Circovirus in Blood of a Febrile Horse with Hepatitis
Abstract
1. Introduction
2. Materials and Methods
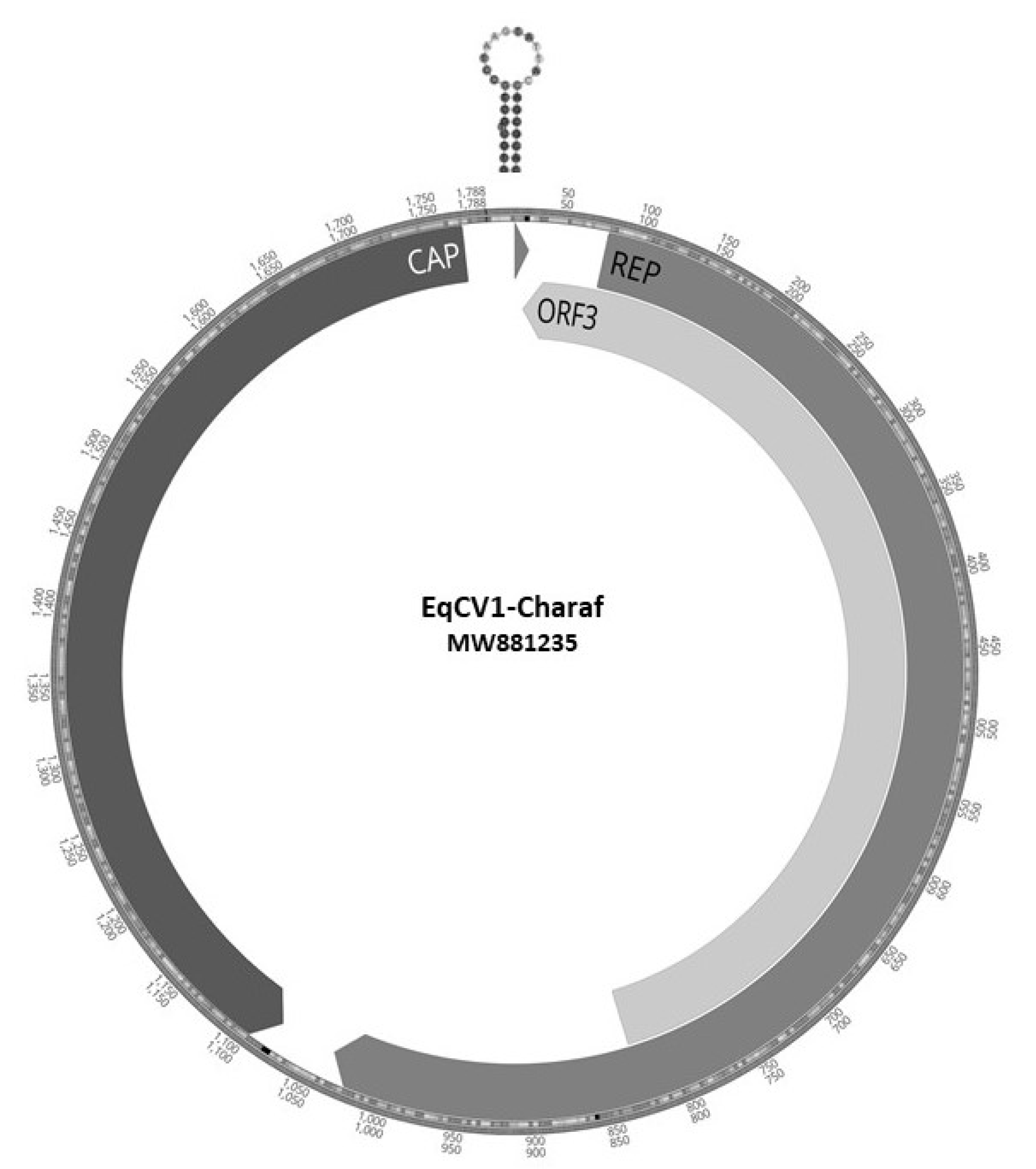
3. Results
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Breitbart, M.; Delwart, E.; Rosario, K.; Segales, J.; Varsani, A.; Ictv Report, C. Ictv virus taxonomy profile: Circoviridae. J. Gen. Virol. 2017, 98, 1997–1998. [Google Scholar] [CrossRef] [PubMed]
- Alex, C.E.; Fahsbender, E.; Altan, E.; Bildfell, R.; Wolff, P.; Jin, L.; Black, W.; Jackson, K.; Woods, L.; Munk, B.; et al. Viruses in unexplained encephalitis cases in american black bears (ursus americanus). PLoS ONE 2020, 15, e0244056. [Google Scholar] [CrossRef] [PubMed]
- Altan, E.; Kubiski, S.V.; Burchell, J.; Bicknese, E.; Deng, X.; Delwart, E. The first reptilian circovirus identified infects gut and liver tissues of black-headed pythons. Vet. Res. 2019, 50, 35. [Google Scholar] [CrossRef]
- Bexton, S.; Wiersma, L.C.; Getu, S.; van Run, P.R.; Verjans, G.M.; Schipper, D.; Schapendonk, C.M.; Bodewes, R.; Oldroyd, L.; Haagmans, B.L.; et al. Detection of circovirus in foxes with meningoencephalitis, united kingdom, 2009–2013. Emerg. Infect. Dis 2015, 21, 1205–1208. [Google Scholar] [CrossRef] [PubMed]
- Chang, W.S.; Li, C.X.; Hall, J.; Eden, J.S.; Hyndman, T.H.; Holmes, E.C.; Rose, K. Meta-transcriptomic discovery of a divergent circovirus and a chaphamaparvovirus in captive reptiles with proliferative respiratory syndrome. Viruses 2020, 12, 1073. [Google Scholar] [CrossRef]
- Chiappetta, C.M.; Cibulski, S.P.; Lima, F.E.S.; Varela, A.P.M.; Amorim, D.B.; Tavares, M.; Roehe, P.M. Molecular detection of circovirus and adenovirus in feces of fur seals (arctocephalus spp.). Ecohealth 2017, 14, 69–77. [Google Scholar] [CrossRef]
- Ellis, J. Porcine circovirus: A historical perspective. Vet. Pathol 2014, 51, 315–327. [Google Scholar] [CrossRef]
- Fisher, M.; Harrison, T.M.R.; Nebroski, M.; Kruczkiewicz, P.; Rothenburger, J.L.; Ambagala, A.; Macbeth, B.; Lung, O. Discovery and comparative genomic analysis of elk circovirus (elkcv), a novel circovirus species and the first reported from a cervid host. Sci. Rep. 2020, 10, 19548. [Google Scholar] [CrossRef]
- Ge, J.; Gu, S.; Cui, X.; Zhao, L.; Ma, D.; Shi, Y.; Wang, Y.; Lu, T.; Chen, H. Genomic characterization of circoviruses associated with acute gastroenteritis in minks in northeastern china. Arch. Virol 2018, 163, 2727–2735. [Google Scholar] [CrossRef]
- Li, L.; McGraw, S.; Zhu, K.; Leutenegger, C.M.; Marks, S.L.; Kubiski, S.; Gaffney, P.; Dela Cruz, F.N., Jr.; Wang, C.; Delwart, E.; et al. Circovirus in tissues of dogs with vasculitis and hemorrhage. Emerg. Infect. Dis 2013, 19, 534–541. [Google Scholar] [CrossRef]
- Lian, H.; Liu, Y.; Li, N.; Wang, Y.; Zhang, S.; Hu, R. Novel circovirus from mink, china. Emerg. Infect. Dis 2014, 20, 1548–1550. [Google Scholar] [CrossRef] [PubMed]
- Lőrincz, M.; Cságola, A.; Farkas, S.L.; Székely, C.; Tuboly, T. First detection and analysis of a fish circovirus. J. Gen. Virol. 2011, 92, 1817–1821. [Google Scholar] [CrossRef] [PubMed]
- Lorincz, M.; Dan, A.; Lang, M.; Csaba, G.; Toth, A.G.; Szekely, C.; Csagola, A.; Tuboly, T. Novel circovirus in european catfish (silurus glanis). Arch. Virol. 2012, 157, 1173–1176. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Nishizawa, T.; Sugimoto, Y.; Takeda, T.; Kodera, Y.; Hatano, Y.; Takahashi, M.; Okamoto, H. Identification and full-genome characterization of novel circoviruses in masked palm civets (paguma larvata). Virus Res. 2018, 258, 50–54. [Google Scholar] [CrossRef]
- Payne, N.; Kraberger, S.; Fontenele, R.S.; Schmidlin, K.; Bergeman, M.H.; Cassaigne, I.; Culver, M.; Varsani, A.; Van Doorslaer, K. Novel circoviruses detected in feces of sonoran felids. Viruses 2020, 12, 1027. [Google Scholar] [CrossRef] [PubMed]
- Tisza, M.J.; Pastrana, D.V.; Welch, N.L.; Stewart, B.; Peretti, A.; Starrett, G.J.; Pang, Y.S.; Krishnamurthy, S.R.; Pesavento, P.A.; McDermott, D.H.; et al. Discovery of several thousand highly diverse circular DNA viruses. Elife 2020, 9, e51971. [Google Scholar] [CrossRef]
- Todd, D. Avian circovirus diseases: Lessons for the study of pmws. Vet. Microbiol. 2004, 98, 169–174. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Lu, L.; Du, J.; Yang, L.; Ren, X.; Liu, B.; Jiang, J.; Yang, J.; Dong, J.; Sun, L.; et al. Comparative analysis of rodent and small mammal viromes to better understand the wildlife origin of emerging infectious diseases. Microbiome 2018, 6, 178. [Google Scholar] [CrossRef]
- Zhang, H.H.; Hu, W.Q.; Li, J.Y.; Liu, T.N.; Zhou, J.Y.; Opriessnig, T.; Xiao, C.T. Novel circovirus species identified in farmed pigs designated as porcine circovirus 4, hunan province, china. Transbound Emerg. Dis. 2020, 67, 1057–1061. [Google Scholar] [CrossRef]
- Zhang, W.; Yang, S.; Shan, T.; Hou, R.; Liu, Z.; Li, W.; Guo, L.; Wang, Y.; Chen, P.; Wang, X.; et al. Virome comparisons in wild-diseased and healthy captive giant pandas. Microbiome 2017, 5, 90. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Altan, E.; Reyes, G.; Halstead, B.; Deng, X.; Delwart, E. Virome of bat guano from nine northern california roosts. J. Virol. 2021, 95, e01713-20. [Google Scholar] [CrossRef]
- Wu, Z.; Ren, X.; Yang, L.; Hu, Y.; Yang, J.; He, G.; Zhang, J.; Dong, J.; Sun, L.; Du, J.; et al. Virome analysis for identification of novel mammalian viruses in bat species from chinese provinces. J. Virol. 2012, 86, 10999–11012. [Google Scholar] [CrossRef] [PubMed]
- Baker, K.S.; Leggett, R.M.; Bexfield, N.H.; Alston, M.; Daly, G.; Todd, S.; Tachedjian, M.; Holmes, C.E.; Crameri, S.; Wang, L.F.; et al. Metagenomic study of the viruses of african straw-coloured fruit bats: Detection of a chiropteran poxvirus and isolation of a novel adenovirus. Virology 2013, 441, 95–106. [Google Scholar] [CrossRef]
- Lima, F.E.; Cibulski, S.P.; Dall Bello, A.G.; Mayer, F.Q.; Witt, A.A.; Roehe, P.M.; d’Azevedo, P.A. A novel chiropteran circovirus genome recovered from a brazilian insectivorous bat species. Genome Announc. 2015, 3, e01393-15. [Google Scholar] [CrossRef] [PubMed]
- Hu, D.; Zhu, C.; Wang, Y.; Ai, L.; Yang, L.; Ye, F.; Ding, C.; Chen, J.; He, B.; Zhu, J.; et al. Virome analysis for identification of novel mammalian viruses in bats from southeast china. Sci. Rep. 2017, 7, 10917. [Google Scholar] [CrossRef]
- Zhu, A.; Jiang, T.; Hu, T.; Mi, S.; Zhao, Z.; Zhang, F.; Feng, J.; Fan, Q.; He, B.; Tu, C. Molecular characterization of a novel bat-associated circovirus with a poly-t tract in the 3’ intergenic region. Virus Res. 2018, 250, 95–103. [Google Scholar] [CrossRef] [PubMed]
- Matsumoto, T.; Sato, M.; Nishizono, A.; Ahmed, K. A novel bat-associated circovirus identified in northern hokkaido, japan. Arch. Virol. 2019, 164, 2179–2182. [Google Scholar] [CrossRef]
- Lecis, R.; Mucedda, M.; Pidinchedda, E.; Zobba, R.; Pittau, M.; Alberti, A. Genomic characterization of a novel bat-associated circovirus detected in european miniopterus schreibersii bats. Virus Genes 2020, 56, 325–328. [Google Scholar] [CrossRef]
- Li, L.; Deng, X.; Mee, E.T.; Collot-Teixeira, S.; Anderson, R.; Schepelmann, S.; Minor, P.D.; Delwart, E. Comparing viral metagenomics methods using a highly multiplexed human viral pathogens reagent. J. Virol. Methods 2015, 213, 139–146. [Google Scholar] [CrossRef]
- Altan, E.; Delaney, M.A.; Colegrove, K.M.; Spraker, T.R.; Wheeler, E.A.; Deng, X.; Li, Y.; Gulland, F.M.D.; Delwart, E. Complex virome in a mesenteric lymph node from a californian sea lion (zalophus californianus) with polyserositis and steatitis. Viruses 2020, 12, 793. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. Mafft multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol Evol 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Zuker, M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003, 31, 3406–3415. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. Mega x: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Altan, E.; Li, Y.; Sabino-Santos, G., Jr.; Sawaswong, V.; Barnum, S.; Pusterla, N.; Deng, X.; Delwart, E. Viruses in horses with neurologic and respiratory diseases. Viruses 2019, 11, 942. [Google Scholar] [CrossRef] [PubMed]
- Badenhorst, M.; de Heus, P.; Auer, A.; Tegtmeyer, B.; Stang, A.; Dimmel, K.; Tichy, A.; Kubacki, J.; Bachofen, C.; Steinmann, E.; et al. Active equine parvovirus-hepatitis infection is most frequently detected in austrian horses of advanced age. Equine Vet. J. 2021. [Google Scholar] [CrossRef]
- Divers, T.J.; Tennant, B.C.; Kumar, A.; McDonough, S.; Cullen, J.; Bhuva, N.; Jain, K.; Chauhan, L.S.; Scheel, T.K.H.; Lipkin, W.I.; et al. New parvovirus associated with serum hepatitis in horses after inoculation of common biological product. Emerg. Infect. Dis. 2018, 24, 303–310. [Google Scholar] [CrossRef] [PubMed]
- Tomlinson, J.E.; Jager, M.; Struzyna, A.; Laverack, M.; Fortier, L.A.; Dubovi, E.; Foil, L.D.; Burbelo, P.D.; Divers, T.J.; Van de Walle, G.R. Tropism, pathology, and transmission of equine parvovirus-hepatitis. Emerg. Microbes Infect. 2020, 9, 651–663. [Google Scholar] [CrossRef]
- Tomlinson, J.E.; Tennant, B.C.; Struzyna, A.; Mrad, D.; Browne, N.; Whelchel, D.; Johnson, P.J.; Jamieson, C.; Lohr, C.V.; Bildfell, R.; et al. Viral testing of 10 cases of theiler’s disease and 37 in-contact horses in the absence of equine biologic product administration: A prospective study (2014–2018). J. Vet. Intern. Med. 2019, 33, 258–265. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, X.; Cheng, A.; Wang, M.; Yin, Z.; Huang, J.; Jia, R. Apoptosis triggered by orf3 proteins of the circoviridae family. Front. Cell Infect. Microbiol. 2020, 10, 609071. [Google Scholar] [CrossRef]
- Rosario, K.; Duffy, S.; Breitbart, M. A field guide to eukaryotic circular single-stranded DNA viruses: Insights gained from metagenomics. Arch. Virol. 2012, 157, 1851–1871. [Google Scholar] [CrossRef]
- Le, S.Q.; Gascuel, O. An improved general amino acid replacement matrix. Mol. Biol. Evol. 2008, 25, 1307–1320. [Google Scholar] [CrossRef] [PubMed]
- Vargas-Bermúdez, D.S.; Vargas-Pinto, M.A.; Mogollón, J.D.; Jaime, J. Field infection of a gilt and its litter demonstrates vertical transmission and effect on reproductive failure caused by porcine circovirus type 3 (pcv3). BMC Vet. Res. 2021, 17, 150. [Google Scholar] [CrossRef] [PubMed]
- Klaumann, F.; Correa-Fiz, F.; Franzo, G.; Sibila, M.; Núñez, J.I.; Segalés, J. Current knowledge on porcine circovirus 3 (pcv-3): A novel virus with a yet unknown impact on the swine industry. Front. Vet. Sci 2018, 5, 315. [Google Scholar] [CrossRef] [PubMed]
- Opriessnig, T.; Karuppannan, A.K.; Castro, A.; Xiao, C.T. Porcine circoviruses: Current status, knowledge gaps and challenges. Virus Res. 2020, 286, 198044. [Google Scholar] [CrossRef]
- Zhai, S.L.; Lu, S.S.; Wei, W.K.; Lv, D.H.; Wen, X.H.; Zhai, Q.; Chen, Q.L.; Sun, Y.W.; Xi, Y. Reservoirs of porcine circoviruses: A mini review. Front. Vet. Sci 2019, 6, 319. [Google Scholar] [CrossRef]
- Klaumann, F.; Dias-Alves, A.; Cabezón, O.; Mentaberre, G.; Castillo-Contreras, R.; López-Béjar, M.; Casas-Díaz, E.; Sibila, M.; Correa-Fiz, F.; Segalés, J. Porcine circovirus 3 is highly prevalent in serum and tissues and may persistently infect wild boar (sus scrofa scrofa). Transbound Emerg. Dis 2019, 66, 91–101. [Google Scholar] [CrossRef]
- Franzo, G.; Tucciarone, C.M.; Drigo, M.; Cecchinato, M.; Martini, M.; Mondin, A.; Menandro, M.L. First report of wild boar susceptibility to porcine circovirus type 3: High prevalence in the colli euganei regional park (italy) in the absence of clinical signs. Transbound Emerg. Dis 2018, 65, 957–962. [Google Scholar] [CrossRef]
- Zhai, S.L.; He, D.S.; Qi, W.B.; Chen, S.N.; Deng, S.F.; Hu, J.; Li, X.P.; Li, L.; Chen, R.A.; Luo, M.L.; et al. Complete genome characterization and phylogenetic analysis of three distinct buffalo-origin pcv2 isolates from china. Infect. Genet. Evol. 2014, 28, 278–282. [Google Scholar] [CrossRef]
- Zhang, W.; Li, L.; Deng, X.; Kapusinszky, B.; Delwart, E. What is for dinner? Viral metagenomics of us store bought beef, pork, and chicken. Virology 2014, 468–470, 303–310. [Google Scholar] [CrossRef]
- Czyżewska-Dors, E.; Núñez, J.I.; Saporiti, V.; Huerta, E.; Riutord, C.; Cabezón, O.; Segalés, J.; Sibila, M. Detection of porcine circovirus 3 in wildlife species in spain. Pathogens 2020, 9, 341. [Google Scholar] [CrossRef]
- Franzo, G.; Grassi, L.; Tucciarone, C.M.; Drigo, M.; Martini, M.; Pasotto, D.; Mondin, A.; Menandro, M.L. A wild circulation: High presence of porcine circovirus 3 in different mammalian wild hosts and ticks. Transbound Emerg. Dis. 2019, 66, 1548–1557. [Google Scholar] [CrossRef] [PubMed]
- Song, T.; Hao, J.; Zhang, R.; Tang, M.; Li, W.; Hui, W.; Fu, Q.; Wang, C.; Xin, S.; Zhang, S.; et al. First detection and phylogenetic analysis of porcine circovirus type 2 in raccoon dogs. BMC Vet. Res. 2019, 15, 107. [Google Scholar] [CrossRef] [PubMed]
- Zhai, S.L.; Chen, S.N.; Liu, W.; Li, X.P.; Deng, S.F.; Wen, X.H.; Luo, M.L.; Lv, D.H.; Wei, W.K.; Chen, R.A. Molecular detection and genome characterization of porcine circovirus type 2 in rats captured on commercial swine farms. Arch. Virol. 2016, 161, 3237–3244. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Liu, Z.; Zou, Y.; Zhang, N.; Wang, D.; Tu, D.; Yang, L.; Deng, Z.; Yang, Y.; Jiang, P.; et al. First molecular detection of porcine circovirus type 3 in dogs in china. Virus Genes 2018, 54, 140–144. [Google Scholar] [CrossRef]
- Halami, M.Y.; Freick, M.; Shehata, A.A.; Müller, H.; Vahlenkamp, T.W. Susceptibility of calves to porcine circovirus-2 (pcv2). Vet. Microbiol. 2014, 173, 125–131. [Google Scholar] [CrossRef]
- Burbelo, P.D.; Ragheb, J.A.; Kapoor, A.; Zhang, Y. The serological evidence in humans supports a negligible risk of zoonotic infection from porcine circovirus type 2. Biologicals 2013, 41, 430–434. [Google Scholar] [CrossRef] [PubMed]
- Ellis, J.A.; Konoby, C.; West, K.H.; Allan, G.M.; Krakowka, S.; McNeilly, F.; Meehan, B.; Walker, I. Lack of antibodies to porcine circovirus type 2 virus in beef and dairy cattle and horses in western canada. Can. Vet. J. 2001, 42, 461–464. [Google Scholar]
- Vidigal, P.M.; Mafra, C.L.; Silva, F.M.; Fietto, J.L.; Silva Júnior, A.; Almeida, M.R. Tripping over emerging pathogens around the world: A phylogeographical approach for determining the epidemiology of porcine circovirus-2 (pcv-2), considering global trading. Virus Res. 2012, 163, 320–327. [Google Scholar] [CrossRef]
- Firth, C.; Charleston, M.A.; Duffy, S.; Shapiro, B.; Holmes, E.C. Insights into the evolutionary history of an emerging livestock pathogen: Porcine circovirus 2. J. Virol. 2009, 83, 12813–12821. [Google Scholar] [CrossRef]
- Zhai, S.L.; Chen, R.A.; Chen, S.N.; Wen, X.H.; Lv, D.H.; Wu, D.C.; Yuan, J.; Huang, Z.; Zhou, X.R.; Luo, M.L.; et al. First molecular detection of porcine circovirus type 2 in bovids in china. Virus Genes 2014, 49, 507–511. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hui, A.; Altan, E.; Slovis, N.; Fletcher, C.; Deng, X.; Delwart, E. Circovirus in Blood of a Febrile Horse with Hepatitis. Viruses 2021, 13, 944. https://doi.org/10.3390/v13050944
Hui A, Altan E, Slovis N, Fletcher C, Deng X, Delwart E. Circovirus in Blood of a Febrile Horse with Hepatitis. Viruses. 2021; 13(5):944. https://doi.org/10.3390/v13050944
Chicago/Turabian StyleHui, Alvin, Eda Altan, Nathan Slovis, Caitlin Fletcher, Xutao Deng, and Eric Delwart. 2021. "Circovirus in Blood of a Febrile Horse with Hepatitis" Viruses 13, no. 5: 944. https://doi.org/10.3390/v13050944
APA StyleHui, A., Altan, E., Slovis, N., Fletcher, C., Deng, X., & Delwart, E. (2021). Circovirus in Blood of a Febrile Horse with Hepatitis. Viruses, 13(5), 944. https://doi.org/10.3390/v13050944






