Immunological Responses to Seoul Orthohantavirus in Experimentally and Naturally Infected Brown Rats (Rattus norvegicus)
Abstract
1. Introduction
2. Materials and Methods
2.1. Virus and Cells
2.2. Experimental Infection in Laboratory Rats
2.3. Collection and Analyses of Brown Rats of SEOV
2.4. Antibody Detection in Rat Sera
2.5. Quantification of Viral RNA
2.6. Detection of SEOV-Specific Cytotoxic T Lymphocytes (CTLs) in Inbred WKAH/hkm Rats
2.7. Detection of SEOV-Specific CTLs in Experimentally Inoculated Outbred Rats and Naturally Infected Wild Brown Rats
2.8. Statistical Analysis
3. Results
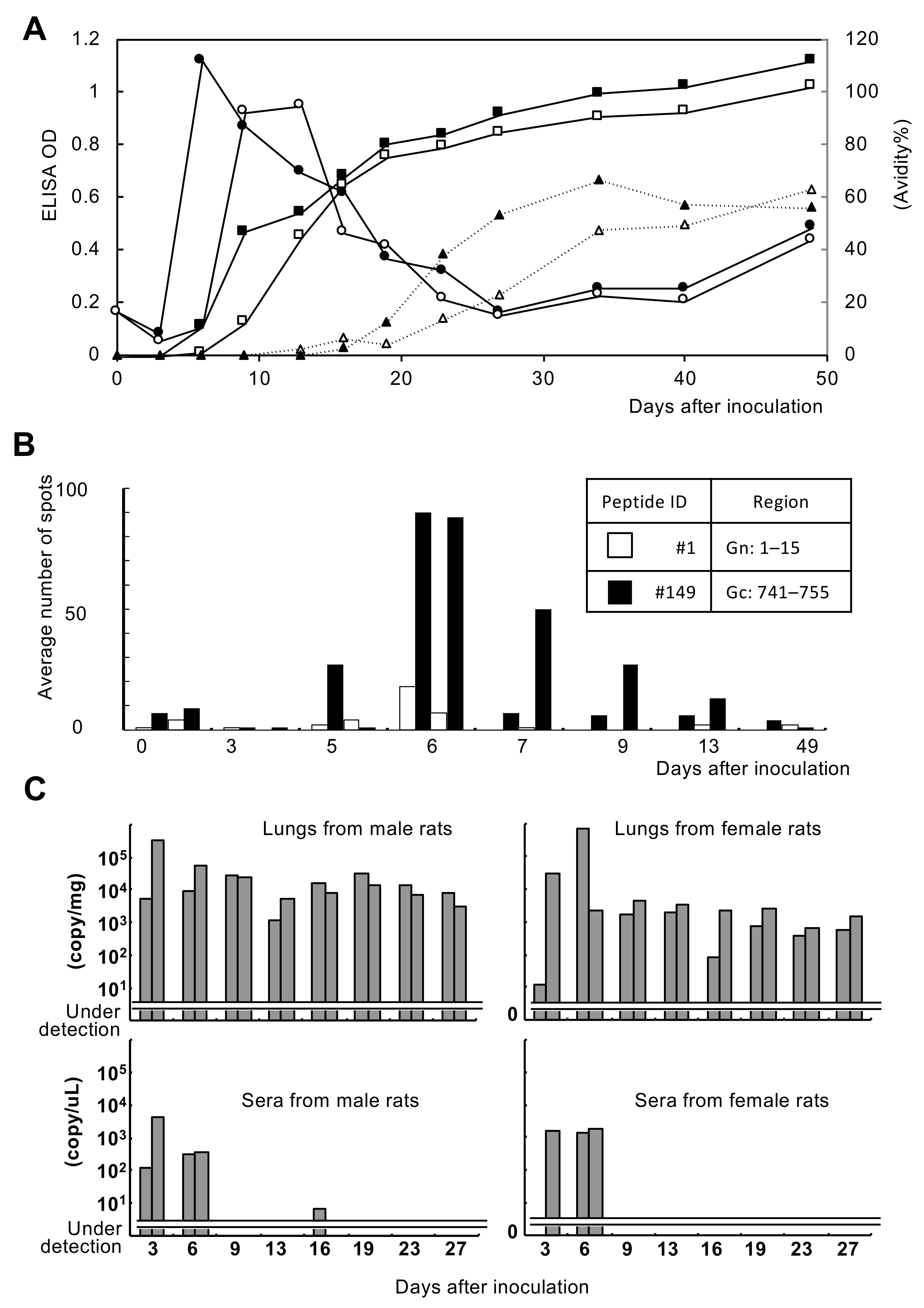
3.1. Immune Response of Experimentally Infected Rats
3.2. Immune Response of Naturally Infected Rats
3.2.1. Antibody Production
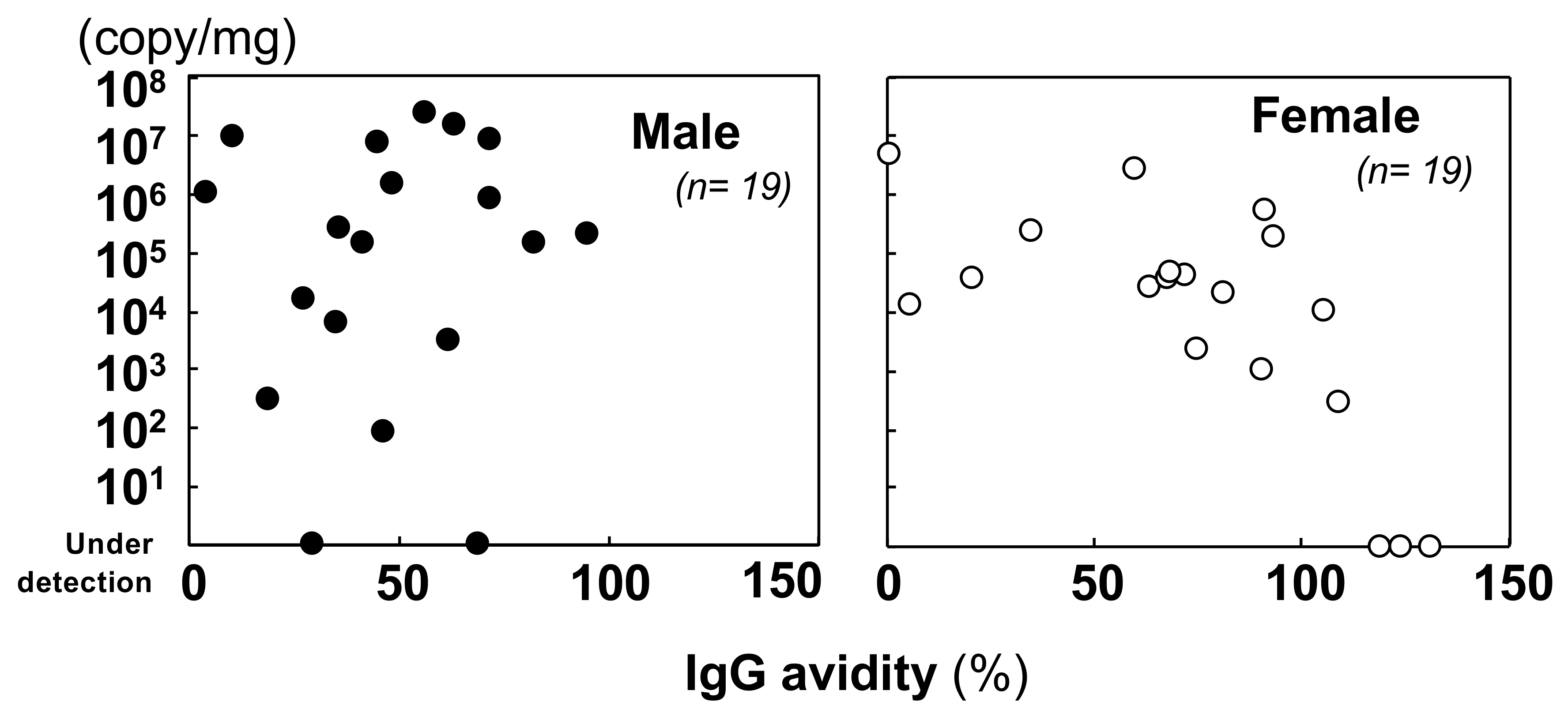
3.2.2. Quantification of the Viral Genome and Comparison with the Immunological Status
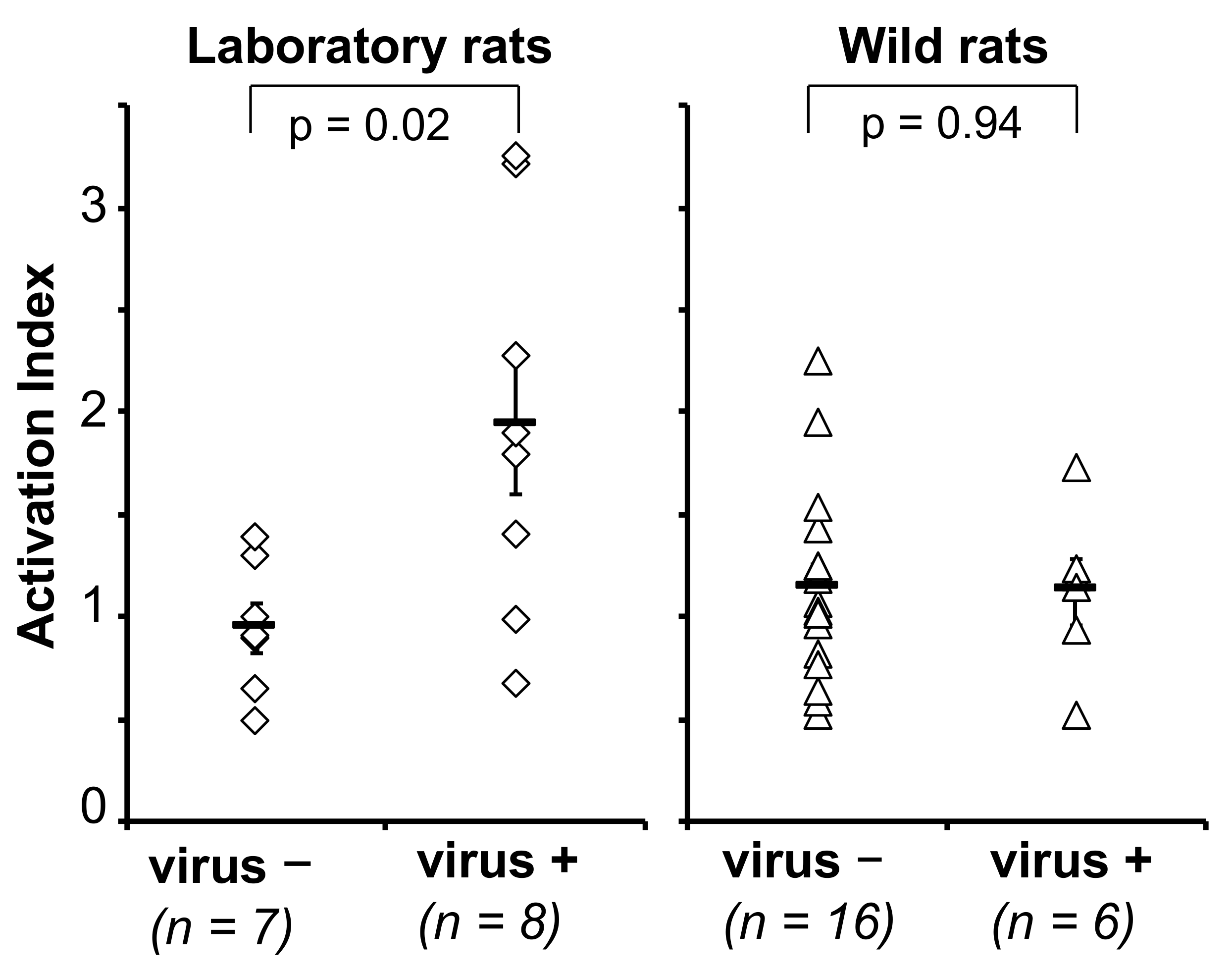
3.2.3. Cellular Immune Response
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Yanagihara, R.; Gu, S.H.; Arai, S.; Kang, H.J.; Song, J.W. Hantaviruses: Rediscovery and new beginnings. Virus Res. 2014, 187, 6–14. [Google Scholar] [CrossRef]
- Lee, H.W.; Lee, P.W.; Johnson, K.M. Isolation of the etiologic agent of Korean hemorrhagic fever. J. Infect. Dis. 1978, 137, 298–308. [Google Scholar] [CrossRef]
- Nichol, S.T.; Spiropoulou, C.F.; Morzunov, S.; Rollin, P.E.; Ksiazek, T.G.; Feldmann, H.; Sanchez, A.; Childs, J.; Zaki, S.; Peters, C.J. Genetic identification of a hantavirus associated with an outbreak of acute respiratory illness. Science 1993, 262, 914–917. [Google Scholar] [CrossRef]
- Plyusnin, A.; Vapalahti, O.; Lundkvist, A. Hantaviruses: Genome structure, expression and evolution. J. Gen. Virol. 1996, 77, 2677–2687. [Google Scholar] [CrossRef]
- Ramsden, C.; Holmes, E.C.; Charleston, M.A. Hantavirus evolution in relation to its rodent and insectivore hosts: No evidence for codivergence. Mol. Biol. Evol. 2009, 26, 143–153. [Google Scholar] [CrossRef] [PubMed]
- Plyusnin, A.; Sironen, T. Evolution of hantaviruses: Co-speciation with reservoir hosts for more than 100 MYR. Virus Res. 2014, 187, 22–26. [Google Scholar] [CrossRef]
- Lee, H.W.; Baek, L.J.; Johnson, K.M. Isolation of Hantaan virus, the etiologic agent of Korean hemorrhagic fever, from wild urban rats. J. Infect. Dis. 1982, 146, 638–644. [Google Scholar] [CrossRef]
- Knust, B.; Brown, S.; de St Maurice, A.; Whitmer, S.; Koske, S.E.; Ervin, E.; Patel, K.; Graziano, J.; Morales-Betoulle, M.E.; House, J.; et al. Seoul virus Infection and spread in United States home-based ratteries: Rat and human testing results from a multistate outbreak investigation. J. Infect. Dis. 2020, 222, 1311–1319. [Google Scholar] [CrossRef] [PubMed]
- Fill, M.A. Multistate outbreak of Seoul virus: Implications for the One Health movement and pandemic preparedness. J. Infect. Dis. 2020, 222, 1247–1249. [Google Scholar] [CrossRef] [PubMed]
- Lie, K.C.; Aziz, M.H.; Kosasih, H.; Neal, A.; Halim, C.L.; Wulan, W.N.; Karyana, M.; Hadi, U. Case report: Two confirmed cases of human Seoul virus infections in Indonesia. BMC Infect. Dis. 2018, 18, 578. [Google Scholar] [CrossRef] [PubMed]
- Swanink, C.; Reimerink, J.; Gisolf, J.; de Vries, A.; Claassen, M.; Martens, L.; Waegemaekers, T.; Rozendaal, H.; Valkenburgh, S.; Hoornweg, T.; et al. Autochthonous human case of Seoul virus infection, the Netherlands. Emerg. Infect. Dis. 2018, 24, 2158–2163. [Google Scholar] [CrossRef] [PubMed]
- Kerins, J.L.; Koske, S.E.; Kazmierczak, J.; Austin, C.; Gowdy, K.; Dibernardo, A.; Seoul Virus Working Group; Canadian Seoul Virus Investigation Group (Federal); Canadian Seoul Virus Investigation Group (Provincial). Outbreak of Seoul virus among rats and rat owners—United States and Canada, 2017. Morb. Mortal. Wkly. Rep. 2018, 67, 131–134. [Google Scholar] [CrossRef] [PubMed]
- Reynes, J.M.; Carli, D.; Bour, J.B.; Boudjeltia, S.; Dewilde, A.; Gerbier, G.; Nussbaumer, T.; Jacomo, V.; Rapt, M.P.; Rollin, P.E.; et al. Seoul virus infection in humans, France, 2014–2016. Emerg. Infect. Dis. 2017, 23, 973–977. [Google Scholar] [CrossRef] [PubMed]
- Jonsson, C.B.; Figueiredo, L.T.; Vapalahti, O. A global perspective on hantavirus ecology, epidemiology, and disease. Clin. Microbiol. Rev. 2010, 23, 412–441. [Google Scholar] [CrossRef]
- Kariwa, H.; Fujiki, M.; Yoshimatsu, K.; Arikawa, J.; Takashima, I.; Hashimoto, N. Urine-Associated Horizontal Transmission of Seoul Virus Among Rats. Arch. Virol. 1998, 143, 365–374. [Google Scholar] [CrossRef]
- Maas, M.; van Heteren, M.; de Vries, A.; Kuiken, T.; Hoornweg, T.; Kroeze, E.V.; Rockx, B. Seoul virus tropism and pathology in naturally infected feeder rats. Viruses 2019, 11, 531. [Google Scholar] [CrossRef]
- Arikawa, J.; Takashima, I.; Hashimoto, N.; Takahashi, K.; Yagi, K.; Hattori, K. Epidemiological studies of hemorrhagic fever with renal syndrome (HFRS) related virus infection among urban rats in Hokkaido, Japan. Arch. Virol. 1986, 88, 231–240. [Google Scholar] [CrossRef]
- Kariwa, H.; Lokugamage, K.; Lokugamage, N.; Miyamoto, H.; Yoshii, K.; Nakauchi, M.; Yoshimatsu, K.; Arikawa, J.; Ivanov, L.I.; Iwasaki, T.; et al. A comparative epidemiological study of hantavirus infection in Japan and Far East Russia. Jpn. J. Vet. Res. 2007, 54, 145–161. [Google Scholar]
- Dohmae, K.; Okabe, M.; Nishimune, Y. Experimental transmission of hantavirus infection in laboratory rats. J. Infect. Dis. 1994, 170, 1589–1592. [Google Scholar] [CrossRef]
- Kitamura, T.; Morita, C.; Komatsu, T.; Sugiyama, K.; Arikawa, J.; Shiga, S.; Takeda, H.; Akao, Y.; Imaizumi, K.; Oya, A.; et al. Isolation of virus causing hemorrhagic fever with renal syndrome (HFRS) through a cell culture system. Jpn. J. Med. Sci. Biol. 1983, 36, 17–25. [Google Scholar] [CrossRef]
- Kariwa, H.; Kimura, M.; Yoshizumi, S.; Arikawa, J.; Yoshimatsu, K.; Takashima, I.; Hashimoto, N. Modes of Seoul virus infections: Persistency in newborn rats and transiency in adult rats. Arch. Virol. 1996, 141, 2327–2338. [Google Scholar] [CrossRef] [PubMed]
- Kawamura, K.; Zhang, X.K.; Arikawa, J.; Takashima, I.; Dempo, K.; Hashimoto, N. Susceptibility of laboratory and wild rodents to Rattus or Apodemus-type hantaviruses. Acta Virol. 1991, 35, 54–63. [Google Scholar]
- Truong, T.-T.; Yoshimatsu, K.; Araki, K.; Lee, B.-H.; Nakamura, I.; Endo, R.; Shimizu, K.; Yasuda, P.S.; Koma, T.; Taruishi, M.; et al. Molecular epidemiological and serological studies of hantavirus infection in Northern Vietnam. J. Vet. Med. Sci. 2009, 71, 1357–1363. [Google Scholar] [CrossRef]
- Okamoto, N. Age determination by eye lens weight in the Norway rat. Jpn. Soc. Med. Entomol. Zool. 1980, 31, 193–200. [Google Scholar] [CrossRef]
- Yasuda, S.P.; Yoshimatsu, K.; Koma, T.; Shimizu, K.; Endo, R.; Isozumi, R.; Arikawa, J. Application of truncated nucleocapsid protein (N) for serotyping ELISA of murinae-asociated hantavirus Infection in rats. J. Vet. Med. Sci. 2012, 74, 215–219. [Google Scholar] [CrossRef]
- Araki, K.; Yoshimatsu, K.; Ogino, M.; Ebihara, H.; Lundkvist, A.; Kariwa, H.; Takashima, I.; Arikawa, J. Truncated hantavirus nucleocapsid proteins for serotyping Hantaan, Seoul, and Dobrava hantavirus infections. J. Clin. Microbiol. 2001, 39, 2397–2404. [Google Scholar] [CrossRef] [PubMed]
- Yoshimatsu, K.; Arikawa, J.; Yoshida, R.; Li, H.; Yoo, Y.C.; Kariwa, H.; Hashimoto, N.; Kakinuma, M.; Nobunaga, T.; Azuma, I. Production of recombinant hantavirus nucleocapsid protein expressed in silkworm larvae and its use as a diagnostic antigen in detecting antibodies in serum from infected rats. Lab. Anim. Sci. 1995, 45, 641–646. [Google Scholar]
- R Core Team. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing: Vienna, Austria, 2021; Available online: https://www.R-project.org/ (accessed on 7 April 2021).
- Araki, K.; Yoshimatsu, K.; Lee, B.-H.; Kariwa, H.; Takashima, I.; Arikawa, J. Hantavirus-specific CD8+ T cell responses in newborn mice persistently infected with Hantaan virus. J. Virol. 2003, 77, 8408–8417. [Google Scholar] [CrossRef]
- Araki, K.; Yoshimatsu, K.; Lee, B.H.; Kariwa, H.; Takashima, I.; Arikawa, J. A new model of Hantaan virus persistence in mice: The balance between HTNV infection and CD8+ T-cell responses. Virology 2004, 322, 318–327. [Google Scholar] [CrossRef]
- Luan, V.D.; Yoshimatsu, K.; Endo, R.; Taruishi, M.; Huong, V.T.; Dat, D.T.; Tien, P.C.; Shimizu, K.; Koma, T.; Yasuda, S.P.; et al. Studies on hantavirus infection in small mammals captured in southern and central highland area of Vietnam. J. Vet. Med. Sci. 2012, 74, 1155–1162. [Google Scholar] [CrossRef]
- Hedman, K.; Vaheri, A.; Brummer, K.M. Rapid diagnosis of hantavirus disease with an IgG-avidity assay. Lancet 1991, 338, 1353–1356. [Google Scholar] [CrossRef]
- Klein, S.L.; Bird, B.H.; Glass, G.E. Sex differences in Seoul virus infection are not related to adult sex steroid concentrations in Norway rats. J. Virol. 2000, 74, 8213–8217. [Google Scholar] [CrossRef] [PubMed]
- Klein, S.L.; Marson, A.L.; Scott, A.L.; Ketner, G.; Glass, G.E. Neonatal sex steroids affect responses to Seoul virus infection in male but not female Norway rats. Brain Behav. Immun. 2002, 16, 736–746. [Google Scholar] [CrossRef]
- Hannah, M.F.; Bajic, V.B.; Klein, S.L. Sex differences in the recognition of and innate antiviral responses to Seoul virus in Norway rats. Brain Behav. Immun. 2008, 22, 503–516. [Google Scholar] [CrossRef] [PubMed]
- Hinson, E.R.; Shone, S.M.; Zink, M.C.; Glass, G.E.; Klein, S.L. Wounding: The primary mode of Seoul virus transmission among male Norway rats. Am. J. Trop. Med. Hyg. 2004, 70, 310–317. [Google Scholar] [CrossRef] [PubMed]
- Mc, E.L.; Marston, D.A.; Pounder, K.C.; Goharriz, H.; Wise, E.L.; Verner-Carlsson, J.; Jennings, D.; Johnson, N.; Civello, A.; Nunez, A.; et al. High prevalence of Seoul hantavirus in a breeding colony of pet rats. Epidemiol. Infect. 2017, 145, 3115–3124. [Google Scholar]
- Yamanishi, K.; Dantas, J.R.; Takahashi, M.; Yamanouchi, T.; Domae, K.; Kawamata, J.; Kurata, T. Isolation of hemorrhagic fever with renal syndrome (HFRS) virus from a tumor specimen in a rat. Biken J. 1983, 26, 155–160. [Google Scholar]
- Lundkvist, A.; Cheng, Y.; Sjolander, K.B.; Niklasson, B.; Vaheri, A.; Plyusnin, A. Cell culture adaptation of Puumala hantavirus changes the infectivity for its natural reservoir, Clethrionomys glareolus, and leads to accumulation of mutants with altered genomic RNA S segment. J. Virol. 1997, 71, 9515–9523. [Google Scholar] [CrossRef] [PubMed]
- Warner, B.M.; Stein, D.R.; Griffin, B.D.; Tierney, K.; Leung, A.; Sloan, A.; Kobasa, D.; Poliquin, G.; Kobinger, G.P.; Safronetz, D. Development and characterization of a Sin Nombre Virus transmission model in Peromyscus maniculatus. Viruses 2019, 11, 183. [Google Scholar] [CrossRef]
- Kariwa, H.; Isegawa, Y.; Arikawa, J.; Takashima, I.; Ueda, S.; Yamanishi, K.; Hashimoto, N. Comparison of nucleotide sequences of M genome segments among Seoul virus strains isolated from eastern Asia. Virus Res. 1994, 33, 27–38. [Google Scholar] [CrossRef]
- Easterbrook, J.D.; Zink, M.C.; Klein, S.L. Regulatory T cells enhance persistence of the zoonotic pathogen Seoul virus in its reservoir host. Proc. Natl. Acad. Sci. USA 2007, 104, 15502–15507. [Google Scholar] [CrossRef] [PubMed]

| All Specimens | IgG + | IgG − | Total |
|---|---|---|---|
| IgM + | 28 | 1 | 29 |
| IgM − | 15 | 155 | 170 |
| Total | 43 | 156 | 199 |
| Phase | IgG Avidity (%) | IgM Positive Rate (%) | |
|---|---|---|---|
| Male | Female * | ||
| Acute | ≤30 | 1/5 (20%) | 3/3 (100%) |
| Chronic | <30–≤60 | 5/7 (71%) | 2/2 (100%) |
| <60–≤90 | 6/7 (86%) | 6/7 (86%) | |
| >90 | 1/1 (100%) | 4/9 (44%) | |
| Viral Genome in Lung | IgM Antibody in Sera * | |
|---|---|---|
| + | − | |
| + | 24 | 9 |
| − | 3 | 4 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yasuda, S.P.; Shimizu, K.; Koma, T.; Hoa, N.T.; Le, M.Q.; Wei, Z.; Muthusinghe, D.S.; Lokupathirage, S.M.W.; Hasebe, F.; Yamashiro, T.; et al. Immunological Responses to Seoul Orthohantavirus in Experimentally and Naturally Infected Brown Rats (Rattus norvegicus). Viruses 2021, 13, 665. https://doi.org/10.3390/v13040665
Yasuda SP, Shimizu K, Koma T, Hoa NT, Le MQ, Wei Z, Muthusinghe DS, Lokupathirage SMW, Hasebe F, Yamashiro T, et al. Immunological Responses to Seoul Orthohantavirus in Experimentally and Naturally Infected Brown Rats (Rattus norvegicus). Viruses. 2021; 13(4):665. https://doi.org/10.3390/v13040665
Chicago/Turabian StyleYasuda, Shumpei P., Kenta Shimizu, Takaaki Koma, Nguyen Thuy Hoa, Mai Quynh Le, Zhuoxing Wei, Devinda S. Muthusinghe, Sithumini M. W. Lokupathirage, Futoshi Hasebe, Tetsu Yamashiro, and et al. 2021. "Immunological Responses to Seoul Orthohantavirus in Experimentally and Naturally Infected Brown Rats (Rattus norvegicus)" Viruses 13, no. 4: 665. https://doi.org/10.3390/v13040665
APA StyleYasuda, S. P., Shimizu, K., Koma, T., Hoa, N. T., Le, M. Q., Wei, Z., Muthusinghe, D. S., Lokupathirage, S. M. W., Hasebe, F., Yamashiro, T., Arikawa, J., & Yoshimatsu, K. (2021). Immunological Responses to Seoul Orthohantavirus in Experimentally and Naturally Infected Brown Rats (Rattus norvegicus). Viruses, 13(4), 665. https://doi.org/10.3390/v13040665






