Identification of SARS-CoV-2 and Enteroviruses in Sewage Water—A Pilot Study
Abstract
1. Introduction
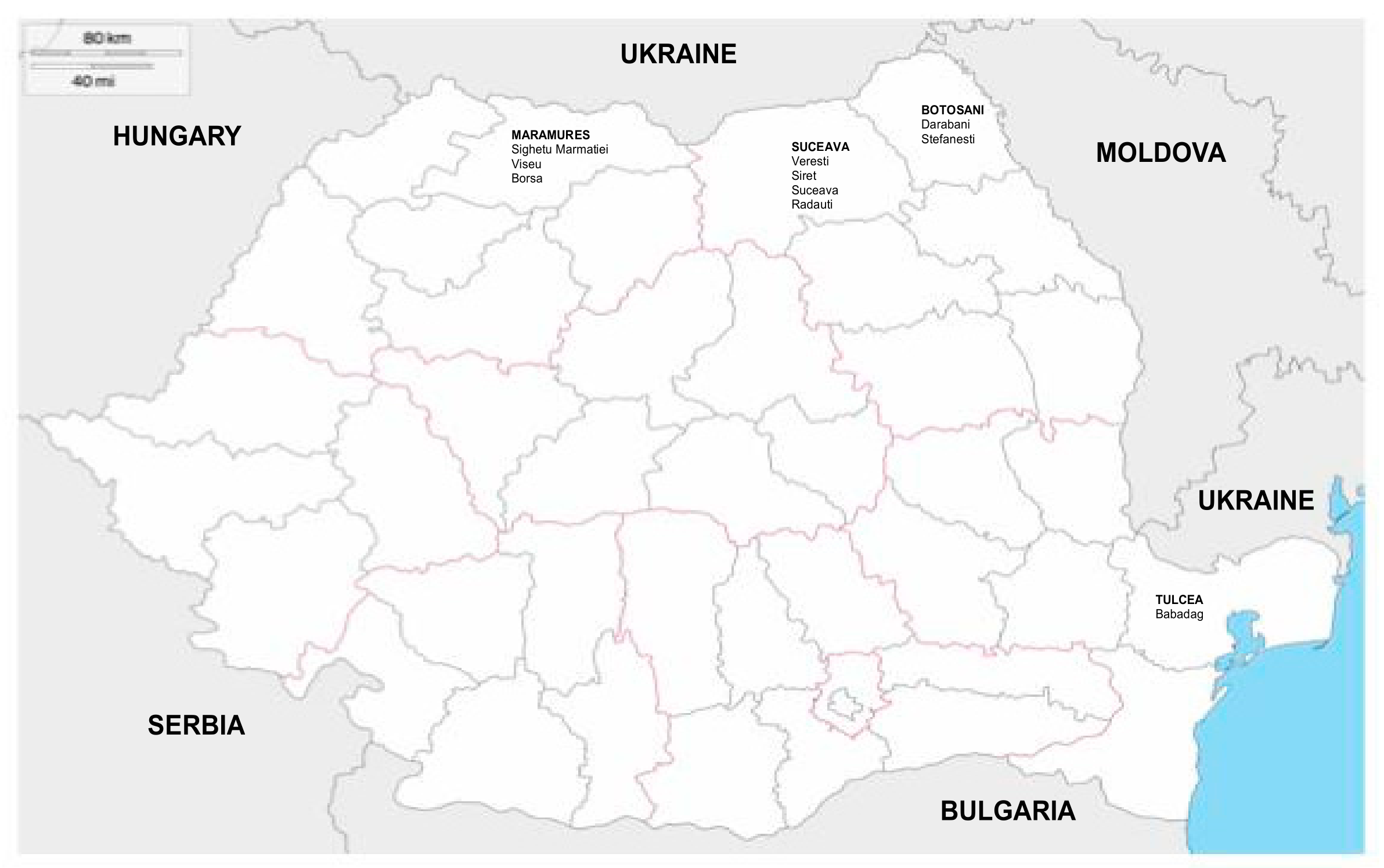
2. Materials and Methods
3. Results
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kew, O.; Pallansch, M. Breaking the Last Chains of Poliovirus Transmission: Progress and Challenges in Global Polio Eradication. Annu. Rev. Virol. 2018, 5, 427–451. [Google Scholar] [CrossRef] [PubMed]
- Anonymous. Certification of Poliomyelitis Eradication—Certification of Poliomyelitis Eradication Fifteenth Meeting of the European Regional Certification Commission Copenhagen, Denmark, 19–21 June 2002; World Health Organization—Regional Office for Europe: Copenhagen, Denmark, 2005. [Google Scholar]
- European Centre for Disease Prevention and Control. Outbreak of Vaccine-Derived Poliovirus Type 1 (cVDPV1) in Ukraine, August 2015—2 September, Stockholm. 2015. Available online: https://www.ecdc.europa.eu/en/publications-data/rapid-risk-assessment-outbreak-circulating-vaccine-derived-poliovirus-type-1 (accessed on 4 January 2021).
- World Health Organization. Guidelines for Environmental Surveillance of Poliovirus Circulation; World Health Organization: Geneva, Switzerland, 2003; Available online: https://apps.who.int/iris/handle/10665/67854 (accessed on 4 January 2021).
- Băicuş, A. The integration of the molecular methods in the diagnosis algorithm for the poliovirus detection in the sewage water: Comparing concentration and detection methods. A Pilot Study. Rom. J. Intern. Med. 2017, 55, 245–248. [Google Scholar] [CrossRef][Green Version]
- Băicuș, A. Monitoring Enterovirus and Norovirus circulation in sewage water using isolation on cell culture lines and GeneXpert system. Romanian Biotechnol. Lett. 2019, 24, 820–825. [Google Scholar] [CrossRef]
- Băicuș, A.; Joffret, M.L.; Bessaud, M.; Delpeyroux, F.; Oprisan, G. Reinforced poliovirus and enterovirus surveillance in Ro-mania, 2015–2016. Arch. Virol. 2020, 165, 2627–2632. [Google Scholar] [CrossRef] [PubMed]
- Hu, B.; Guo, H.; Zhou, P.; Shi, Z.-L. Characteristics of SARS-CoV-2 and COVID-19. Nat. Rev. Microbiol. 2021, 19, 141–154. [Google Scholar] [CrossRef] [PubMed]
- Gorbalenya, A.E. The species Severe acute respiratory syndrome-related coronavirus: Classifying 2019-nCoV and naming it SARS-CoV-2. Nat. Microbiol. 2020, 5, 536–544. [Google Scholar]
- Lu, R.X.; Li, J.; Niu, P.; Yang, B.; Wu, H.; Wang, W.; Song, H.; Huang, B.; Zhu, N.; Bi, Y.; et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: Implications for virus origins and receptor binding. Lancet 2020, 395, 565–574. [Google Scholar] [CrossRef]
- Zhou, P.; Yang, X.-L.; Wang, X.-G.; Hu, B.; Zhang, L.; Zhang, W.; Si, H.-R.; Zhu, Y.; Li, B.; Huang, C.-L.; et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 2020, 579, 270–273. [Google Scholar] [CrossRef] [PubMed]
- Zheng, S.; Fan, J.; Yu, F.; Feng, B.; Lou, B.; Zou, Q.; Xie, G.; Lin, S.; Wang, R.; Yang, X.; et al. Viral load dynamics and disease severity in patients infected with SARS-CoV-2 in Zhejiang province, China, January–March 2020: Retrospective cohort study. BMJ 2020, 369, m1443. [Google Scholar] [CrossRef]
- Tran, H.N.; Le, G.T.; Nguyen, D.T.; Juang, R.-S.; Rinklebe, J.; Bhatnagar, A.; Lima, E.C.; Iqbal, H.M.; Sarmah, A.K.; Chao, H.-P. SARS-CoV-2 coronavirus in water and wastewater: A critical review about presence and concern. Environ. Res. 2021, 193, 110265. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Polio Laboratory Manual, 4th ed.; World Health Organization: Geneva, Switzerland, 2004; Available online: https://apps.who.int/iris/handle/10665/68762 (accessed on 4 January 2021).
- World Health Organisation. WHO Polio Laboratory Manual/Supplement 1: An Alternative Test algorithm for Poliovirus Isolation and Characterization. 2007. Available online: http://polioeradication.org/tools-and-library/policy-reports/gpln-publications/ (accessed on 4 January 2021).
- Wong, K.; Onan, B.M.; Xagoraraki, I. Quantification of Enteric Viruses, Pathogen Indicators, and Salmonella Bacteria in Class B Anaerobically Digested Biosolids by Culture and Molecular Methods. Appl. Environ. Microbiol. 2010, 76, 6441–6448. [Google Scholar] [CrossRef] [PubMed]
- Viau, E.; Bibby, K.; Paez-Rubio, T.; Peccia, J. Toward a Consensus View on the Infectious Risks Associated with Land Application of Sewage Sludge. Environ. Sci. Technol. 2011, 45, 5459–5469. [Google Scholar] [CrossRef] [PubMed]
| No. | Region | Collection Site and Date–Laboratory Arrival Date | NG Test SARS-CoV-2 Ag | Siemens Clinitest Rapid COVID-19 Antigen Test | Respiratory 2.1 Panel Plus Biofire FILM Array | Enterovirus Isolation on RD and L20B Cell Culture Lines | E ** Gene | RdPR ** Gene | N ** Gene |
|---|---|---|---|---|---|---|---|---|---|
| 1 | SV | VERESTI 4.11–5.11.2020 | Positive | Positive | Adenovirus | Negative | ND | D | D |
| 2 | SIRET 4.11–5.11.2020 | Positive | Positive | Adenovirus Human Rhinovirus/EV * SARS-CoV-2 | Negative | D | D | D | |
| 3 | SUCEAVA 4.11–5.11.2020 | Positive | Positive | Adenovirus, Human Rhinovirus/EV | Negative | ND | ND | ND | |
| 4 | RADAUTI 4.11–5.11.2020 | Positive | Positive | Negative | Negative | ND | ND | D | |
| 5 | BT | DARABANI 25.08–26.08.2020 | Positive | Positive | Adenovirus Human Rhinovirus/EV | Non-polio enterovirus | ND | ND | ND |
| 6 | DARABANI 3.11–4.11.2020 | Positive | Positive | Adenovirus Human Rhinovirus/EV | Non-polio enterovirus | ND | D | ND | |
| 7 | DARABANI 16.02–17.02.2021 | Negative | Positive | Adenovirus Human Rhinovirus/EV | Negative | ND | ND | ND | |
| 8 | STEFANESTI 3.11–4.11.2020 | Positive | Positive | Adenovirus Human Rhinovirus/EV | Non-polio enterovirus | ND | ND | ND | |
| 9 | STEFANESTI 25.08–26.08.2020 | Positive | Positive | Negative | Negative | ND | D | D | |
| 10 | BABADAG 15.01–16.01.2020 | Positive | Negative | Adenovirus Human Rhinovirus/EV | Negative | ND | ND | ND | |
| 11 | TL | BABADAG 28.10–29.10.2020 | Positive | Positive | Adenovirus Human Rhinovirus/EV SARS-CoV-2 | Negative | ND | ND | ND |
| 12 | BABADAG 20.01–25.01.2021 | Negative | Positive | Adenovirus Human Rhinovirus/EV | Negative | ND | ND | ND | |
| 13 | SIGHETUL MARMATIEI 14.01–15.01.2020 | Positive | Negative | Adenovirus Human Rhinovirus/EV Coronavirus HKU1 | Non-polio enterovirus | ND | ND | ND | |
| 14 | MM | SIGHETUL MARMATIEI 10.11–11.11.2020 | Positive | Positive | Adenovirus Human Rhinovirus/EV SARS-CoV-2 | Negative | D | D | ND |
| 15 | SIGHETUL MARMATIEI 8.12–9.12.2020 | Positive | Positive | Adenovirus, Human Rhinovirus/EV SARS-CoV-2 | Negative | ND | D | D | |
| 16 | SIGHETUL MARMATIEI 12.01–13.01.2021 | Positive | Positive | Adenovirus, Human Rhinovirus/EV | Negative | ND | ND | ND | |
| 17 | VISEU 14.01–15.01.2020 | Positive | Positive | Adenovirus, Human Rhinovirus/EV | Negative | ND | ND | ND | |
| 18 | VISEU 13.10–14.10.2020 | Positive | Positive | Adenovirus, Human Rhinovirus/EV | Non-polio enterovirus | ND | D | ND | |
| 19 | VISEU 10.11–11.11.2020 | Positive | Positive | Adenovirus Human Rhinovirus/EV SARS-CoV-2 | Negative | ND | D | ND | |
| 20 | VISEU 8.12–9.12.2020 | Positive | Positive | Adenovirus Human Rhinovirus/EV SARS-CoV-2 | Non-polio enterovirus | ND | ND | ND | |
| 21 | VISEU 12.01–13.01.2021 | Positive | Positive | Adenovirus Human Rhinovirus/EV | Negative | ND | ND | ND | |
| 22 | BORSA 14.01–15.01.2020 | Positive | Positive | Adenovirus Human Rhinovirus/EV | Negative | ND | ND | ND | |
| 23 | BORSA 13.10–14.10.2020 | Positive | Positive | Adenovirus Human Rhinovirus/EV | Non-polio enterovirus | ND | D | ND | |
| 24 | BORSA 10.11–11.11.2020 | Positive | Positive | Adenovirus Human Rhinovirus/EV SARS-CoV-2 | Negative | ND | D | ND | |
| 25 | BORSA 8.12–9.12.2020 | Positive | Positive | Adenovirus, Human Rhinovirus/EV SARS-CoV-2 | Non-polio enterovirus | ND | D | D |
| Number samples | 27 | 77 | 26 | 8 |
|---|---|---|---|---|
| Virus searched for | Polio surveillance Biofire respiratory panel+SARS-CoV-2 | Polio surveillance Biofire respiratory panel (detecting MERS and 4 coronavirus strains) | Polio surveillance Biofire respiratory panel+SARS-CoV-2 | Polio surveillance Biofire respiratory panel+SARS-CoV-2 |
| Virus found | 8 NPEV/27 samples 4 Adenovirus/ 4 samples 4 Human Rhinovirus/Enterovirus/4 samples | 7 NPEV */77 samples 34 Adenovirus/53 samples 17 Human Rhinovirus/Enterovirus/53 samples | 10 NPEV/26 samples 15 Adenovirus/17samples 14 Human Rhinovirus/Enterovirus/ 17 samples 8 SARS-CoV-2/17 samples | 2 NPEV//8 samples 4 Adenovirus/ 4 samples 4 Human Rhinovirus/Enterovirus/4 samples |
| Period | January–February ** | March–September | October–December | January 2021 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Băicuș, A.; Cherciu, C.M.; Lazăr, M. Identification of SARS-CoV-2 and Enteroviruses in Sewage Water—A Pilot Study. Viruses 2021, 13, 844. https://doi.org/10.3390/v13050844
Băicuș A, Cherciu CM, Lazăr M. Identification of SARS-CoV-2 and Enteroviruses in Sewage Water—A Pilot Study. Viruses. 2021; 13(5):844. https://doi.org/10.3390/v13050844
Chicago/Turabian StyleBăicuș, Anda, Carmen Maria Cherciu, and Mihaela Lazăr. 2021. "Identification of SARS-CoV-2 and Enteroviruses in Sewage Water—A Pilot Study" Viruses 13, no. 5: 844. https://doi.org/10.3390/v13050844
APA StyleBăicuș, A., Cherciu, C. M., & Lazăr, M. (2021). Identification of SARS-CoV-2 and Enteroviruses in Sewage Water—A Pilot Study. Viruses, 13(5), 844. https://doi.org/10.3390/v13050844






