus and negatively regulates viral replication by disrupting NS4B self-interaction. Cell. Microbiol. 2014, 16, 1603–1618. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Jäntti, J. Functional characterization of the trans-membrane domain interactions of the Sec61 protein translocation complex beta-subunit. BMC Cell Biol. 2009, 10, 76. [Google Scholar] [CrossRef] [Green Version]

| Assay | Virus | Sequence (5′–3′) |
|---|---|---|
| cDNA synthesis | WNVKUN | AATATGCTGTGTTGTTGTGG |
| ZIKV | GATCTTGGTGAATGTGAACG | |
| LGTV | CTCCCTGTGAGTTCATAATTGG | |
| qPCR assay | WNVKUN | CAGACCACGCCATGGCG |
| CTAGGGCCGCGTGGG | ||
| FAM-TCTGCGGAGAGTGCAGTCTGCGA-NFQ | ||
| ZIKV | CCGCTGCCCAACACAAG | |
| CCACTAACGTTCTTTTGCAGACAT | ||
| FAM-AGCCTACCTTGACAAGCAGTCAGACACTCAA-NFQ | ||
| LGTV | ACTGAACTGGAGAAGGAGGA | |
| CCACAGTCCCATGACGATAAG | ||
| FAM-TAGGCTTGATTGCCTCGGCCTTTC-NFQ |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tran, P.-T.-H.; Asghar, N.; Johansson, M.; Melik, W. Roles of the Endogenous Lunapark Protein during Flavivirus Replication. Viruses 2021, 13, 1198. https://doi.org/10.3390/v13071198
Tran P-T-H, Asghar N, Johansson M, Melik W. Roles of the Endogenous Lunapark Protein during Flavivirus Replication. Viruses. 2021; 13(7):1198. https://doi.org/10.3390/v13071198
Chicago/Turabian StyleTran, Pham-Tue-Hung, Naveed Asghar, Magnus Johansson, and Wessam Melik. 2021. "Roles of the Endogenous Lunapark Protein during Flavivirus Replication" Viruses 13, no. 7: 1198. https://doi.org/10.3390/v13071198
APA StyleTran, P.-T.-H., Asghar, N., Johansson, M., & Melik, W. (2021). Roles of the Endogenous Lunapark Protein during Flavivirus Replication. Viruses, 13(7), 1198. https://doi.org/10.3390/v13071198





