Pathogenic Role of Epstein–Barr Virus in Lung Cancers
Abstract
1. Lung Cancers
2. Epstein–Barr Virus
3. Presence of EBV in Lung Cancers
4. Etiological Role of EBV in LC
4.1. EBV Transcriptome
4.2. EBV-Associated Immunoevasion
4.3. EBV-Associated Alteration of Tumor Pathways
4.4. EBV-Associated Genomic Changes
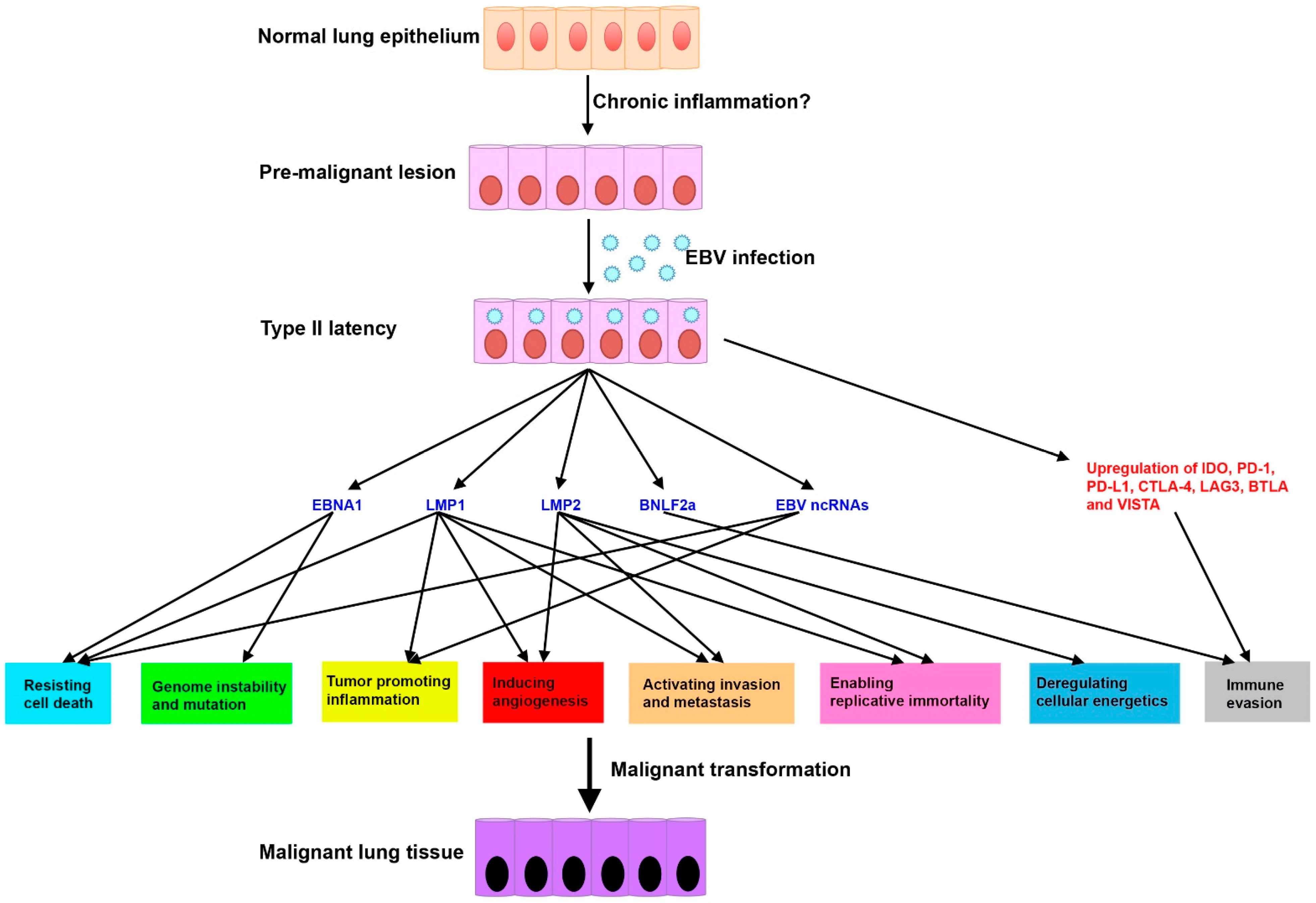
4.5. Proposed Disease Model of EBV-Associated LCs
5. Experimental Models for EBV(+) LC
6. Challenges and Future Directions
Author Contributions
Funding
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer statistics, 2021. CA Cancer J. Clin. 2021, 71, 7–33. [Google Scholar] [CrossRef] [PubMed]
- Travis, W.D.; Brambilla, E.; Nicholson, A.G.; Yatabe, Y.; Austin, J.H.M.; Beasley, M.B.; Chirieac, L.R.; Dacic, S.; Duhig, E.; Flieder, D.B.; et al. The 2015 world health organization classification of lung tumors: Impact of genetic, clinical and radiologic advances since the 2004 classification. J. Thorac. Oncol. 2015, 10, 1243–1260. [Google Scholar] [CrossRef]
- Zappa, C.; Mousa, S.A. Non-small cell lung cancer: Current treatment and future advances. Transl. Lung Cancer Res. 2016, 5, 288–300. [Google Scholar] [CrossRef]
- Thun, M.J.; Henley, S.J.; Calle, E.E. Tobacco use and cancer: An epidemiologic perspective for geneticists. Oncogene 2002, 21, 7307–7325. [Google Scholar] [CrossRef] [PubMed]
- Sun, S.; Schiller, J.H.; Gazdar, A.F. Lung cancer in never smokers—A different disease. Nat. Rev. Cancer 2007, 7, 778–790. [Google Scholar] [CrossRef]
- Cheng, T.Y.; Cramb, S.M.; Baade, P.D.; Youlden, D.R.; Nwogu, C.; Reid, M.E. The international epidemiology of lung cancer: Latest trends, disparities, and tumor characteristics. J. Thorac. Oncol. 2016, 11, 1653–1671. [Google Scholar] [CrossRef] [PubMed]
- Young, L.S.; Yap, L.F.; Murray, P.G. Epstein-barr virus: More than 50 years old and still providing surprises. Nat. Rev. Cancer 2016, 16, 789–802. [Google Scholar] [CrossRef] [PubMed]
- Rickinson, A.; Kieff, E. Epstein-barr virus. In Fields Virology, 5th ed.; Knipe, D., Howley, P., Eds.; Lippincott Williams & Wilkins: Philadelphia, PA, USA, 2007; pp. 2655–2700. [Google Scholar]
- Shannon-Lowe, C.; Rickinson, A. The global landscape of ebv-associated tumors. Front. Oncol. 2019, 9, 713. [Google Scholar] [CrossRef]
- de Martel, C.; Ferlay, J.; Franceschi, S.; Vignat, J.; Bray, F.; Forman, D.; Plummer, M. Global burden of cancers attributable to infections in 2008: A review and synthetic analysis. Lancet Oncol. 2012, 13, 607–615. [Google Scholar] [CrossRef]
- Khan, G.; Hashim, M.J. Global burden of deaths from epstein-barr virus attributable malignancies 1990–2010. Infect. Agent. Cancer 2014, 9, 38. [Google Scholar] [CrossRef]
- Kheir, F.; Zhao, M.; Strong, M.J.; Yu, Y.; Nanbo, A.; Flemington, E.K.; Morris, G.F.; Reiss, K.; Li, L.; Lin, Z. Detection of epstein-barr virus infection in non-small cell lung cancer. Cancers 2019, 11, 759. [Google Scholar] [CrossRef]
- De Sanjose, S.; Bosch, R.; Schouten, T.; Verkuijlen, S.; Nieters, A.; Foretova, L.; Maynadie, M.; Cocco, P.L.; Staines, A.; Becker, N.; et al. Epstein-barr virus infection and risk of lymphoma: Immunoblot analysis of antibody responses against ebv-related proteins in a large series of lymphoma subjects and matched controls. Int. J. Cancer 2007, 121, 1806–1812. [Google Scholar] [CrossRef]
- Strong, M.J.; O’Grady, T.; Lin, Z.; Xu, G.; Baddoo, M.; Parsons, C.; Zhang, K.; Taylor, C.M.; Flemington, E.K. Epstein-barr virus and human herpesvirus 6 detection in a non-hodgkin’s diffuse large b-cell lymphoma cohort by using rna sequencing. J. Virol. 2013, 87, 13059–13062. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Strong, M.J.; Xu, G.; Coco, J.; Baribault, C.; Vinay, D.S.; Lacey, M.R.; Strong, A.L.; Lehman, T.A.; Seddon, M.B.; Lin, Z.; et al. Differences in gastric carcinoma microenvironment stratify according to ebv infection intensity: Implications for possible immune adjuvant therapy. PLoS Pathog. 2013, 9, e1003341. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.; Nanbo, A.; Becnel, D.; Qin, Z.; Morris, G.F.; Li, L.; Lin, Z. Ubiquitin modification of the epstein-barr virus immediate early transactivator zta. J. Virol. 2020, 94. [Google Scholar] [CrossRef]
- Lin, Z.; Flemington, E. Regulation of ebv latency by viral lytic proteins. In Epstein-Barr Virus: Latency and Transformation; Robertson, E.S., Ed.; Caister Academic Press: London, UK, 2010; pp. 167–192. [Google Scholar]
- Tugizov, S.M.; Herrera, R.; Palefsky, J.M. Epstein-barr virus transcytosis through polarized oral epithelial cells. J. Virol. 2013, 87, 8179–8194. [Google Scholar] [CrossRef]
- Laichalk, L.L.; Thorley-Lawson, D.A. Terminal differentiation into plasma cells initiates the replicative cycle of epstein-barr virus in vivo. J. Virol. 2005, 79, 1296–1307. [Google Scholar] [CrossRef]
- Sun, C.C.; Thorley-Lawson, D.A. Plasma cell-specific transcription factor xbp-1s binds to and transactivates the epstein-barr virus bzlf1 promoter. J. Virol. 2007, 81, 13566–13577. [Google Scholar] [CrossRef]
- Wilson, J.B.; Manet, E.; Gruffat, H.; Busson, P.; Blondel, M.; Fahraeus, R. Ebna1: Oncogenic activity, immune evasion and biochemical functions provide targets for novel therapeutic strategies against epstein-barr virus- associated cancers. Cancers 2018, 10, 109. [Google Scholar] [CrossRef] [PubMed]
- Moss, W.N.; Steitz, J.A. Genome-wide analyses of epstein-barr virus reveal conserved rna structures and a novel stable intronic sequence rna. BMC Genom. 2013, 14, 543. [Google Scholar] [CrossRef]
- Hutzinger, R.; Feederle, R.; Mrazek, J.; Schiefermeier, N.; Balwierz, P.J.; Zavolan, M.; Polacek, N.; Delecluse, H.J.; Huttenhofer, A. Expression and processing of a small nucleolar rna from the epstein-barr virus genome. PLoS Pathog. 2009, 5, e1000547. [Google Scholar] [CrossRef] [PubMed]
- O’Grady, T.; Cao, S.; Strong, M.J.; Concha, M.; Wang, X.; Splinter Bondurant, S.; Adams, M.; Baddoo, M.; Srivastav, S.K.; Lin, Z.; et al. Global bidirectional transcription of the epstein-barr virus genome during reactivation. J. Virol. 2014, 88, 1604–1616. [Google Scholar] [CrossRef] [PubMed]
- O’Grady, T.; Wang, X.; Honer Zu Bentrup, K.; Baddoo, M.; Concha, M.; Flemington, E.K. Global transcript structure resolution of high gene density genomes through multi-platform data integration. Nucleic Acids Res. 2016, 44, e145. [Google Scholar] [CrossRef] [PubMed]
- Cao, S.; Strong, M.J.; Wang, X.; Moss, W.N.; Concha, M.; Lin, Z.; O’Grady, T.; Baddoo, M.; Fewell, C.; Renne, R.; et al. High-throughput rna sequencing-based virome analysis of 50 lymphoma cell lines from the cancer cell line encyclopedia project. J. Virol. 2015, 89, 713–729. [Google Scholar] [CrossRef]
- Concha, M.; Wang, X.; Cao, S.; Baddoo, M.; Fewell, C.; Lin, Z.; Hulme, W.; Hedges, D.; McBride, J.; Flemington, E.K. Identification of new viral genes and transcript isoforms during epstein-barr virus reactivation using rna-seq. J. Virol. 2012, 86, 1458–1467. [Google Scholar] [CrossRef]
- Pfeffer, S.; Zavolan, M.; Grasser, F.A.; Chien, M.; Russo, J.J.; Ju, J.; John, B.; Enright, A.J.; Marks, D.; Sander, C.; et al. Identification of virus-encoded micrornas. Science 2004, 304, 734–736. [Google Scholar] [CrossRef]
- Cai, X.; Schafer, A.; Lu, S.; Bilello, J.P.; Desrosiers, R.C.; Edwards, R.; Raab-Traub, N.; Cullen, B.R. Epstein-barr virus micrornas are evolutionarily conserved and differentially expressed. PLoS Pathog. 2006, 2, e23. [Google Scholar] [CrossRef]
- Cao, S.; Moss, W.; O’Grady, T.; Concha, M.; Strong, M.J.; Wang, X.; Yu, Y.; Baddoo, M.; Zhang, K.; Fewell, C.; et al. New noncoding lytic transcripts derived from the epstein-barr virus latency origin of replication, orip, are hyperedited, bind the paraspeckle protein, nono/p54nrb, and support viral lytic transcription. J. Virol. 2015, 89, 7120–7132. [Google Scholar] [CrossRef]
- Khatiwada, S.; Subedi, A. Lung microbiome and coronavirus disease 2019 (COVID-19): Possible link and implications. Hum. Microb. J. 2020, 17, 100073. [Google Scholar] [CrossRef]
- Engels, E.A. Inflammation in the development of lung cancer: Epidemiological evidence. Expert Rev. Anticancer Ther. 2008, 8, 605–615. [Google Scholar] [CrossRef]
- Leroux, C.; Girard, N.; Cottin, V.; Greenland, T.; Mornex, J.F.; Archer, F. Jaagsiekte sheep retrovirus (jsrv): From virus to lung cancer in sheep. Vet. Res. 2007, 38, 211–228. [Google Scholar] [CrossRef] [PubMed]
- Desgranges, C.; de-The, G. Epstein-barr virus specific iga serum antibodies in nasopharyngeal and other respiratory carcinomas. Int. J. Cancer 1979, 24, 555–559. [Google Scholar] [CrossRef]
- Lung, M.L.; Lam, W.K.; So, S.Y.; Lam, W.P.; Chan, K.H.; Ng, M.H. Evidence that respiratory tract is major reservoir for epstein-barr virus. Lancet 1985, 1, 889–892. [Google Scholar] [CrossRef]
- Begin, L.R.; Eskandari, J.; Joncas, J.; Panasci, L. Epstein-barr virus related lymphoepithelioma-like carcinoma of lung. J. Surg. Oncol. 1987, 36, 280–283. [Google Scholar] [CrossRef]
- Wockel, W.; Hofler, G.; Popper, H.H.; Morresi, A. Lymphoepithelioma-like carcinoma of the lung. Pathol. Res. Pract. 1995, 191, 1170–1174. [Google Scholar] [CrossRef]
- Higashiyama, M.; Doi, O.; Kodama, K.; Yokouchi, H.; Tateishi, R.; Horiuchi, K.; Mishima, K. Lymphoepithelioma-like carcinoma of the lung: Analysis of two cases for epstein-barr virus infection. Hum. Pathol. 1995, 26, 1278–1282. [Google Scholar] [CrossRef]
- Chan, J.K.; Hui, P.K.; Tsang, W.Y.; Law, C.K.; Ma, C.C.; Yip, T.T.; Poon, Y.F. Primary lymphoepithelioma-like carcinoma of the lung. A clinicopathologic study of 11 cases. Cancer 1995, 76, 413–422. [Google Scholar] [CrossRef]
- Han, A.J.; Xiong, M.; Zong, Y.S. Association of epstein-barr virus with lymphoepithelioma-like carcinoma of the lung in southern china. Am. J. Clin. Pathol. 2000, 114, 220–226. [Google Scholar] [CrossRef]
- Gomez-Roman, J.J.; Martinez, M.N.; Fernandez, S.L.; Val-Bernal, J.F. Epstein-barr virus-associated adenocarcinomas and squamous-cell lung carcinomas. Mod. Pathol. 2009, 22, 530–537. [Google Scholar] [CrossRef]
- Chen, F.F.; Yan, J.J.; Lai, W.W.; Jin, Y.T.; Su, I.J. Epstein-barr virus-associated nonsmall cell lung carcinoma: Undifferentiated “lymphoepithelioma-like” carcinoma as a distinct entity with better prognosis. Cancer 1998, 82, 2334–2342. [Google Scholar] [CrossRef]
- Li, C.M.; Han, G.L.; Zhang, S.J. Detection of epstein-barr virus in lung carcinoma tissue by in situ hybridization. Zhonghua Shi Yan He Lin Chuang Bing Du Xue Za Zhi 2007, 21, 288–290. [Google Scholar]
- Kasai, K.; Sato, Y.; Kameya, T.; Inoue, H.; Yoshimura, H.; Kon, S.; Kikuchi, K. Incidence of latent infection of epstein-barr virus in lung cancers--an analysis of eber1 expression in lung cancers by in situ hybridization. J. Pathol. 1994, 174, 257–265. [Google Scholar] [CrossRef]
- Huber, M.; Pavlova, B.; Muhlberger, H.; Hollaus, P.; Lintner, F. Detection of the epstein-barr virus in primary adenocarcinoma of the lung with signet-ring cells. Virchows Arch. 2002, 441, 25–30. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Xiong, H.; Yan, S.; Wu, N.; Lu, Z. Identification and characterization of epstein-barr virus genomes in lung carcinoma biopsy samples by next-generation sequencing technology. Sci. Rep. 2016, 6, 26156. [Google Scholar] [CrossRef]
- Jafarian, A.H.; Omidi-Ashrafi, A.; Mohamadian-Roshan, N.; Karimi-Shahri, M.; Ghazvini, K.; Boroumand-Noughabi, S. Association of epstein barr virus deoxyribonucleic acid with lung carcinoma. Indian J. Pathol. Microbiol. 2013, 56, 359–364. [Google Scholar] [PubMed]
- Chu, P.G.; Cerilli, L.; Chen, Y.Y.; Mills, S.E.; Weiss, L.M. Epstein-barr virus plays no role in the tumorigenesis of small-cell carcinoma of the lung. Mod. Pathol. 2004, 17, 158–164. [Google Scholar] [CrossRef] [PubMed]
- Butler, A.E.; Colby, T.V.; Weiss, L.; Lombard, C. Lymphoepithelioma-like carcinoma of the lung. Am. J. Surg. Pathol. 1989, 13, 632–639. [Google Scholar] [CrossRef] [PubMed]
- Gal, A.A.; Unger, E.R.; Koss, M.N.; Yen, T.S. Detection of epstein-barr virus in lymphoepithelioma-like carcinoma of the lung. Mod. Pathol. 1991, 4, 264–268. [Google Scholar] [PubMed]
- Miller, B.; Montgomery, C.; Watne, A.L.; Johnson, D.; Bailey, T.; Kowalski, R. Lymphoepithelioma-like carcinoma of the lung. J. Surg. Oncol. 1991, 48, 62–68. [Google Scholar] [CrossRef] [PubMed]
- Pittaluga, S.; Wong, M.P.; Chung, L.P.; Loke, S.L. Clonal epstein-barr virus in lymphoepithelioma-like carcinoma of the lung. Am. J. Surg. Pathol. 1993, 17, 678–682. [Google Scholar] [CrossRef]
- Wong, M.P.; Chung, L.P.; Yuen, S.T.; Leung, S.Y.; Chan, S.Y.; Wang, E.; Fu, K.H. In situ detection of epstein-barr virus in non-small cell lung carcinomas. J Pathol. 1995, 177, 233–240. [Google Scholar] [CrossRef] [PubMed]
- Ferrara, G.; Nappi, O. Lymphoepithelioma-like carcinoma of the lung. Two cases diagnosed in caucasian patients. Tumori 1995, 81, 144–147. [Google Scholar] [CrossRef] [PubMed]
- Conway, E.J.; Hudnall, S.D.; Lazarides, A.; Bahler, A.; Fraire, A.E.; Cagle, P.T. Absence of evidence for an etiologic role for epstein-barr virus in neoplasms of the lung and pleura. Mod. Pathol. 1996, 9, 491–495. [Google Scholar] [PubMed]
- Frank, M.W.; Shields, T.W.; Joob, A.W.; Kies, M.S.; Sturgis, C.D.; Yeldandi, A.; Cribbins, A.J.; Fullerton, D.A. Lymphoepithelioma-like carcinoma of the lung. Ann. Thorac. Surg. 1997, 64, 1162–1164. [Google Scholar] [CrossRef]
- Curcio, L.D.; Cohen, J.S.; Grannis, F.W., Jr.; Paz, I.B.; Chilcote, R.; Weiss, L.M. Primary lymphoepithelioma-like carcinoma of the lung in a child. Report of an epstein-barr virus-related neoplasm. Chest 1997, 111, 250–251. [Google Scholar] [CrossRef]
- Kasai, K.; Kon, S.; Sato, N.; Muraishi, K.; Yoshida, H.; Nakai, N.; Hamakawa, H.; Itoh, C.; Yamaoka, S. Case report of lymphoepithelioma-like carcinoma of the lung--lymphoid population consisting of cytotoxic t cells in resting state. Pathol. Res. Pract. 1999, 195, 773–779. [Google Scholar] [CrossRef]
- Muraishi, K.; Kon, S.; Yosida, H.; Hamakawa, H.; Nakai, N.; Itoh, C.; Yamaoka, S.; Kasai, K. lymphoepithelioma-like carcinoma of the lung. Nihon Kokyuki Gakkai Zasshi 1999, 37, 565–570. [Google Scholar] [PubMed]
- Castro, C.Y.; Ostrowski, M.L.; Barrios, R.; Green, L.K.; Popper, H.H.; Powell, S.; Cagle, P.T.; Ro, J.Y. Relationship between epstein-barr virus and lymphoepithelioma-like carcinoma of the lung: A clinicopathologic study of 6 cases and review of the literature. Hum. Pathol. 2001, 32, 863–872. [Google Scholar] [CrossRef] [PubMed]
- Ngan, R.K.; Yip, T.T.; Cheng, W.W.; Chan, J.K.; Cho, W.C.; Ma, V.W.; Wan, K.K.; Au, S.K.; Law, C.K.; Lau, W.H. Circulating epstein-barr virus DNA in serum of patients with lymphoepithelioma-like carcinoma of the lung: A potential surrogate marker for monitoring disease. Clin. Cancer Res. 2002, 8, 986–994. [Google Scholar]
- Chang, Y.L.; Wu, C.T.; Shih, J.Y.; Lee, Y.C. New aspects in clinicopathologic and oncogene studies of 23 pulmonary lymphoepithelioma-like carcinomas. Am. J. Surg. Pathol. 2002, 26, 715–723. [Google Scholar] [CrossRef]
- Brouchet, L.; Valmary, S.; Dahan, M.; Didier, A.; Galateau-Salle, F.; Brousset, P.; Degano, B. Detection of oncogenic virus genomes and gene products in lung carcinoma. Br. J. Cancer 2005, 92, 743–746. [Google Scholar] [CrossRef][Green Version]
- Chau, S.L.; Tong, J.H.; Chow, C.; Kwan, J.S.; Lung, R.W.; Chung, L.Y.; Tin, E.K.; Wong, S.S.; Cheung, A.H.; Lau, R.W.; et al. Distinct molecular landscape of epstein-barr virus associated pulmonary lymphoepithelioma-like carcinoma revealed by genomic sequencing. Cancers 2020, 12, 2065. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.; Chen, X.; Zhou, P.; Yang, L.; Ren, J.; Yang, X.; Li, W. Primary pulmonary lymphoepithelioma-like carcinoma: A rare type of lung cancer with a favorable outcome in comparison to squamous carcinoma. Respir. Res. 2019, 20, 262. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.; Zhang, Y.; Dai, S.; Zhou, P.; Luo, W.; Wang, Z.; Chen, X.; Cheng, P.; Zheng, G.; Ren, J.; et al. Molecular characteristics of primary pulmonary lymphoepithelioma-like carcinoma based on integrated genomic analyses. Signal. Transduct. Target. Ther. 2021, 6, 6. [Google Scholar] [CrossRef] [PubMed]
- Firincioglulari, A.; Akinci Ozyurek, B.; Erdogan, Y.; Incekara, F.; Yilmaz, E.; Ozaydin, H.E. Lymphoepithelioma-like carcinoma of the lung: A rare case report and review of the literature. Tuberk. Toraks 2020, 68, 453–457. [Google Scholar] [CrossRef] [PubMed]
- Hong, S.; Liu, D.; Luo, S.; Fang, W.; Zhan, J.; Fu, S.; Zhang, Y.; Wu, X.; Zhou, H.; Chen, X.; et al. The genomic landscape of epstein-barr virus-associated pulmonary lymphoepithelioma-like carcinoma. Nat. Commun. 2019, 10, 3108. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.C.; Hsueh, C.; Ho, S.Y.; Liao, C.Y. Lymphoepithelioma-like carcinoma of the lung: An unusual case and literature review. Case Rep. Pulmonol. 2013, 2013, 143405. [Google Scholar] [CrossRef]
- Kobayashi, M.; Ito, M.; Sano, K.; Honda, T.; Nakayama, J. Pulmonary lymphoepithelioma-like carcinoma: Predominant infiltration of tumor-associated cytotoxic t lymphocytes might represent the enhanced tumor immunity. Intern. Med. 2004, 43, 323–326. [Google Scholar] [CrossRef][Green Version]
- Koshiol, J.; Gulley, M.L.; Zhao, Y.; Rubagotti, M.; Marincola, F.M.; Rotunno, M.; Tang, W.; Bergen, A.W.; Bertazzi, P.A.; Roy, D.; et al. Epstein-barr virus micrornas and lung cancer. Br. J. Cancer 2011, 105, 320–326. [Google Scholar] [CrossRef] [PubMed]
- Lim, W.T.; Chuah, K.L.; Leong, S.S.; Tan, E.H.; Toh, C.K. Assessment of human papillomavirus and epstein-barr virus in lung adenocarcinoma. Oncol. Rep. 2009, 21, 971–975. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Sasaki, A.; Kato, T.; Ujiie, H.; Cho, Y.; Sato, M.; Kaji, M. Primary pulmonary lymphoepithelioma-like carcinoma with positive expression of epstein-barr virus and pd-l1: A case report. Int. J. Surg. Case Rep. 2021, 79, 431–435. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, S.; Chen, F.; Date, H. Pulmonary lymphoepithelioma-like carcinoma with rapid progression. Gen. Thorac. Cardiovasc. Surg. 2012, 60, 164–167. [Google Scholar] [CrossRef] [PubMed]
- Wong, J.F.; Teo, M.C. Case report: Lymphoepithelial-like carcinoma of the lung—A chronic disease? World J. Surg. Oncol. 2012, 10, 91. [Google Scholar] [CrossRef]
- Yoshino, N.; Kubokura, H.; Yamauchi, S.; Ohaki, Y.; Koizumi, K.; Shimizu, K. Lymphoepithelioma-like carcinoma of the lung: Case in which the patient has been followed up for 7 years postoperatively. JPN J. Thorac. Cardiovasc. Surg. 2005, 53, 653–656. [Google Scholar] [CrossRef]
- Lin, Z.; Xu, G.; Deng, N.; Taylor, C.; Zhu, D.; Flemington, E.K. Quantitative and qualitative rna-seq-based evaluation of epstein-barr virus transcription in type i latency burkitt’s lymphoma cells. J. Virol. 2010, 84, 13053–13058. [Google Scholar] [CrossRef]
- Lin, Z.; Puetter, A.; Coco, J.; Xu, G.; Strong, M.J.; Wang, X.; Fewell, C.; Baddoo, M.; Taylor, C.; Flemington, E.K. Detection of murine leukemia virus in the epstein-barr virus-positive human b-cell line jy, using a computational rna-seq-based exogenous agent detection pipeline, parses. J. Virol. 2012, 86, 2970–2977. [Google Scholar] [CrossRef]
- Lin, Z.; Wang, X.; Strong, M.J.; Concha, M.; Baddoo, M.; Xu, G.; Baribault, C.; Fewell, C.; Hulme, W.; Hedges, D.; et al. Whole-genome sequencing of the akata and mutu epstein-barr virus strains. J. Virol. 2013, 87, 1172–1182. [Google Scholar] [CrossRef] [PubMed]
- Strong, M.J.; Baddoo, M.; Nanbo, A.; Xu, M.; Puetter, A.; Lin, Z. Comprehensive high-throughput rna sequencing analysis reveals contamination of multiple nasopharyngeal carcinoma cell lines with hela cell genomes. J. Virol. 2014, 88, 10696–10704. [Google Scholar] [CrossRef]
- Strong, M.J.; Laskow, T.; Nakhoul, H.; Blanchard, E.; Liu, Y.; Wang, X.; Baddoo, M.; Lin, Z.; Yin, Q.; Flemington, E.K. Latent expression of the epstein-barr virus (ebv)-encoded major histocompatibility complex class i tap inhibitor, bnlf2a, in ebv-positive gastric carcinomas. J. Virol. 2015, 89, 10110–10114. [Google Scholar] [CrossRef]
- Strong, M.J.; Blanchard, E.t.; Lin, Z.; Morris, C.A.; Baddoo, M.; Taylor, C.M.; Ware, M.L.; Flemington, E.K. A comprehensive next generation sequencing-based virome assessment in brain tissue suggests no major virus-tumor association. Acta Neuropathol. Commun. 2016, 4, 71. [Google Scholar] [CrossRef]
- Feng, H.; Shuda, M.; Chang, Y.; Moore, P.S. Clonal integration of a polyomavirus in human merkel cell carcinoma. Science 2008, 319, 1096–1100. [Google Scholar] [CrossRef]
- Castellarin, M.; Warren, R.L.; Freeman, J.D.; Dreolini, L.; Krzywinski, M.; Strauss, J.; Barnes, R.; Watson, P.; Allen-Vercoe, E.; Moore, R.A.; et al. Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome Res. 2012, 22, 299–306. [Google Scholar] [CrossRef] [PubMed]
- Kostic, A.D.; Gevers, D.; Pedamallu, C.S.; Michaud, M.; Duke, F.; Earl, A.M.; Ojesina, A.I.; Jung, J.; Bass, A.J.; Tabernero, J.; et al. Genomic analysis identifies association of fusobacterium with colorectal carcinoma. Genome Res. 2012, 22, 292–298. [Google Scholar] [CrossRef] [PubMed]
- Ambinder, R.F. Gammaherpesviruses and “hit-and-run” oncogenesis. Am. J. Pathol. 2000, 156, 1–3. [Google Scholar] [CrossRef]
- Hu, H.; Luo, M.L.; Desmedt, C.; Nabavi, S.; Yadegarynia, S.; Hong, A.; Konstantinopoulos, P.A.; Gabrielson, E.; Hines-Boykin, R.; Pihan, G.; et al. Epstein-barr virus infection of mammary epithelial cells promotes malignant transformation. EBioMedicine 2016, 9, 148–160. [Google Scholar] [CrossRef]
- Al-Mozaini, M.; Bodelon, G.; Karstegl, C.E.; Jin, B.; Al-Ahdal, M.; Farrell, P.J. Epstein-barr virus bart gene expression. J. Gen. Virol. 2009, 90, 307–316. [Google Scholar] [CrossRef]
- Smith, P.R.; de Jesus, O.; Turner, D.; Hollyoake, M.; Karstegl, C.E.; Griffin, B.E.; Karran, L.; Wang, Y.; Hayward, S.D.; Farrell, P.J. Structure and coding content of cst (bart) family rnas of epstein-barr virus. J. Virol. 2000, 74, 3082–3092. [Google Scholar] [CrossRef]
- Marquitz, A.R.; Mathur, A.; Edwards, R.H.; Raab-Traub, N. Host gene expression is regulated by two types of noncoding rnas transcribed from the epstein-barr virus bamhi a rightward transcript region. J. Virol. 2015, 89, 11256–11268. [Google Scholar] [CrossRef]
- Lin, Z.; Flemington, E.K. Mirnas in the pathogenesis of oncogenic human viruses. Cancer Lett. 2011, 305, 186–199. [Google Scholar] [CrossRef]
- Fox, C.P.; Haigh, T.A.; Taylor, G.S.; Long, H.M.; Lee, S.P.; Shannon-Lowe, C.; O’Connor, S.; Bollard, C.M.; Iqbal, J.; Chan, W.C.; et al. A novel latent membrane 2 transcript expressed in epstein-barr virus-positive nk- and t-cell lymphoproliferative disease encodes a target for cellular immunotherapy. Blood 2010, 116, 3695–3704. [Google Scholar] [CrossRef] [PubMed]
- Cen, O.; Longnecker, R. Latent membrane protein 2 (lmp2). Curr. Top. Microbiol. Immunol. 2015, 391, 151–180. [Google Scholar]
- Hwu, P.; Du, M.X.; Lapointe, R.; Do, M.; Taylor, M.W.; Young, H.A. Indoleamine 2,3-dioxygenase production by human dendritic cells results in the inhibition of t cell proliferation. J. Immunol. 2000, 164, 3596–3599. [Google Scholar] [CrossRef] [PubMed]
- Munn, D.H.; Shafizadeh, E.; Attwood, J.T.; Bondarev, I.; Pashine, A.; Mellor, A.L. Inhibition of t cell proliferation by macrophage tryptophan catabolism. J. Exp. Med. 1999, 189, 1363–1372. [Google Scholar] [CrossRef] [PubMed]
- Uyttenhove, C.; Pilotte, L.; Theate, I.; Stroobant, V.; Colau, D.; Parmentier, N.; Boon, T.; Van den Eynde, B.J. Evidence for a tumoral immune resistance mechanism based on tryptophan degradation by indoleamine 2,3-dioxygenase. Nat. Med. 2003, 9, 1269–1274. [Google Scholar] [CrossRef]
- Bell, M.J.; Abbott, R.J.; Croft, N.P.; Hislop, A.D.; Burrows, S.R. An hla-a2-restricted t-cell epitope mapped to the bnlf2a immune evasion protein of epstein-barr virus that inhibits tap. J. Virol. 2009, 83, 2783–2788. [Google Scholar] [CrossRef]
- Horst, D.; van Leeuwen, D.; Croft, N.P.; Garstka, M.A.; Hislop, A.D.; Kremmer, E.; Rickinson, A.B.; Wiertz, E.J.; Ressing, M.E. Specific targeting of the ebv lytic phase protein bnlf2a to the transporter associated with antigen processing results in impairment of hla class i-restricted antigen presentation. J. Immunol. 2009, 182, 2313–2324. [Google Scholar] [CrossRef]
- Croft, N.P.; Shannon-Lowe, C.; Bell, A.I.; Horst, D.; Kremmer, E.; Ressing, M.E.; Wiertz, E.J.; Middeldorp, J.M.; Rowe, M.; Rickinson, A.B.; et al. Stage-specific inhibition of mhc class i presentation by the epstein-barr virus bnlf2a protein during virus lytic cycle. PLoS Pathog. 2009, 5, e1000490. [Google Scholar] [CrossRef] [PubMed]
- Horst, D.; Favaloro, V.; Vilardi, F.; van Leeuwen, H.C.; Garstka, M.A.; Hislop, A.D.; Rabu, C.; Kremmer, E.; Rickinson, A.B.; High, S.; et al. Ebv protein bnlf2a exploits host tail-anchored protein integration machinery to inhibit tap. J. Immunol. 2011, 186, 3594–3605. [Google Scholar] [CrossRef] [PubMed]
- Wycisk, A.I.; Lin, J.; Loch, S.; Hobohm, K.; Funke, J.; Wieneke, R.; Koch, J.; Skach, W.R.; Mayerhofer, P.U.; Tampe, R. Epstein-barr viral bnlf2a protein hijacks the tail-anchored protein insertion machinery to block antigen processing by the transport complex tap. J. Biol. Chem. 2011, 286, 41402–41412. [Google Scholar] [CrossRef]
- Thorley-Lawson, D.A.; Gross, A. Persistence of the epstein-barr virus and the origins of associated lymphomas. N. Engl. J. Med. 2004, 350, 1328–1337. [Google Scholar] [CrossRef]
- Dutta, D.; Dutta, S.; Veettil, M.V.; Roy, A.; Ansari, M.A.; Iqbal, J.; Chikoti, L.; Kumar, B.; Johnson, K.E.; Chandran, B. Brca1 regulates ifi16 mediated nuclear innate sensing of herpes viral DNA and subsequent induction of the innate inflammasome and interferon-beta responses. PLoS Pathog. 2015, 11, e1005030. [Google Scholar] [CrossRef] [PubMed]
- Liao, G.; Huang, J.; Fixman, E.D.; Hayward, S.D. The epstein-barr virus replication protein bblf2/3 provides an origin-tethering function through interaction with the zinc finger DNA binding protein zbrk1 and the kap-1 corepressor. J. Virol. 2005, 79, 245–256. [Google Scholar] [CrossRef]
- Chang, Y.L.; Yang, C.Y.; Lin, M.W.; Wu, C.T.; Yang, P.C. Pd-l1 is highly expressed in lung lymphoepithelioma-like carcinoma: A potential rationale for immunotherapy. Lung Cancer 2015, 88, 254–259. [Google Scholar] [CrossRef] [PubMed]
- Yin, K.; Feng, H.B.; Li, L.L.; Chen, Y.; Xie, Z.; Lv, Z.Y.; Guo, W.B.; Lu, D.X.; Yang, X.N.; Yan, W.Q.; et al. Low frequency of mutation of epidermal growth factor receptor (egfr) and arrangement of anaplastic lymphoma kinase (alk) in primary pulmonary lymphoepithelioma-like carcinoma. Thorac. Cancer 2020, 11, 346–352. [Google Scholar] [CrossRef] [PubMed]
- Tam, I.Y.; Chung, L.P.; Suen, W.S.; Wang, E.; Wong, M.C.; Ho, K.K.; Lam, W.K.; Chiu, S.W.; Girard, L.; Minna, J.D.; et al. Distinct epidermal growth factor receptor and kras mutation patterns in non-small cell lung cancer patients with different tobacco exposure and clinicopathologic features. Clin. Cancer Res. 2006, 12, 1647–1653. [Google Scholar] [CrossRef]
- Xie, Z.; Liu, L.; Lin, X.; Xie, X.; Gu, Y.; Liu, M.; Zhang, J.; Ouyang, M.; Lizaso, A.; Zhang, H.; et al. A multicenter analysis of genomic profiles and pd-l1 expression of primary lymphoepithelioma-like carcinoma of the lung. Mod. Pathol. 2020, 33, 626–638. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. The hallmarks of cancer. Cell 2000, 100, 57–70. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef]
- Barretina, J.; Caponigro, G.; Stransky, N.; Venkatesan, K.; Margolin, A.A.; Kim, S.; Wilson, C.J.; Lehar, J.; Kryukov, G.V.; Sonkin, D.; et al. The cancer cell line encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 2012, 483, 603–607. [Google Scholar] [CrossRef]
- Zhao, M.; Nanbo, A.; Sun, L.; Lin, Z. Extracellular vesicles in epstein-barr virus’ life cycle and pathogenesis. Microorganisms 2019, 7, 48. [Google Scholar] [CrossRef]
- Lin, Z.; Swan, K.; Zhang, X.; Cao, S.; Brett, Z.; Drury, S.; Strong, M.J.; Fewell, C.; Puetter, A.; Wang, X.; et al. Secreted oral epithelial cell membrane vesicles induce epstein-barr virus reactivation in latently infected b cells. J. Virol. 2016, 90, 3469–3479. [Google Scholar] [CrossRef] [PubMed]
- Ni, C.; Chen, Y.; Zeng, M.; Pei, R.; Du, Y.; Tang, L.; Wang, M.; Hu, Y.; Zhu, H.; He, M.; et al. In-cell infection: A novel pathway for epstein-barr virus infection mediated by cell-in-cell structures. Cell Res. 2015, 25, 785–800. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.B.; Zhang, H.; Zhang, J.P.; Li, Y.; Zhao, B.; Feng, G.K.; Du, Y.; Xiong, D.; Zhong, Q.; Liu, W.L.; et al. Neuropilin 1 is an entry factor that promotes ebv infection of nasopharyngeal epithelial cells. Nat. Commun. 2015, 6, 6240. [Google Scholar] [CrossRef] [PubMed]
- Xiong, D.; Du, Y.; Wang, H.B.; Zhao, B.; Zhang, H.; Li, Y.; Hu, L.J.; Cao, J.Y.; Zhong, Q.; Liu, W.L.; et al. Nonmuscle myosin heavy chain iia mediates epstein-barr virus infection of nasopharyngeal epithelial cells. Proc. Natl. Acad. Sci. USA 2015, 112, 11036–11041. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Sathiyamoorthy, K.; Zhang, X.; Schaller, S.; Perez White, B.E.; Jardetzky, T.S.; Longnecker, R. Ephrin receptor a2 is a functional entry receptor for epstein-barr virus. Nat. Microbiol. 2018, 3, 172–180. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Li, Y.; Wang, H.B.; Zhang, A.; Chen, M.L.; Fang, Z.X.; Dong, X.D.; Li, S.B.; Du, Y.; Xiong, D.; et al. Ephrin receptor a2 is an epithelial cell receptor for epstein-barr virus entry. Nat. Microbiol. 2018, 3, 1–8. [Google Scholar] [CrossRef]
- Feng, F.T.; Cui, Q.; Liu, W.S.; Guo, Y.M.; Feng, Q.S.; Chen, L.Z.; Xu, M.; Luo, B.; Li, D.J.; Hu, L.F.; et al. A single nucleotide polymorphism in the epstein-barr virus genome is strongly associated with a high risk of nasopharyngeal carcinoma. Chin. J. Cancer 2015, 34, 563–572. [Google Scholar] [CrossRef]
| Latency Types | EBV Genes | Examples of EBV Associated Cancers |
|---|---|---|
| 0 | EBER1, EBER2, RPMS1, viral miRNAs | Memory B cells in EBV(+) individuals |
| I | EBER1, EBER2, RPMS1, viral miRNAs, and EBNA1 | Burkitt’s lymphoma |
| II | EBER1, EBER2, RPMS1, viral miRNAs, EBNA1, LMP1, LMP2A, and LMP2B | Nasopharyngeal carcinoma Lung cancer |
| III | EBER1, EBER2, RPMS1, viral miRNAs, EBNA1, EBNA2, EBNA3A, EBNA3B, EBNA3C, EBNA-LP, LMP1, LMP2A, and LMP2B | AIDS-associated lymphoma |
| Tumor Types | Number of Total Cases | Number of EBV(+) Cases | EBV Incidence Rates (%) | Detection Methods | Geographical Sites | References |
|---|---|---|---|---|---|---|
| LELC | 1 | 1 | 100.0 | Serology | America | [36] |
| LELC | 4 | 3 | 75.0 | ISH, Serology | America | [49] |
| LELC | 1 | 1 | 100.0 | ISH, Serology, PCR | America | [50] |
| LELC | 1 | 0 | 0.0 | ISH | America | [51] |
| LELC | 5 | 5 | 100.0 | ISH, Southern Blot | Asia | [52] |
| NSCLC/SCLC | 80 | 5 | 6.3 | ISH, IHC, PCR | Asia | [44] |
| NSCLC | 167 | 9 | 5.4 | ISH, Southern Blot, IHC | Asia | [53] |
| LELC | 1 | 1 | 100.0 | ISH, Serology, PCR | Europe | [37] |
| LELC | 11 | 11 | 100.0 | ISH | Asia | [39] |
| LELC | 2 | 2 | 100.0 | ISH, PCR, IHC | Asia | [38] |
| LELC | 2 | 0 | 0.0 | ISH | Europe | [54] |
| NSCLC | 130 | 0 | 0.0 | ISH | America | [55] |
| LELC | 1 | 0 | 0.0 | IHC | America | [56] |
| LELC | 1 | 1 | 100.0 | ISH | America | [57] |
| NSCLC | 127 | 11 | 8.7 | ISH, IHC | Asia | [42] |
| NSCLC | 5 | 5 | 100.0 | ISH | Asia | [58] |
| LELC | 1 | 1 | 100.0 | ISH, PCR | Asia | [59] |
| NSCLC | 51 | 30 | 58.8 | ISH, IHC | Asia | [40] |
| LELC | 6 | 0 | 0.0 | ISH | America | [60] |
| LUAD | 3 | 1 | 33.3 | ISH, PCR, IHC | Europe | [45] |
| LELC | 11 | 11 | 100.0 | ISH, Serology, PCR | Asia | [61] |
| LELC | 23 | 23 | 100.0 | Serology | Asia | [62] |
| SCLC | 23 | 1 | 4.3 | ISH, IHC | America | [48] |
| LELC | 1 | 1 | 100.0 | ISH | Asia | [70] |
| NSCLC/SCLC | 122 | 0 | 0.0 | ISH, PCR, IHC | Europe | [63] |
| LELC | 1 | 0 | 0.0 | ISH | Asia | [76] |
| NSCLC | 108 | 36 | 33.3 | ISH | Asia | [43] |
| NSCLC | 19 | 12 | 63.2 | ISH, PCR, IHC | Europe | [41] |
| LUAD | 110 | 0 | 0.0 | ISH | Asia | [72] |
| NSCLC | 48 | 7 | 14.6 | PCR, Microarray | America | [71] |
| LELC | 1 | 1 | 100.0 | ISH, PCR, Serology | Asia | [74] |
| LELC | 1 | 1 | 100.0 | ISH | Asia | [75] |
| LELC | 1 | 1 | 100.0 | ISH | Asia | [69] |
| NSCLC/SCLC | 48 | 5 | 10.4 | PCR, IHC | Asia | [47] |
| NSCLC | 66 | 4 | 6.1 | ISH, NGS | Asia | [46] |
| NSCLC | 1127 | 7 | 0.6 | NGS, ISH | America, Europe, Asia | [12] |
| NSCLC | 176 | 33 | 18.8 | ISH, PCR | Asia | [65] |
| LELC | 150 | 150 | 100.0 | ISH | Asia | [68] |
| LELC | 57 | 57 | 100.0 | ISH | Asia | [64] |
| LELC | 1 | 1 | 100.0 | ISH, PCR | Asia, Europe | [67] |
| LELC | 8 | 8 | 100.0 | NGS | Asia | [66] |
| LELC | 1 | 1 | 100.0 | ISH | Asia | [73] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Becnel, D.; Abdelghani, R.; Nanbo, A.; Avilala, J.; Kahn, J.; Li, L.; Lin, Z. Pathogenic Role of Epstein–Barr Virus in Lung Cancers. Viruses 2021, 13, 877. https://doi.org/10.3390/v13050877
Becnel D, Abdelghani R, Nanbo A, Avilala J, Kahn J, Li L, Lin Z. Pathogenic Role of Epstein–Barr Virus in Lung Cancers. Viruses. 2021; 13(5):877. https://doi.org/10.3390/v13050877
Chicago/Turabian StyleBecnel, David, Ramsy Abdelghani, Asuka Nanbo, Janardhan Avilala, Jacob Kahn, Li Li, and Zhen Lin. 2021. "Pathogenic Role of Epstein–Barr Virus in Lung Cancers" Viruses 13, no. 5: 877. https://doi.org/10.3390/v13050877
APA StyleBecnel, D., Abdelghani, R., Nanbo, A., Avilala, J., Kahn, J., Li, L., & Lin, Z. (2021). Pathogenic Role of Epstein–Barr Virus in Lung Cancers. Viruses, 13(5), 877. https://doi.org/10.3390/v13050877







