Imaging Techniques to Study Plant Virus Replication and Vertical Transmission
Abstract
1. Introduction
2. Imaging Techniques Used in Plant Virology
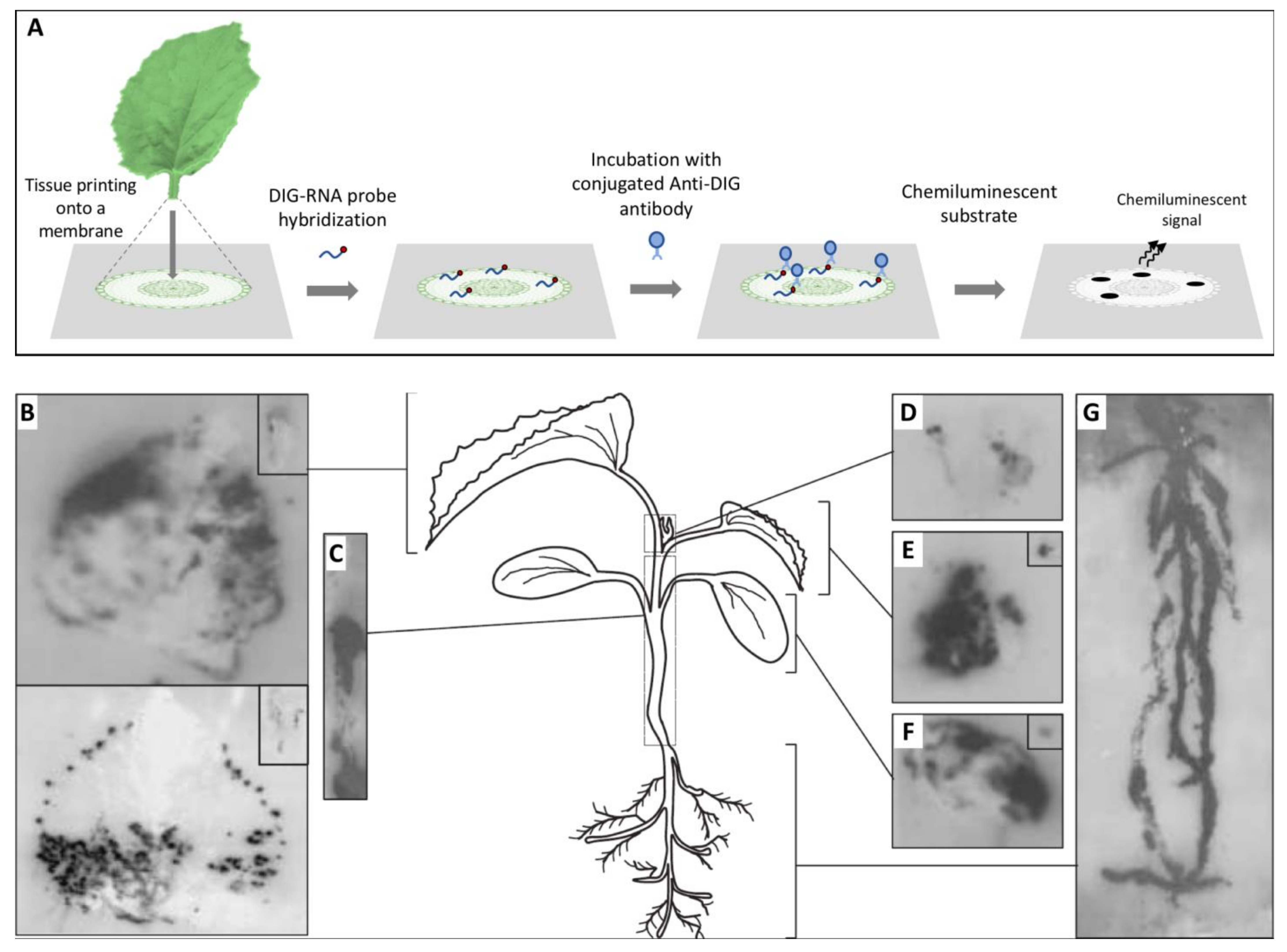
2.1. Macroscopic Techniques
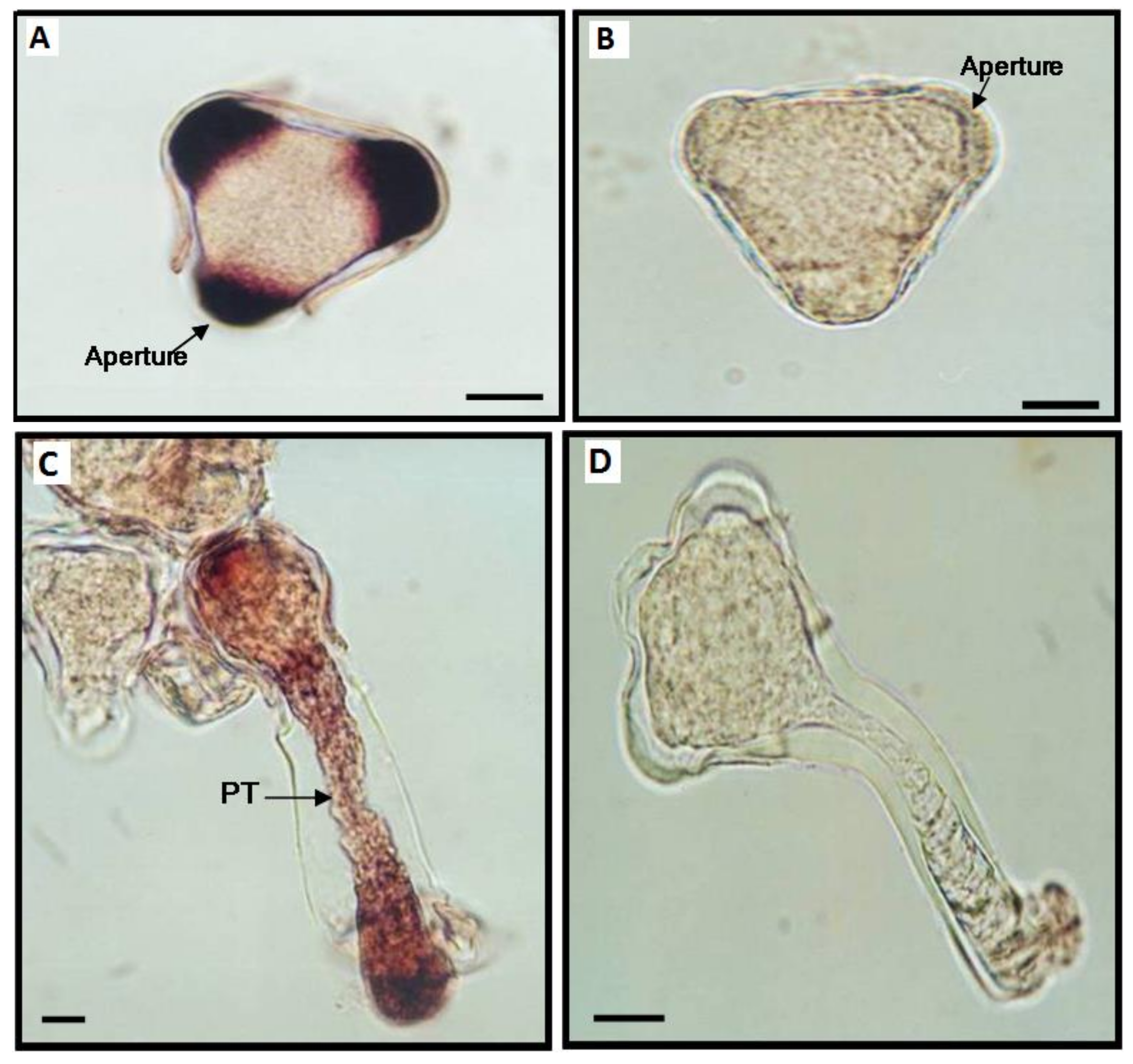
2.2. Light Microscopy
2.3. Electron Microscopy
2.4. Fluorescence Microscopy
2.5. Three-Dimensional (3D) Imaging Techniques
2.6. Correlative Light Electron Microscopy (CLEM)
3. Use of Imaging Techniques to Study Plant Virus Replication
3.1. Genesis of Plant ss(+)RNA Virus Replication Factories
3.2. Replication Factories of Viruses of The Family Tombusviridae
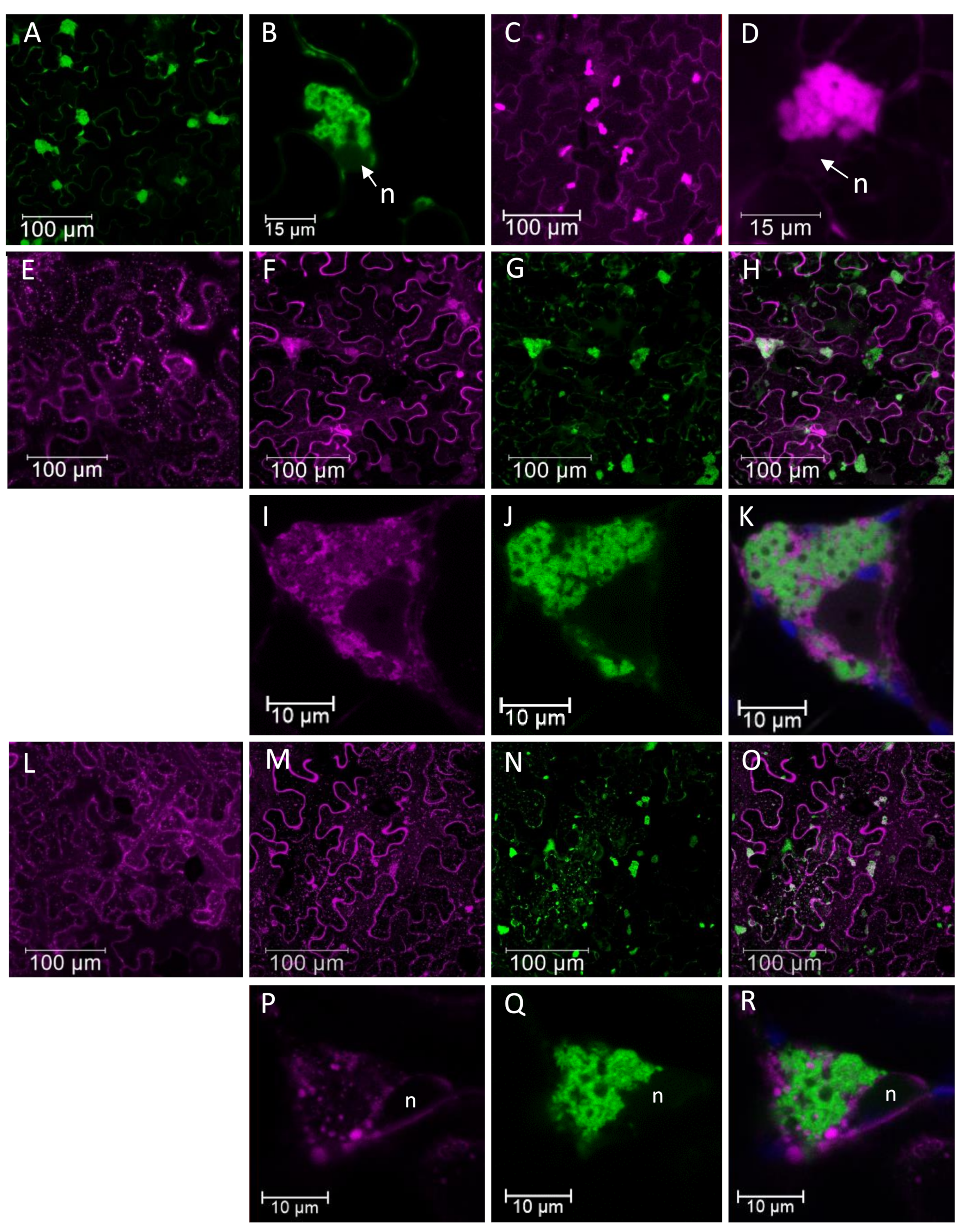
3.3. Potexvirus Replication Factories
3.4. Potyvirus Replication Factories
3.5. Spatial Distribution of Active Replication Sites in Tissues and Whole Plants
4. Use of Imaging Techniques to Study Vertical Transmission of Plant Viruses
5. Conclusions and Future Trends
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Más, P.; Pallás, V. Non-isotopic tissue-printing hybridization: A new technique to study long-distance plant virus movement. J. Virol. Methods 1995, 52, 317–326. [Google Scholar] [CrossRef]
- García-Castillo, S.; Sánchez-Pina, M.A.; Pallás, V.; Kien, F.; Abraham, J.-D.; Schuster, C.; Kieny, M.P. Spatio-temporal analysis of the RNAs, coat and movement (p7) proteins of Carnation mottle virus in Chenopodium quinoa plants. J. Gen. Virol. 2003, 84, 745–749. [Google Scholar] [CrossRef]
- Marco, C.F.; Aguilar, J.M.; Abad, J.; Gómez-Guillamón, M.L.; Aranda, M.A. Melon Resistance to Cucurbit yellow stunting disorder virus Is Characterized by Reduced Virus Accumulation. Phytopathology 2003, 93, 844–852. [Google Scholar] [CrossRef]
- Gosalvez-Bernal, B.; Garcia-Castillo, S.; Pallás, V.; Sanchez-Pina, M. Distribution of carnation viruses in the shoot tip: Exclusion from the shoot apical meristem. Physiol. Mol. Plant Pathol. 2006, 69, 43–51. [Google Scholar] [CrossRef]
- Gosalvez-Bernal, B.; Genoves, A.; Navarro, J.A.; Pallas, V.; Pina, M.A.S. Distribution and pathway for phloem-dependent movement of Melon necrotic spot virus in melon plants. Mol. Plant Pathol. 2008, 9, 447–461. [Google Scholar] [CrossRef]
- Amari, K.; Burgos, L.; Pallás, V.; Sánchez-Pina, M.A. Vertical transmission of Prunus necrotic ringspot virus: Hitch-hiking from gametes to seedling. J. Gen. Virol. 2009, 90, 1767–1774. [Google Scholar] [CrossRef] [PubMed]
- Guiu-Aragonés, C.; Sánchez-Pina, M.A.; Díaz-Pendón, J.A.; Pena, E.J.; Heinlein, M.; Martín-Hernández, A.M. cmv1is a gate forCucumber mosaic virus transport from bundle sheath cells to phloem in melon. Mol. Plant Pathol. 2016, 17, 973–984. [Google Scholar] [CrossRef] [PubMed]
- Balsalobre, J.M.; Más, P.; Sánchez-Pina, M.A.; Pallás, V. Spatial Distribution of Acidic Chitinases and Their Messenger RNAs in Tobacco Plants Infected with Cherry Leaf Roll Virus. Mol. Plant-Microbe Interact. 1997, 10, 784–788. [Google Scholar] [CrossRef]
- Goldman, V.; Czosnek, H. Whiteflies (Bemisia tabaci) issued from eggs bombarded with infectious DNA clones of Tomato yellow leaf curl virus from Israel (TYLCV) are able to infect tomato plants. Arch. Virol. 2002, 147, 787–801. [Google Scholar] [CrossRef]
- Gómez, G.; Pallás, V. A Long-Distance Translocatable Phloem Protein from Cucumber Forms a Ribonucleoprotein Complex In Vivo with Hop Stunt Viroid RNA. J. Virol. 2004, 78, 10104–10110. [Google Scholar] [CrossRef]
- Peter, K.A.; Gildow, F.; Palukaitis, P.; Gray, S.M. The C Terminus of the Polerovirus P5 Readthrough Domain Limits Virus Infection to the Phloem. J. Virol. 2009, 83, 5419–5429. [Google Scholar] [CrossRef]
- Collum, T.D.; Culver, J.N. Tobacco mosaic virus infection disproportionately impacts phloem associated translatomes in Arabidopsis thaliana and Nicotiana benthamiana. Virology 2017, 510, 76–89. [Google Scholar] [CrossRef]
- García-Castillo, S.; Marcos, J.F.; Pallas, V.; Pina, M.A.S. Influence of the plant growing conditions on the translocation routes and systemic infection of carnation mottle virus in Chenopodium quinoa plants. Physiol. Mol. Plant Pathol. 2001, 58, 229–238. [Google Scholar] [CrossRef]
- Amari, K.; Burgos, L.; Pallas, V.; Sanchez-Pina, M.A. Prunus necrotic ringspot virus Early Invasion and Its Effects on Apricot Pollen Grain Performance. Phytopathology 2007, 97, 892–899. [Google Scholar] [CrossRef] [PubMed]
- Rajamäki, M.-L.; Valkonen, J.P.T. Viral Genome-Linked Protein (VPg) Controls Accumulation and Phloem-Loading of a Potyvirus in Inoculated Potato Leaves. Mol. Plant-Microbe Interact. 2002, 15, 138–149. [Google Scholar] [CrossRef]
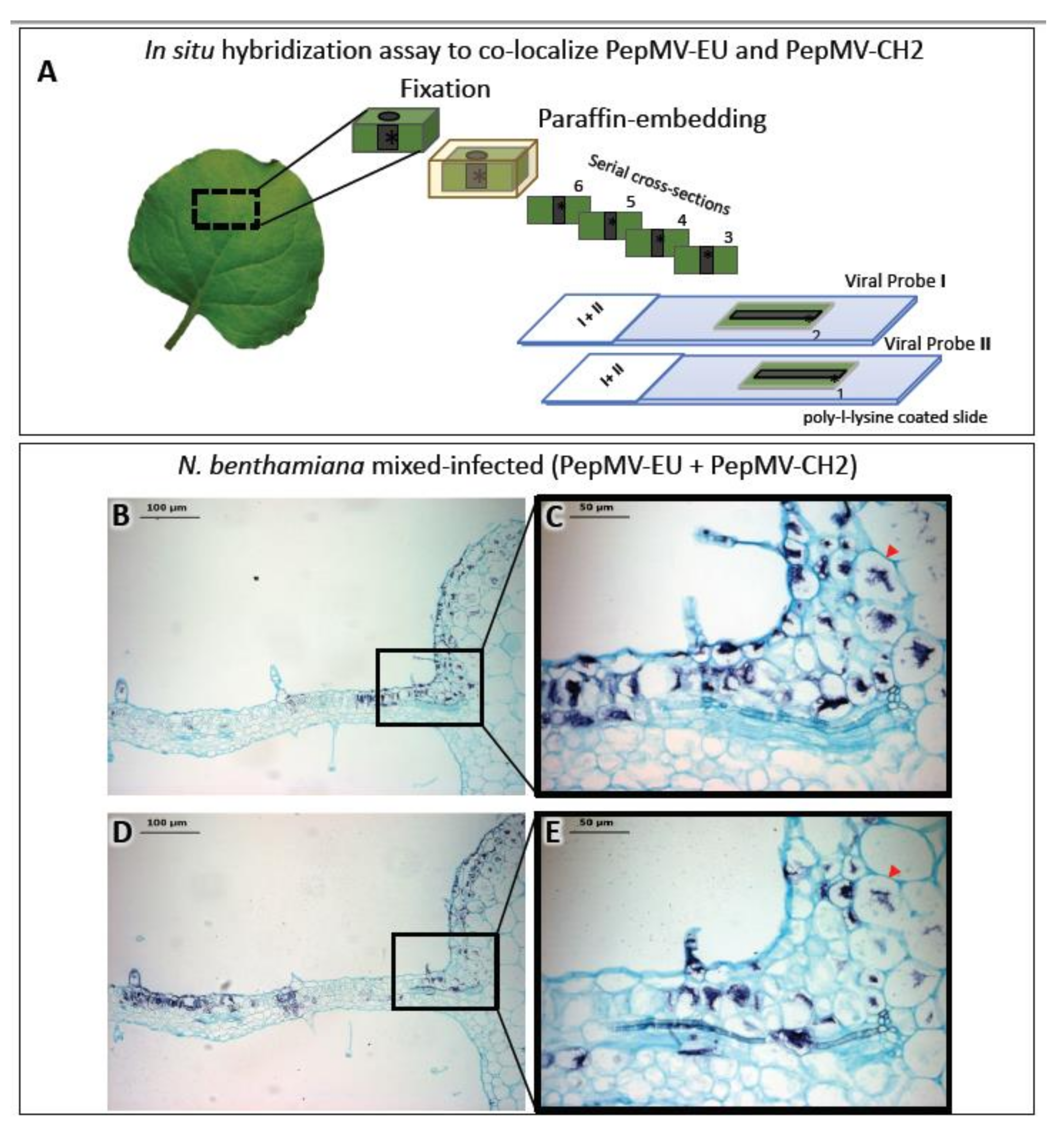
- Gómez-Aix, C.; Alcaide, C.; Gómez, P.; Aranda, M.; Sánchez-Pina, M. In situ hybridization for the localization of two pepino mosaic virus isolates in mixed infections. J. Virol. Methods 2019, 267, 42–47. [Google Scholar] [CrossRef]
- Wan, J.; Basu, K.; Mui, J.; Vali, H.; Zheng, H.; Laliberté, J.-F. Ultrastructural Characterization of Turnip Mosaic Virus-Induced Cellular Rearrangements Reveals Membrane-Bound Viral Particles Accumulating in Vacuoles. J. Virol. 2015, 89, 12441–12456. [Google Scholar] [CrossRef] [PubMed]
- Aranda, M.A.; Escaler, M.; Wang, D.; Maule, A.J. Induction of HSP70 and polyubiquitin expression associated with plant virus replication. Proc. Natl. Acad. Sci. USA 1996, 93, 15289–15293. [Google Scholar] [CrossRef] [PubMed]
- Aranda, M.A.; Maule, A. Virus-Induced Host Gene Shutoff in Animals and Plants. Virology 1998, 243, 261–267. [Google Scholar] [CrossRef] [PubMed]
- Amari, K.; Díaz-Vivancos, P.; Pallás, V.; Sánchez-Pina, M.A.; Hernández, J.A. Oxidative stress induction by Prunus necrotic ringspot virus infection in apricot seeds. Physiol. Plant. 2007, 131, 302–310. [Google Scholar] [CrossRef] [PubMed]
- Jackson, D. In situ hybridization in plants. In Molecular Plant Pathology: A Practical Approach; Gurr, S.J., McPherson, M.J., Eds.; IRL Press: Oxford, UK, 1991; pp. 163–174. [Google Scholar]
- Lim, H.-S.; Vaira, A.M.; Bae, H.; Bragg, J.N.; Ruzin, S.E.; Bauchan, G.R.; Dienelt, M.M.; Owens, R.A.; Hammond, J. Mutation of a chloroplast-targeting signal in Alternanthera mosaic virus TGB3 impairs cell-to-cell movement and eliminates long-distance virus movement. J. Gen. Virol. 2010, 91, 2102–2115. [Google Scholar] [CrossRef] [PubMed]
- Sicard, A.; Pirolles, E.; Gallet, R.; Vernerey, M.-S.; Yvon, M.; Urbino, C.; Peterschmitt, M.; Gutierrez, S.; Michalakis, Y.; Blanc, S. A multicellular way of life for a multipartite virus. eLife 2019, 8, 1–14. [Google Scholar] [CrossRef]
- Silva, G.; Souza, M. Review Genomic in situ hybridization in plants. Genet. Mol. Res. 2013, 12, 2953–2965. [Google Scholar] [CrossRef]
- Johansen, B. In situ PCR on Plant Material with Sub-cellular Resolution. Ann. Bot. 1997, 80, 697–700. [Google Scholar] [CrossRef][Green Version]
- Rojas, M.R.; Jiang, H.; Salati, R.; Xoconostle-Cázares, B.; Sudarshana, M.; Lucas, W.J.; Gilbertson, R.L. Functional Analysis of Proteins Involved in Movement of the Monopartite Begomovirus, Tomato Yellow Leaf Curl Virus. Virology 2001, 291, 110–125. [Google Scholar] [CrossRef] [PubMed]
- Silva, C.; Tereso, S.; Nolasco, G.; Oliveira, M.M. Cellular Location of Prune dwarf virus in Almond Sections by In Situ Reverse Transcription-Polymerase Chain Reaction. Phytopathology 2003, 93, 278–285. [Google Scholar] [CrossRef] [PubMed]
- Pesquet, E.; Barbier, O.; Ranocha, P.; Jauneau, A.; Goffner, D. Multiple gene detection by in situ RT-PCR in isolated plant cells and tissues. Plant J. 2004, 39, 947–959. [Google Scholar] [CrossRef]
- Yoo, B.-C.; Kragler, F.; Varkonyi-Gasic, E.; Haywood, V.; Archer-Evans, S.; Lee, Y.M.; Lough, T.J.; Lucas, W.J. A Systemic Small RNA Signaling System in Plants. Plant Cell 2004, 16, 1979–2000. [Google Scholar] [CrossRef]
- Ham, B.-K.; Brandom, J.L.; Xoconostle-Cázares, B.; Ringgold, V.; Lough, T.J.; Lucas, W.J. A Polypyrimidine Tract Binding Protein, Pumpkin RBP50, Forms the Basis of a Phloem-Mobile Ribonucleoprotein Complex. Plant Cell 2009, 21, 197–215. [Google Scholar] [CrossRef]
- Chang, C.-H.; Hsu, F.-C.; Lee, S.-C.; Lo, Y.-S.; Wang, J.-D.; Shaw, J.; Taliansky, M.; Chang, B.-Y.; Hsu, Y.-H.; Lin, N.-S. The Nucleolar Fibrillarin Protein Is Required for Helper Virus-Independent Long-Distance Trafficking of a Subviral Satellite RNA in Plants. Plant Cell 2016, 28, 2586–2602. [Google Scholar] [CrossRef]
- Carroll, T.W.; Mayhew, D.E. Anther and pollen infection in relation to the pollen and seed transmissibility of two strains of barley stripe mosaic virus in barley. Can. J. Bot. 1976, 54, 1604–1621. [Google Scholar] [CrossRef]
- Carroll, T.W.; Mayhew, D.E. Occurrence of virions in developing ovules and embryo sacs of barley in relation to the seed transmissibility of barley stripe mosaic virus. Can. J. Bot. 1976, 54, 2497–2512. [Google Scholar] [CrossRef]
- Hunter, D.G.; Bowyer, J.W. Cytopathology of Developing Anthers and Pollen Mother Cells from Lettuce Plants Infected by Lettuce Mosaic Potyvirus. J. Phytopathol. 1997, 145, 521–524. [Google Scholar] [CrossRef]
- Roberts, I.M.; Wang, D.; Thomas, C.L.; Maule, A.J. Pea seed-borne mosaic virus seed transmission exploits novel symplastic pathways to infect the pea embryo and is, in part, dependent upon chance. Protoplasma 2003, 222, 31–43. [Google Scholar] [CrossRef]
- Minicka, J.; Otulak, K.; Garbaczewska, G.; Pospieszny, H.; Hasiów-Jaroszewska, B. Ultrastructural insights into tomato infections caused by three different pathotypes of Pepino mosaic virus and immunolocalization of viral coat proteins. Micron 2015, 79, 84–92. [Google Scholar] [CrossRef]
- Sempere, R.N.; Gómez-Aix, C.; Ruíz-Ramón, F.; Gómez, P.; Hasiów-Jaroszewska, B.; Sánchez-Pina, M.A.; Aranda, M.A. Pepino mosaic virus RNA-Dependent RNA Polymerase POL Domain Is a Hypersensitive Response-Like Elicitor Shared by Necrotic and Mild Isolates. Phytopathology 2016, 106, 395–406. [Google Scholar] [CrossRef] [PubMed]
- Tilsner, J.; Linnik, O.; Christensen, N.M.; Bell, K.; Roberts, I.M.; Lacomme, C.; Oparka, K.J. Live-cell imaging of viral RNA genomes using a Pumilio-based reporter. Plant J. 2009, 57, 758–770. [Google Scholar] [CrossRef] [PubMed]
- Tilsner, J.; Linnik, O.; Wright, K.M.; Bell, K.; Roberts, A.G.; Lacomme, C.; Cruz, S.S.; Oparka, K.J. The TGB1 Movement Protein of Potato virus X Reorganizes Actin and Endomembranes into the X-Body, a Viral Replication Factory. Plant Physiol. 2012, 158, 1359–1370. [Google Scholar] [CrossRef]
- Tilsner, J.; Linnik, O.; Louveaux, M.; Roberts, I.M.; Chapman, S.N.; Oparka, K.J. Replication and trafficking of a plant virus are coupled at the entrances of plasmodesmata. J. Cell Biol. 2013, 201, 981–995. [Google Scholar] [CrossRef] [PubMed]
- Zechmann, B.; Zellnig, G. Cytological modifications in zucchini yellow mosaic virus (ZYMV)-infected Styrian pumpkin plants. Arch. Virol. 2003, 148, 1119–1133. [Google Scholar] [CrossRef]
- Brandizzi, F.; Frangne, N.; Marc-Martin, S.; Hawes, C.; Neuhaus, J.-M.; Paris, N. The Destination for Single-Pass Membrane Proteins Is Influenced Markedly by the Length of the Hydrophobic Domain. Plant Cell 2002, 14, 1077–1092. [Google Scholar] [CrossRef] [PubMed]
- Ju, H.-J.; Samuels, T.D.; Wang, Y.-S.; Blancaflor, E.; Payton, M.; Mitra, R.; Krishnamurthy, K.; Nelson, R.S.; Verchot-Lubicz, J. The Potato Virus X TGBp2 Movement Protein Associates with Endoplasmic Reticulum-Derived Vesicles during Virus Infection1. Plant Physiol. 2005, 138, 1877–1895. [Google Scholar] [CrossRef] [PubMed]
- Wan, J.; Cabanillas, D.G.; Zheng, H.; Laliberté, J.-F. Turnip mosaic virus Moves Systemically through Both Phloem and Xylem as Membrane-Associated Complexes. Plant Physiol. 2015, 167, 1374–1388. [Google Scholar] [CrossRef]
- Milne, R.; Lesemann, D.-E. Immunosorbent Electron Microscopy in Plant Virus Studies. Methods Virol. 1984, VIII, 85–101. [Google Scholar] [CrossRef]
- Ritzenthaler, C.; Laporte, C.; Gaire, F.; Dunoyer, P.; Schmitt, C.; Duval, S.; Pieéquet, A.M.; Loudes, O.; Rohfritsch, C.; Stussi-Garaud, P.; et al. Grapevine Fanleaf Virus Replication Occurs on Endoplasmic Reticulum-Derived Membranes. J. Virol. 2002, 76, 8808–8819. [Google Scholar] [CrossRef]
- Kaplan, I.B.; Lee, L.; Ripoll, D.R.; Palukaitis, P.; Gildow, F.; Gray, S.M. Point mutations in the potato leafroll virus major capsid protein alter virion stability and aphid transmission. J. Gen. Virol. 2007, 88, 1821–1830. [Google Scholar] [CrossRef]
- Smirnova, E.; Firth, A.E.; Miller, W.A.; Scheidecker, D.; Brault, V.; Reinbold, C.; Rakotondrafara, A.M.; Chung, B.Y.-W.; Ziegler-Graff, V. Discovery of a Small Non-AUG-Initiated ORF in Poleroviruses and Luteoviruses That Is Required for Long-Distance Movement. PLoS Pathog. 2015, 11, e1004868. [Google Scholar] [CrossRef] [PubMed]
- Laue, M. Electron Microscopy of Viruses; Elsevier BV: Amsterdam, The Netherlands, 2010; Volume 96, pp. 1–20. [Google Scholar]
- Lough, T.J.; Netzler, N.E.; Emerson, S.J.; Sutherland, P.; Carr, F.; Beck, D.L.; Lucas, W.J.; Forster, R.L.S. Cell-to-Cell Movement of Potexviruses: Evidence for a Ribonucleoprotein Complex Involving the Coat Protein and First Triple Gene Block Protein. Mol. Plant-Microbe Interact. 2000, 13, 962–974. [Google Scholar] [CrossRef]
- De, S.; Pollari, M.; Varjosalo, M.; Mäkinen, K. Association of host protein VARICOSE with HCPro within a multiprotein complex is crucial for RNA silencing suppression, translation, encapsidation and systemic spread of potato virus A infection. PLoS Pathog. 2020, 16, e1008956. [Google Scholar] [CrossRef] [PubMed]
- Shaw, S.L. Imaging the live plant cell. Plant J. 2006, 45, 573–598. [Google Scholar] [CrossRef]
- Tsien, R.Y. The Green Fluorescent Protein. Annu. Rev. Biochem. 1998, 67, 509–544. [Google Scholar] [CrossRef]
- Miyawaki, A.; Sawano, A.; Kogure, T. Lighting up cells: Labelling proteins with fluorophores. Nat. Rev. Mol. Cell Biol 2003, 4, 1–7. [Google Scholar]
- Shaner, N.C.; E Campbell, R.; A Steinbach, P.; Giepmans, B.N.G.; E Palmer, A.; Tsien, R.Y. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat. Biotechnol. 2004, 22, 1567–1572. [Google Scholar] [CrossRef] [PubMed]
- Dixit, R.; Cyr, R.; Gilroy, S. Using intrinsically fluorescent proteins for plant cell imaging. Plant J. 2006, 45, 599–615. [Google Scholar] [CrossRef] [PubMed]
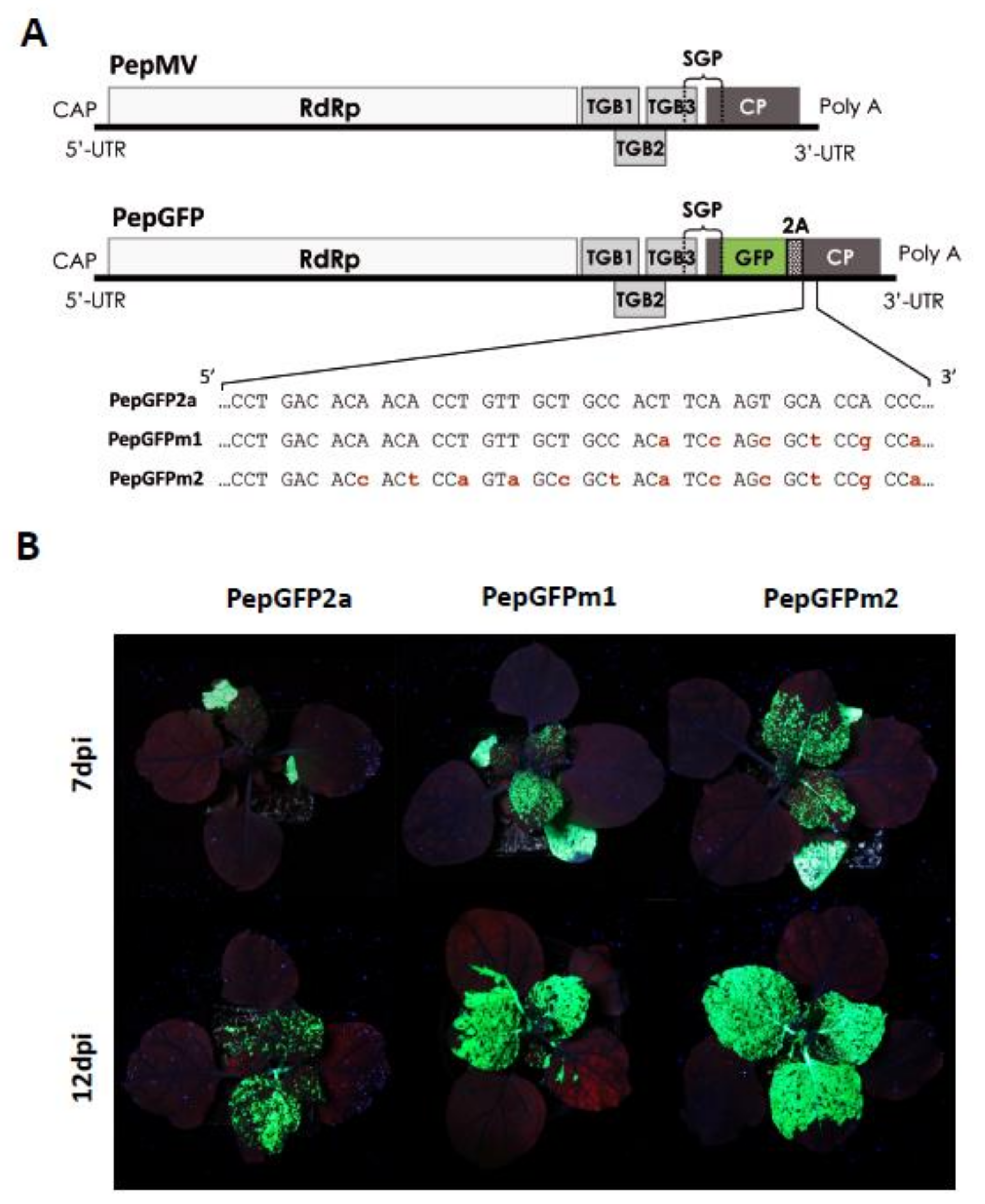
- Sempere, R.N.; Gómez, P.; Truniger, V.; Aranda, M.A. Development of expression vectors based on pepino mosaic virus. Plant Methods 2011, 7, 6–14. [Google Scholar] [CrossRef]
- Ruiz-Ramón, F.; Sempere, R.N.; Méndez-López, E.; Sánchez-Pina, M.A.; Aranda, M.A. Second generation of pepino mosaic virus vectors: Improved stability in tomato and a wide range of reporter genes. Plant Methods 2019, 15, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Ala-Poikela, M.; Goytia, E.; Haikonen, T.; Rajamäki, M.-L.; Valkonen, J.P.T. Helper Component Proteinase of the Genus Potyvirus Is an Interaction Partner of Translation Initiation Factors eIF(iso)4E and eIF4E and Contains a 4E Binding Motif. J. Virol. 2011, 85, 6784–6794. [Google Scholar] [CrossRef]
- Martin, K.M.; Dietzgen, R.G.; Wang, R.; Goodin, M.M. Lettuce necrotic yellows cytorhabdovirus protein localization and interaction map, and comparison with nucleorhabdoviruses. J. Gen. Virol. 2012, 93, 906–914. [Google Scholar] [CrossRef]
- Vijayapalani, P.; Maeshima, M.; Nagasaki-Takekuchi, N.; Miller, W.A. Interaction of the Trans-Frame Potyvirus Protein P3N-PIPO with Host Protein PCaP1 Facilitates Potyvirus Movement. PLoS Pathog. 2012, 8, e1002639. [Google Scholar] [CrossRef] [PubMed]
- Hung, C.-J.; Huang, Y.-W.; Liou, M.-R.; Lee, Y.-C.; Lin, N.-S.; Meng, M.; Tsai, C.-H.; Hu, C.-C.; Hsu, Y.-H. Phosphorylation of Coat Protein by Protein Kinase CK2 Regulates Cell-to-Cell Movement of Bamboo mosaic virus Through Modulating RNA Binding. Mol. Plant-Microbe Interact. 2014, 27, 1211–1225. [Google Scholar] [CrossRef]
- Peiró, A.; Martínez-Gil, L.; Tamborero, S.; Pallás, V.; Sánchez-Navarro, J.A.; Mingarro, I.; Simon, A. The Tobacco mosaic virus Movement Protein Associates with but Does Not Integrate into Biological Membranes. J. Virol. 2014, 88, 3016–3026. [Google Scholar] [CrossRef]
- Pouwels, J.; van der Velden, T.; Willemse, J.; Borst, J.W.; van Lent, J.; Bisseling, T.; Wellink, J. Studies on the origin and structure of tubules made by the movement protein of Cowpea mosaic virus. J. Gen. Virol. 2004, 85, 3787–3796. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.-H.; Tian, G.-W.; Gafni, Y.; Citovsky, V. Effects of Calreticulin on Viral Cell-to-Cell Movement. Plant Physiol. 2005, 138, 1866–1876. [Google Scholar] [CrossRef] [PubMed]
- Snippe, M.; Borst, J.W.; Goldbach, R.; Kormelink, R. The use of fluorescence microscopy to visualise homotypic interactions of tomato spotted wilt virus nucleocapsid protein in living cells. J. Virol. Methods 2005, 125, 15–22. [Google Scholar] [CrossRef]
- Brandner, K.; Sambade, A.; Boutant, E.; Didier, P.; Mély, Y.; Ritzenthaler, C.; Heinlein, M. Tobacco Mosaic Virus Movement Protein Interacts with Green Fluorescent Protein-Tagged Microtubule End-Binding Protein 1. Plant Physiol. 2008, 147, 611–623. [Google Scholar] [CrossRef] [PubMed]
- Amari, K.; Boutant, E.; Hofmann, C.; Schmitt-Keichinger, C.; Fernandez-Calvino, L.; Didier, P.; Lerich, A.; Mutterer, J.; Thomas, C.L.; Heinlein, M.; et al. A Family of Plasmodesmal Proteins with Receptor-Like Properties for Plant Viral Movement Proteins. PLOS Pathog. 2010, 6, e1001119. [Google Scholar] [CrossRef]
- Kwok, E.Y.; Hanson, M.R. Stromules and the dynamic nature of plastid morphology. J. Microsc. 2004, 214, 124–137. [Google Scholar] [CrossRef]
- Amari, K.; di Donato, M.; Dolja, V.V.; Heinlein, M. Myosins VIII and XI Play Distinct Roles in Reproduction and Transport of Tobacco Mosaic Virus. PLoS Pathog. 2014, 10, e1004448. [Google Scholar] [CrossRef]
- Serra-Soriano, M.; Pallas, V.; Navarro, J.A. A model for transport of a viral membrane protein through the early secretory pathway: Minimal sequence and endoplasmic reticulum lateral mobility requirements. Plant J. 2014, 77, 863–879. [Google Scholar] [CrossRef]
- Lippincott-Schwartz, J.; Patterson, G.H. Development and Use of Fluorescent Protein Markers in Living Cells. Science 2003, 300, 87–91. [Google Scholar] [CrossRef] [PubMed]
- Baumeister, W. Electron tomography: Towards visualizing the molecular organization of the cytoplasm. Curr. Opin. Struct. Biol. 2002, 12, 679–684. [Google Scholar] [CrossRef]
- Romero-Brey, I.; Bartenschlager, R. Viral Infection at High Magnification: 3D Electron Microscopy Methods to Analyze the Architecture of Infected Cells. Viruses 2015, 7, 6316–6345. [Google Scholar] [CrossRef]
- McIntosh, R.; Nicastro, D.; Mastronarde, D. New views of cells in 3D: An introduction to electron tomography. Trends Cell Biol. 2005, 15, 43–51. [Google Scholar] [CrossRef]
- Wang, P.; Liang, Z.; Kang, B. Electron tomography of plant organelles and the outlook for correlative microscopic approaches. New Phytol. 2019, 223, 1756–1761. [Google Scholar] [CrossRef]
- Lin, J.; Nicastro, D. Asymmetric distribution and spatial switching of dynein activity generates ciliary motility. Sciences 2018, 360, eaar1968. [Google Scholar] [CrossRef] [PubMed]
- Kang, B.-H. Electron Microscopy and High-Pressure Freezing of Arabidopsis; Elsevier BV: Amsteradam, The Netherlands, 2010; Volume 96, pp. 259–283. [Google Scholar]
- Arkill, K.P.; Qvortrup, K.; Starborg, T.; Mantell, J.M.; Knupp, C.; Michel, C.C.; Harper, S.J.; Salmon, A.H.J.; Squire, J.M.; O Bates, D.; et al. Resolution of the three dimensional structure of components of the glomerular filtration barrier. BMC Nephrol. 2014, 15, 24. [Google Scholar] [CrossRef]
- Peddie, C.J.; Collinson, L.M. Exploring the third dimension: Volume electron microscopy comes of age. Micron 2014, 61, 9–19. [Google Scholar] [CrossRef] [PubMed]
- Bennett, A.E.; Narayan, K.; Shi, D.; Hartnell, L.M.; Gousset, K.; He, H.; Lowekamp, B.C.; Yoo, T.S.; Bliss, D.; Freed, E.O.; et al. Ion-Abrasion Scanning Electron Microscopy Reveals Surface-Connected Tubular Conduits in HIV-Infected Macrophages. PLOS Pathog. 2009, 5, e1000591. [Google Scholar] [CrossRef]
- Do, T.; Murphy, G.; Earl, L.A.; del Prete, G.Q.; Grandinetti, G.; Li, G.-H.; Estes, J.D.; Rao, P.; Trubey, C.M.; A Thomas, J.; et al. Three-Dimensional Imaging of HIV-1 Virological Synapses Reveals Membrane Architectures Involved in Virus Transmission. J. Virol. 2014, 88, 10327–10339. [Google Scholar] [CrossRef] [PubMed]
- Gómez-Aix, C.; García-García, M.; Aranda, M.A.; Sánchez-Pina, M.A. Melon necrotic spot virus Replication Occurs in Association with Altered Mitochondria. Mol. Plant-Microbe Interact. 2015, 28, 387–397. [Google Scholar] [CrossRef]
- Jahn, K.; Barton, D.; Kobayashi, K.; Ratinac, K.; Overall, R.; Braet, F. Correlative microscopy: Providing new understanding in the biomedical and plant sciences. Micron 2012, 43, 565–582. [Google Scholar] [CrossRef]
- Giepmans, B.N.G. Bridging fluorescence microscopy and electron microscopy. Histochem. Cell Biol. 2008, 130, 211–217. [Google Scholar] [CrossRef]
- Van Rijnsoever, C.; Oorschot, V.; Klumperman, J. Correlative light-electron microscopy (CLEM) combining live-cell imaging and immunolabeling of ultrathin cryosections. Nat. Chem. Biol. 2008, 5, 973–980. [Google Scholar] [CrossRef]
- Sosinsky, G.E.; Giepmans, B.N.; Deerinck, T.J.; Gaietta, G.M.; Ellisman, M.H. Markers for Correlated Light and Electron Microscopy. Methods Cell Biol. 2007, 79, 575–591. [Google Scholar] [CrossRef]
- Barton, D.A.; Vantard, M.; Overall, R.L. Analysis of Cortical Arrays from Tradescantia virginiana at High Resolution Reveals Discrete Microtubule Subpopulations and Demonstrates That Confocal Images of Arrays Can Be Misleading. Plant Cell 2008, 20, 982–994. [Google Scholar] [CrossRef] [PubMed]
- Barton, D.A.; Cole, L.; Collings, D.A.; Liu, D.Y.T.; Smith, P.M.C.; Day, D.A.; Overall, R.L. Cell-to-cell transport via the lumen of the endoplasmic reticulum. Plant J. 2011, 66, 806–817. [Google Scholar] [CrossRef] [PubMed]
- Pfeiffer, S.; Beese, M.; Boettcher, M.; Kawaschinski, K.; Krupinska, K. Combined use of confocal laser scanning microscopy and transmission electron microscopy for visualisation of identical cells processed by cryotechniques. Protoplasma 2003, 222, 129–137. [Google Scholar] [CrossRef]
- Corredor, E.; Testillano, P.S.; Coronado, M.-J.; González-Melendi, P.; Fernández-Pacheco, R.; Marquina, C.; Ibarra, M.R.; de la Fuente, J.M.; Rubiales, D.; Pérez-De-Luque, A.; et al. Nanoparticle penetration and transport in living pumpkin plants: In situ subcellular identification. BMC Plant Biol. 2009, 9, 45. [Google Scholar] [CrossRef] [PubMed]
- Mullendore, D.L.; Windt, C.W.; van As, H.; Knoblauch, M. Sieve Tube Geometry in Relation to Phloem Flow. Plant Cell 2010, 22, 579–593. [Google Scholar] [CrossRef] [PubMed]
- González-Melendi, P.; Ramirez, C.; Testillano, P.S.; Kumlehn, J.; Risueño, M.C. Three dimensional confocal and electron microscopy imaging define the dynamics and mechanisms of diploidisation at early stages of barley microspore-derived embryogenesis. Planta 2005, 222, 47–57. [Google Scholar] [CrossRef]
- Clode, P.L.; Kilburn, M.R.; Jones, D.L.; Stockdale, E.A.; Cliff, J.B.; Herrmann, A.M.; Murphy, D.V. In Situ Mapping of Nutrient Uptake in the Rhizosphere Using Nanoscale Secondary Ion Mass Spectrometry. Plant Physiol. 2009, 151, 1751–1757. [Google Scholar] [CrossRef]
- Barton, D.; Gardiner, J.; Overall, R. Towards correlative imaging of plant cortical microtubule arrays: Combining ultrastructure with real-time microtubule dynamics. J. Microsc. 2009, 235, 241–251. [Google Scholar] [CrossRef] [PubMed]
- Linnik, O.; Liesche, J.; Tilsner, J.; Oparka, K.J. Unraveling the Structure of Viral Replication Complexes at Super-Resolution. Front. Plant Sci. 2013, 4, 6. [Google Scholar] [CrossRef] [PubMed]
- Risco, C.; Sanmartín-Conesa, E.; Tzeng, W.-P.; Frey, T.K.; Seybold, V.; de Groot, R.J. Specific, Sensitive, High-Resolution Detection of Protein Molecules in Eukaryotic Cells Using Metal-Tagging Transmission Electron Microscopy. Structure 2012, 20, 759–766. [Google Scholar] [CrossRef] [PubMed]
- Mandahar, C.L. Multiplication of RNA Plant Viruses; Springer: Berlin/Heidelberg, Germany, 2006. [Google Scholar]
- Nagy, P.D.; Pogany, J. Multiple Roles of Viral Replication Proteins in Plant RNA Virus Replication. In Plant Virology Protocols. Methods in Molecular Biology; Foster, G.D., Johansen, I.E., Hong, Y., Eds.; Humana Press: Totowa, NJ, USA, 2008. [Google Scholar]
- Verchot, J. Wrapping membranes around plant virus infection. Curr. Opin. Virol. 2011, 1, 388–395. [Google Scholar] [CrossRef] [PubMed]
- Romero-Brey, I.; Bartenschlager, R. Membranous Replication Factories Induced by Plus-Strand RNA Viruses. Viruses 2014, 6, 2826–2857. [Google Scholar] [CrossRef]
- Schaad, M.C.; Jensen, P.E.; Carrington, J.C. Formation of plant RNA virus replication complexes on membranes: Role of an endoplasmic reticulum-targeted viral protein. EMBO J. 1997, 16, 4049–4059. [Google Scholar] [CrossRef]
- Yi, G.; Kao, C. cis- and trans-Acting Functions of Brome Mosaic Virus Protein 1a in Genomic RNA1 Replication. J. Virol. 2007, 82, 3045–3053. [Google Scholar] [CrossRef][Green Version]
- Carette, J.E.; Stuiver, M.; van Lent, J.; Wellink, J.; van Kammen, A. Cowpea Mosaic Virus Infection Induces a Massive Proliferation of Endoplasmic Reticulum but Not Golgi Membranes and Is Dependent on De Novo Membrane Synthesis. J. Virol. 2000, 74, 6556–6563. [Google Scholar] [CrossRef]
- Han, S.; Sanfaçon, H. Tomato Ringspot Virus Proteins Containing the Nucleoside Triphosphate Binding Domain Are Transmembrane Proteins That Associate with the Endoplasmic Reticulum and Cofractionate with Replication Complexes. J. Virol. 2003, 77, 523–534. [Google Scholar] [CrossRef][Green Version]
- Bamunusinghe, D.; Hemenway, C.L.; Nelson, R.S.; Sanderfoot, A.A.; Ye, C.M.; Silva, M.A.; Payton, M.; Verchot-Lubicz, J. Analysis of potato virus X replicase and TGBp3 subcellular locations. Virology 2009, 393, 272–285. [Google Scholar] [CrossRef]
- Kawakami, S.; Watanabe, Y.; Beachy, R.N. Tobacco mosaic virus infection spreads cell to cell as intact replication complexes. Proc. Natl. Acad. Sci. USA 2004, 101, 6291–6296. [Google Scholar] [CrossRef]
- Nishikiori, M.; Dohi, K.; Mori, M.; Meshi, T.; Naito, S.; Ishikawa, M. Membrane-Bound Tomato Mosaic Virus Replication Proteins Participate in RNA Synthesis and Are Associated with Host Proteins in a Pattern Distinct from Those That Are Not Membrane Bound. J. Virol. 2006, 80, 8459–8468. [Google Scholar] [CrossRef] [PubMed]
- Restrepo-Hartwig, M.; Ahlquist, P. Brome Mosaic Virus RNA Replication Proteins 1a and 2a Colocalize and 1a Independently Localizes on the Yeast Endoplasmic Reticulum. J. Virol. 1999, 73, 10303–10309. [Google Scholar] [CrossRef] [PubMed]
- Paul, D.; Bartenschlager, R. Architecture and biogenesis of plus-strand RNA virus replication factories. World J. Virol. 2013, 2, 32–48. [Google Scholar] [CrossRef] [PubMed]
- Laliberté, J.-F.; Zheng, H. Viral Manipulation of Plant Host Membranes. Annu. Rev. Virol. 2014, 1, 237–259. [Google Scholar] [CrossRef]
- Kovalev, N.; Martín, I.F.D.C.; Pogany, J.; Barajas, D.; Pathak, K.; Risco, C.; Nagy, P.D. Role of Viral RNA and Co-opted Cellular ESCRT-I and ESCRT-III Factors in Formation of Tombusvirus Spherules Harboring the Tombusvirus Replicase. J. Virol. 2016, 90, 3611–3626. [Google Scholar] [CrossRef]
- Schwartz, M.; Chen, J.; Janda, M.; Sullivan, M.; Boon, J.D.; Ahlquist, P. A Positive-Strand RNA Virus Replication Complex Parallels Form and Function of Retrovirus Capsids. Mol. Cell 2002, 9, 505–514. [Google Scholar] [CrossRef]
- Cao, X.; Jin, X.; Zhang, X.; Li, Y.; Wang, C.; Wang, X.; Hong, J.; Wang, X.; Li, D.; Zhang, Y. Morphogenesis of Endoplasmic Reticulum Membrane-Invaginated Vesicles during Beet Black Scorch Virus Infection: Role of Auxiliary Replication Protein and New Implications of Three-Dimensional Architecture. J. Virol. 2015, 89, 6184–6195. [Google Scholar] [CrossRef]
- Boon, J.A.D.; Diaz, A.; Ahlquist, P. Cytoplasmic Viral Replication Complexes. Cell Host Microbe 2010, 8, 77–85. [Google Scholar] [CrossRef]
- Jin, X.; Cao, X.; Wang, X.; Jiang, J.; Wan, J.; Laliberté, J.-F.; Zhang, Y. Three-Dimensional Architecture and Biogenesis of Membrane Structures Associated with Plant Virus Replication. Front. Plant Sci. 2018, 9, 57. [Google Scholar] [CrossRef] [PubMed]
- Nagy, P.D.; Pogany, J. Yeast as a model host to dissect functions of viral and host factors in tombusvirus replication. Virology 2006, 344, 211–220. [Google Scholar] [CrossRef] [PubMed]
- Panavas, T.; Hawkins, C.M.; Panaviene, Z.; Nagy, P.D. The role of the p33:p33/p92 interaction domain in RNA replication and intracellular localization of p33 and p92 proteins of Cucumber necrosis tombusvirus. Virology 2005, 338, 81–95. [Google Scholar] [CrossRef] [PubMed]
- Serviene, E.; Shapka, N.; Cheng, C.-P.; Panavas, T.; Phuangrat, B.; Baker, J.; Nagy, P.D. Genome-wide screen identifies host genes affecting viral RNA recombination. Proc. Natl. Acad. Sci. USA 2005, 102, 10545–10550. [Google Scholar] [CrossRef]
- Serviene, E.; Jiang, Y.; Cheng, C.-P.; Baker, J.; Nagy, P.D. Screening of the Yeast yTHC Collection Identifies Essential Host Factors Affecting Tombusvirus RNA Recombination. J. Virol. 2006, 80, 1231–1241. [Google Scholar] [CrossRef]
- White, K.; Nagy, P.D. Advances in the Molecular Biology of Tombusviruses: Gene Expression, Genome Replication, and Recombination. Base Excision Repair 2004, 78, 187–226. [Google Scholar] [CrossRef]
- Appiano, A.; D’Agostino, G. Distribution of tomato bushy stunt virus in root tips of systemically infected Gomphrena globosa. J. Ultrastruct. Res. 1983, 85, 239–248. [Google Scholar] [CrossRef]
- Martelli, G.P.; Gallitelli, D.; Russo, M. Tombusviruses. In The Plant Viruses; Springer International Publishing: Cham, Switzerland, 1988; pp. 13–72. [Google Scholar]
- McCartney, A.W.; Greenwood, J.S.; Fabian, M.R.; White, K.A.; Mullen, R.T. Localization of the Tomato Bushy Stunt Virus Replication Protein p33 Reveals a Peroxisome-to-Endoplasmic Reticulum Sorting Pathway. Plant Cell 2005, 17, 3513–3531. [Google Scholar] [CrossRef] [PubMed]
- Barajas, D.; Martín, I.F.D.C.; Pogany, J.; Risco, C.; Nagy, P.D. Noncanonical Role for the Host Vps4 AAA+ ATPase ESCRT Protein in the Formation of Tomato Bushy Stunt Virus Replicase. PLoS Pathog. 2014, 10, e1004087. [Google Scholar] [CrossRef]
- De Castro, I.F.; Fernández, J.J.; Barajas, D.; Nagy, P.D.; Risco, C. Three-dimensional imaging of the intracellular assembly of a functional viral RNA replicase complex. J. Cell Sci. 2016, 130, 260–268. [Google Scholar] [CrossRef]
- Csaki, L.S.; Reue, K. Lipins: Multifunctional Lipid Metabolism Proteins. Annu. Rev. Nutr. 2010, 30, 257–272. [Google Scholar] [CrossRef]
- Chuang, C.; Barajas, D.; Qin, J.; Nagy, P.D. Inactivation of the Host Lipin Gene Accelerates RNA Virus Replication through Viral Exploitation of the Expanded Endoplasmic Reticulum Membrane. PLoS Pathog. 2014, 10, e1003944. [Google Scholar] [CrossRef] [PubMed]
- Lesemann, D.-E.; Adam, G. Electron microscopical and serological studies on four isometrical Pelargonium viruses. Acta Hortic. 1994, 377, 41–54. [Google Scholar] [CrossRef]
- Russo, M.; Martelli, G.P. Ultrastructure of turnip crinkle- and saguaro cactus virus-infected tissues. Virology 1982, 118, 109–116. [Google Scholar] [CrossRef]
- Blake, J.A.; Lee, K.W.; Morris, T.J.; Elthon, T.E. Effects of turnip crinkle virus infection on the structure and function of mitochondria and expression of stress proteins in turnips. Physiol. Plant. 2007, 129, 698–706. [Google Scholar] [CrossRef]
- Martínez-Turiño, S.; Hernández, C. Analysis of the subcellular targeting of the smaller replicase protein of Pelargonium flower break virus. Virus Res. 2012, 163, 580–591. [Google Scholar] [CrossRef]
- Navarro, B.; Rubino, L.; Russo, M. Expression of the Cymbidium Ringspot Virus 33-Kilodalton Protein in Saccharomyces cerevisiae and Molecular Dissection of the Peroxisomal Targeting Signal. J. Virol. 2004, 78, 4744–4752. [Google Scholar] [CrossRef][Green Version]
- Rubino, L.; di Franco, A.; Russo, M. Expression of a plant virus non-structural protein in Saccharomyces cerevisiae causes membrane proliferation and altered mitochondrial morphology. Microbiology 2000, 81, 279–286. [Google Scholar] [CrossRef]
- Rubino, L.; Weber-Lotfi, F.; Dietrich, A.; Stussi-Garaud, C.; Russo, M. The open reading frame 1-encoded (‘36K’) protein of Carnation Italian ringspot virus localizes to mitochondria. J. Gen. Virol. 2001, 82, 29–34. [Google Scholar] [CrossRef]
- Weber-Lotfi, F.; Dietrich, A.; Russo, M.; Rubino, L. Mitochondrial Targeting and Membrane Anchoring of a Viral Replicase in Plant and Yeast Cells. J. Virol. 2002, 76, 10485–10496. [Google Scholar] [CrossRef]
- Mochizuki, T.; Hirai, K.; Kanda, A.; Ohnishi, J.; Ohki, T.; Tsuda, S. Induction of necrosis via mitochondrial targeting of Melon necrotic spot virus replication protein p29 by its second transmembrane domain. Virology 2009, 390, 239–249. [Google Scholar] [CrossRef]
- Mueller, S.J.; Reski, R. Mitochondrial Dynamics and the ER: The Plant Perspective. Front. Cell Dev. Biol. 2015, 3, 78. [Google Scholar] [CrossRef]
- Beck, D.L.; Guilford, P.J.; Voot, D.M.; Andersen, M.T.; Forster, R.L. Triple gene block proteins of white clover mosaic potexvirus are required for transport. Virology 1991, 183, 695–702. [Google Scholar] [CrossRef]
- Wu, X.; Liu, J.; Chai, M.; Wang, J.; Li, D.; Wang, A.; Cheng, X. The Potato Virus X TGBp2 Protein Plays Dual Functional Roles in Viral Replication and Movement. J. Virol. 2018, 93, 1–21. [Google Scholar] [CrossRef] [PubMed]
- Aguilar, J.M.; Hernández-Gallardo, M.D.; Cenis, J.L.; Lacasa, A.; Aranda, M.A. Complete sequence of the Pepino mosaic virus RNA genome. Arch. Virol. 2002, 147, 2009–2015. [Google Scholar] [CrossRef]
- Restrepo-Hartwig, M.A.; Carrington, J.C. The tobacco etch potyvirus 6-kilodalton protein is membrane associated and involved in viral replication. J. Virol. 1994, 68, 2388–2397. [Google Scholar] [CrossRef]
- Cotton, S.; Grangeon, R.; Thivierge, K.; Mathieu, I.; Ide, C.; Wei, T.; Wang, A.; Laliberteé, J.-F. Turnip Mosaic Virus RNA Replication Complex Vesicles Are Mobile, Align with Microfilaments, and Are Each Derived from a Single Viral Genome. J. Virol. 2009, 83, 10460–10471. [Google Scholar] [CrossRef]
- Laliberté, J.-F.; Sanfaçon, H. Cellular Remodeling During Plant Virus Infection. Annu. Rev. Phytopathol. 2010, 48, 69–91. [Google Scholar] [CrossRef]
- Wei, T.; Huang, T.-S.; McNeil, J.; Laliberté, J.-F.; Hong, J.; Nelson, R.S.; Wang, A. Sequential Recruitment of the Endoplasmic Reticulum and Chloroplasts for Plant Potyvirus Replication. J. Virol. 2009, 84, 799–809. [Google Scholar] [CrossRef] [PubMed]
- Beauchemin, C.; Laliberteé, J.-F. The Poly(A) Binding Protein Is Internalized in Virus-Induced Vesicles or Redistributed to the Nucleolus during Turnip Mosaic Virus Infection. J. Virol. 2007, 81, 10905–10913. [Google Scholar] [CrossRef] [PubMed]
- Dufresne, P.J.; Thivierge, K.; Cotton, S.; Beauchemin, C.; Ide, C.; Ubalijoro, E.; Laliberté, J.-F.; Fortin, M.G. Heat shock 70 protein interaction with Turnip mosaic virus RNA-dependent RNA polymerase within virus-induced membrane vesicles. Virology 2008, 374, 217–227. [Google Scholar] [CrossRef] [PubMed]
- Huang, T.-S.; Wei, T.; Laliberté, J.-F.; Wang, A. A Host RNA Helicase-Like Protein, AtRH8, Interacts with the Potyviral Genome-Linked Protein, VPg, Associates with the Virus Accumulation Complex, and Is Essential for Infection. Plant Physiol. 2009, 152, 255–266. [Google Scholar] [CrossRef] [PubMed]
- Lõhmus, A.; Varjosalo, M.; Mäkinen, K. Protein composition of 6K2-induced membrane structures formed during Potato virus Ainfection. Mol. Plant Pathol. 2016, 17, 943–958. [Google Scholar] [CrossRef]
- Wei, T.; Zhang, C.; Hou, X.; Sanfaçon, H.; Wang, A. The SNARE Protein Syp71 Is Essential for Turnip Mosaic Virus Infection by Mediating Fusion of Virus-Induced Vesicles with Chloroplasts. PLoS Pathog. 2013, 9, e1003378. [Google Scholar] [CrossRef] [PubMed]
- Movahed, N.; Cabanillas, D.G.; Wan, J.; Vali, H.; Laliberté, J.-F.; Zheng, H. Turnip Mosaic Virus Components Are Released into the Extracellular Space by Vesicles in Infected Leaves. Plant Physiol. 2019, 180, 1375–1388. [Google Scholar] [CrossRef] [PubMed]
- Hafrén, A.; Lõhmus, A.; Mäkinen, K. Formation of Potato Virus A-Induced RNA Granules and Viral Translation Are Interrelated Processes Required for Optimal Virus Accumulation. PLoS Pathog. 2015, 11, e1005314. [Google Scholar] [CrossRef] [PubMed]
- Hafrén, A.; Üstün, S.; Hochmuth, A.; Svenning, S.; Johansen, T.; Hofius, D. Turnip Mosaic Virus Counteracts Selective Autophagy of the Viral Silencing Suppressor HCpro. Plant Physiol. 2018, 176, 649–662. [Google Scholar] [CrossRef]
- Wang, D.; Maule, A.J. Inhibition of Host Gene Expression Associated with Plant Virus Replication. Sciences 1995, 267, 229–231. [Google Scholar] [CrossRef] [PubMed]
- Escaler, M.; Aranda, M.A.; Thomas, C.L.; Maule, A.J. Pea Embryonic Tissues Show Common Responses to the Replication of a Wide Range of Viruses. Virology 2000, 267, 318–325. [Google Scholar] [CrossRef][Green Version]
- Blanc, S.; Uzest, M.; Drucker, M. New research horizons in vector-transmission of plant viruses. Curr. Opin. Microbiol. 2011, 14, 483–491. [Google Scholar] [CrossRef]
- Blanc, S.; Drucker, M.; Uzest, M. Localizing Viruses in Their Insect Vectors. Annu. Rev. Phytopathol. 2014, 52, 403–425. [Google Scholar] [CrossRef]
- Dietzgen, R.G.; Mann, K.S.; Johnson, K.N. Plant Virus–Insect Vector Interactions: Current and Potential Future Research Directions. Viruses 2016, 8, 303. [Google Scholar] [CrossRef]
- Dáder, B.; Then, C.; Berthelot, E.; Ducousso, M.; Ng, J.C.K.; Drucker, C.M. Insect transmission of plant viruses: Multilayered interactions optimize viral propagation. Insect Sci. 2017, 24, 929–946. [Google Scholar] [CrossRef]
- Hull, R. Mathews, Plant Virology, 4th ed.; Academic Press: Cambridge, MA, USA, 2002. [Google Scholar]
- Mink, G.I. Pollen and seed-transmited viruses and viroids. Annu. Rev. Phytopathol. 1993, 31, 375–402. [Google Scholar] [CrossRef] [PubMed]
- Broadbent, L. The epidemiology of tomato mosaic virus IX. Seed transmission of TMV. Ann. Appl. Biol. 1965, 62, 177–205. [Google Scholar] [CrossRef]
- Maule, A.J.; Wang, D. Seed transmission of plant viruses: A lesson in biological complexity. Trends Microbioliogy 1996, 4, 153–158. [Google Scholar] [CrossRef]
- Card, S.D.; Pearson, M.N.; Clover, G.R.G. Plant pathogens transmitted by pollen. Australas. Plant Pathol. 2007, 36, 455–461. [Google Scholar] [CrossRef]
- Wang, D.; Maule, A.J. Contrasting patterns in the spread of two seed-borne viruses in pea embryos. Plant J. 1997, 11, 1333–1340. [Google Scholar] [CrossRef]
- Aparicio, F.; Sánchez-Pina, M.A.; Sánchez-Navarro, J.A.; Pallás, V. Location of Prunus Necrotic Ringspot Ilarvirus Within Pollen Grains of Infected Nectarine Trees: Evidence from RT-PCR, Dot-blot and in situ Hybridisation. Eur. J. Plant Pathol. 1999, 105, 623–627. [Google Scholar] [CrossRef]
- Wang, D.; Maule, A.J. A Model for Seed Transmission of a Plant Virus: Genetic and Structural Analyses of Pea Embryo Invasion by Pea Seed-Borne Mosaic Virus. Plant Cell 1994, 6, 777–787. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sánchez Pina, M.A.; Gómez-Aix, C.; Méndez-López, E.; Gosalvez Bernal, B.; Aranda, M.A. Imaging Techniques to Study Plant Virus Replication and Vertical Transmission. Viruses 2021, 13, 358. https://doi.org/10.3390/v13030358
Sánchez Pina MA, Gómez-Aix C, Méndez-López E, Gosalvez Bernal B, Aranda MA. Imaging Techniques to Study Plant Virus Replication and Vertical Transmission. Viruses. 2021; 13(3):358. https://doi.org/10.3390/v13030358
Chicago/Turabian StyleSánchez Pina, María Amelia, Cristina Gómez-Aix, Eduardo Méndez-López, Blanca Gosalvez Bernal, and Miguel A. Aranda. 2021. "Imaging Techniques to Study Plant Virus Replication and Vertical Transmission" Viruses 13, no. 3: 358. https://doi.org/10.3390/v13030358
APA StyleSánchez Pina, M. A., Gómez-Aix, C., Méndez-López, E., Gosalvez Bernal, B., & Aranda, M. A. (2021). Imaging Techniques to Study Plant Virus Replication and Vertical Transmission. Viruses, 13(3), 358. https://doi.org/10.3390/v13030358






