A Look into Bunyavirales Genomes: Functions of Non-Structural (NS) Proteins
Abstract
1. Introduction
2. Family Peribunyaviridae
2.1. Functions of the NSs
2.2. Potential Roles of the NSm
3. Family Nairoviridae
3.1. Potential Roles of the NSs
3.2. Nairovirus-Specific NS Proteins, Mucin-Like Domain (MLD) and GP38
3.3. Viral Assembly and Infectivity Impacted by the NSm
4. Family Hantaviridae
Functions of the NSs
5. Family Phenuiviridae
5.1. Overview of RVFV NSs Roles in Virulence and Vaccine Development
5.2. The NSs Across Phenuiviridae
5.3. Potential Roles of the NSm
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Blitvich, B.J.; Beaty, B.J.; Blair, C.D.; Brault, A.C.; Dobler, G.; Drebot, M.A.; Haddow, A.D.; Kramer, L.D.; LaBeaud, A.D.; Monath, T.P.; et al. Bunyavirus Taxonomy: Limitations and Misconceptions Associated with the Current ICTV Criteria Used for Species Demarcation. Am. J. Trop. Med. Hyg. 2018, 99, 11–16. [Google Scholar] [CrossRef] [PubMed]
- Abudurexiti, A.; Adkins, S.; Alioto, D.; Alkhovsky, S.V.; Avsic-Zupanc, T.; Ballinger, M.J.; Bente, D.A.; Beer, M.; Bergeron, E.; Blair, C.D.; et al. Taxonomy of the order Bunyavirales: Update 2019. Arch. Virol. 2019, 164, 1949–1965. [Google Scholar] [CrossRef] [PubMed]
- CDC. Bunyaviridae; Centers for Disease Control and Prevention: Atlanta, GA, USA, 2013. [Google Scholar]
- Walter, C.T.; Barr, J.N. Recent advances in the molecular and cellular biology of bunyaviruses. J. Gen. Virol. 2011, 92, 2467–2484. [Google Scholar] [CrossRef]
- Radoshitzky, S.R.; Buchmeier, M.J.; Charrel, R.N.; Clegg, J.C.S.; Gonzalez, J.J.; Gunther, S.; Hepojoki, J.; Kuhn, J.H.; Lukashevich, I.S.; Romanowski, V.; et al. ICTV Virus Taxonomy Profile: Arenaviridae. J. Gen. Virol. 2019, 100, 1200–1201. [Google Scholar] [CrossRef] [PubMed]
- Wichgers Schreur, P.J.; Kormelink, R.; Kortekaas, J. Genome packaging of the Bunyavirales. Curr. Opin. Virol. 2018, 33, 151–155. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Li, J.; Gao, G.F.; Tien, P.; Liu, W. Bunyavirales ribonucleoproteins: The viral replication and transcription machinery. Crit. Rev. Microbiol. 2018, 44, 522–540. [Google Scholar] [CrossRef] [PubMed]
- Navarro, B.; Zicca, S.; Minutolo, M.; Saponari, M.; Alioto, D.; Di Serio, F. A Negative-Stranded RNA Virus Infecting Citrus Trees: The Second Member of a New Genus Within the Order Bunyavirales. Front. Microbiol. 2018, 9, 2340. [Google Scholar] [CrossRef]
- Kuhn, J.H.; Wiley, M.R.; Rodriguez, S.E.; Bao, Y.; Prieto, K.; Travassos da Rosa, A.P.; Guzman, H.; Savji, N.; Ladner, J.T.; Tesh, R.B.; et al. Genomic Characterization of the Genus Nairovirus (Family Bunyaviridae). Viruses 2016, 8, 164. [Google Scholar] [CrossRef] [PubMed]
- Akopyants, N.S.; Lye, L.F.; Dobson, D.E.; Lukes, J.; Beverley, S.M. A Novel Bunyavirus-Like Virus of Trypanosomatid Protist Parasites. Genome Announc. 2016, 4. [Google Scholar] [CrossRef]
- Herath, V.; Romay, G.; Urrutia, C.D.; Verchot, J. Family Level Phylogenies Reveal Relationships of Plant Viruses within the Order Bunyavirales. Viruses 2020, 12, 10. [Google Scholar] [CrossRef]
- Garrison, A.R.; Alkhovsky, S.V.; Avsic-Zupanc, T.; Bente, D.A.; Bergeron, E.; Burt, F.; Di Paola, N.; Ergunay, K.; Hewson, R.; Kuhn, J.H.; et al. ICTV Virus Taxonomy Profile: Nairoviridae. J. Gen. Virol. 2020, 101, 798–799. [Google Scholar] [CrossRef] [PubMed]
- Hughes, H.R.; Adkins, S.; Alkhovskiy, S.; Beer, M.; Blair, C.; Calisher, C.H.; Drebot, M.; Lambert, A.J.; de Souza, W.M.; Marklewitz, M.; et al. ICTV Virus Taxonomy Profile: Peribunyaviridae. J. Gen. Virol. 2020, 101, 1–2. [Google Scholar] [CrossRef] [PubMed]
- Ejiri, H.; Lim, C.K.; Isawa, H.; Yamaguchi, Y.; Fujita, R.; Takayama-Ito, M.; Kuwata, R.; Kobayashi, D.; Horiya, M.; Posadas-Herrera, G.; et al. Isolation and characterization of Kabuto Mountain virus, a new tick-borne phlebovirus from Haemaphysalis flava ticks in Japan. Virus Res. 2018, 244, 252–261. [Google Scholar] [CrossRef] [PubMed]
- Kafer, S.; Paraskevopoulou, S.; Zirkel, F.; Wieseke, N.; Donath, A.; Petersen, M.; Jones, T.C.; Liu, S.; Zhou, X.; Middendorf, M.; et al. Re-assessing the diversity of negative strand RNA viruses in insects. Plos Pathog. 2019, 15, e1008224. [Google Scholar] [CrossRef] [PubMed]
- Laenen, L.; Vergote, V.; Calisher, C.H.; Klempa, B.; Klingstrom, J.; Kuhn, J.H.; Maes, P. Hantaviridae: Current Classification and Future Perspectives. Viruses 2019, 11, 788. [Google Scholar] [CrossRef]
- Vaheri, A.; Strandin, T.; Hepojoki, J.; Sironen, T.; Henttonen, H.; Makela, S.; Mustonen, J. Uncovering the mysteries of hantavirus infections. Nat. Rev. Microbiol. 2013, 11, 539–550. [Google Scholar] [CrossRef]
- Welch, S.R.; Scholte, F.E.M.; Spengler, J.R.; Ritter, J.M.; Coleman-McCray, J.D.; Harmon, J.R.; Nichol, S.T.; Zaki, S.R.; Spiropoulou, C.F.; Bergeron, E. The Crimean-Congo Hemorrhagic Fever Virus NSm Protein is Dispensable for Growth In Vitro and Disease in Ifnar(-/-) Mice. Microorganisms 2020, 8, 775. [Google Scholar] [CrossRef]
- Wuerth, J.D.; Weber, F. Phleboviruses and the Type I Interferon Response. Viruses 2016, 8, 174. [Google Scholar] [CrossRef]
- Dutuze, M.F.; Nzayirambaho, M.; Mores, C.N.; Christofferson, R.C. A Review of Bunyamwera, Batai, and Ngari Viruses: Understudied Orthobunyaviruses With Potential One Health Implications. Front. Vet. Sci. 2018, 5, 69. [Google Scholar] [CrossRef]
- Li, C.X.; Shi, M.; Tian, J.H.; Lin, X.D.; Kang, Y.J.; Chen, L.J.; Qin, X.C.; Xu, J.; Holmes, E.C.; Zhang, Y.Z. Unprecedented genomic diversity of RNA viruses in arthropods reveals the ancestry of negative-sense RNA viruses. Elife 2015, 4, e05378. [Google Scholar] [CrossRef] [PubMed]
- Marklewitz, M.; Zirkel, F.; Rwego, I.B.; Heidemann, H.; Trippner, P.; Kurth, A.; Kallies, R.; Briese, T.; Lipkin, W.I.; Drosten, C.; et al. Discovery of a unique novel clade of mosquito-associated bunyaviruses. J. Virol. 2013, 87, 12850–12865. [Google Scholar] [CrossRef] [PubMed]
- Kopp, A.; Hubner, A.; Zirkel, F.; Hobelsberger, D.; Estrada, A.; Jordan, I.; Gillespie, T.R.; Drosten, C.; Junglen, S. Detection of Two Highly Diverse Peribunyaviruses in Mosquitoes from Palenque, Mexico. Viruses 2019, 11, 832. [Google Scholar] [CrossRef] [PubMed]
- Elliott, R.M. Orthobunyaviruses: Recent genetic and structural insights. Nat. Rev. Microbiol. 2014, 12, 673–685. [Google Scholar] [CrossRef] [PubMed]
- Bridgen, A.; Weber, F.; Fazakerley, J.K.; Elliott, R.M. Bunyamwera bunyavirus nonstructural protein NSs is a nonessential gene product that contributes to viral pathogenesis. Proc. Natl. Acad. Sci. USA 2001, 98, 664–669. [Google Scholar] [CrossRef] [PubMed]
- Shchetinin, A.M.; Lvov, D.K.; Deriabin, P.G.; Botikov, A.G.; Gitelman, A.K.; Kuhn, J.H.; Alkhovsky, S.V. Genetic and Phylogenetic Characterization of Tataguine and Witwatersrand Viruses and Other Orthobunyaviruses of the Anopheles A, Capim, Guama, Koongol, Mapputta, Tete, and Turlock Serogroups. Viruses 2015, 7, 5987–6008. [Google Scholar] [CrossRef]
- Mohamed, M.; McLees, A.; Elliott, R.M. Viruses in the Anopheles A, Anopheles B, and Tete serogroups in the Orthobunyavirus genus (family Bunyaviridae) do not encode an NSs protein. J. Virol. 2009, 83, 7612–7618. [Google Scholar] [CrossRef] [PubMed]
- Thomas, D.; Blakqori, G.; Wagner, V.; Banholzer, M.; Kessler, N.; Elliott, R.M.; Haller, O.; Weber, F. Inhibition of RNA polymerase II phosphorylation by a viral interferon antagonist. J. Biol. Chem. 2004, 279, 31471–31477. [Google Scholar] [CrossRef]
- Verbruggen, P.; Ruf, M.; Blakqori, G.; Overby, A.K.; Heidemann, M.; Eick, D.; Weber, F. Interferon antagonist NSs of La Crosse virus triggers a DNA damage response-like degradation of transcribing RNA polymerase II. J. Biol. Chem. 2011, 286, 3681–3692. [Google Scholar] [CrossRef]
- Fan, X.; Jin, T. Structures of RIG-I-Like Receptors and Insights into Viral RNA Sensing. Adv. Exp. Med. Biol. 2019, 1172, 157–188. [Google Scholar] [CrossRef]
- Dias Junior, A.G.; Sampaio, N.G.; Rehwinkel, J. A Balancing Act: MDA5 in Antiviral Immunity and Autoinflammation. Trends Microbiol. 2019, 27, 75–85. [Google Scholar] [CrossRef]
- Carlton-Smith, C.; Elliott, R.M. Viperin, MTAP44, and protein kinase R contribute to the interferon-induced inhibition of Bunyamwera Orthobunyavirus replication. J. Virol. 2012, 86, 11548–11557. [Google Scholar] [CrossRef]
- Leonard, V.H.; Kohl, A.; Hart, T.J.; Elliott, R.M. Interaction of Bunyamwera Orthobunyavirus NSs protein with mediator protein MED8: A mechanism for inhibiting the interferon response. J. Virol. 2006, 80, 9667–9675. [Google Scholar] [CrossRef]
- de Melo, A.B.J.; de Souza, W.M.; Acrani, G.O.; Carvalho, V.L.; Romeiro, M.F.; Tolardo, A.L.; da Silva, S.P.; Cardoso, J.F.; de Oliveira Chiang, J.; da Silva Goncalves Vianez, J.L.J.; et al. Genomic characterization and evolution of Tacaiuma orthobunyavirus (Peribunyaviridae family) isolated in Brazil. Infect. Genet. Evol. 2018, 60, 71–76. [Google Scholar] [CrossRef] [PubMed]
- Colon-Ramos, D.A.; Irusta, P.M.; Gan, E.C.; Olson, M.R.; Song, J.; Morimoto, R.I.; Elliott, R.M.; Lombard, M.; Hollingsworth, R.; Hardwick, J.M.; et al. Inhibition of translation and induction of apoptosis by Bunyaviral nonstructural proteins bearing sequence similarity to reaper. Mol. Biol. Cell 2003, 14, 4162–4172. [Google Scholar] [CrossRef] [PubMed]
- Vasudevan, D.; Ryoo, H.D. Regulation of Cell Death by IAPs and Their Antagonists. Curr. Top. Dev. Biol. 2015, 114, 185–208. [Google Scholar] [CrossRef]
- Blakqori, G.; Weber, F. Efficient cDNA-based rescue of La Crosse bunyaviruses expressing or lacking the nonstructural protein NSs. J. Virol. 2005, 79, 10420–10428. [Google Scholar] [CrossRef]
- Kohl, A.; Clayton, R.F.; Weber, F.; Bridgen, A.; Randall, R.E.; Elliott, R.M. Bunyamwera virus nonstructural protein NSs counteracts interferon regulatory factor 3-mediated induction of early cell death. J. Virol. 2003, 77, 7999–8008. [Google Scholar] [CrossRef]
- Fazakerley, J.K.; Gonzalez-Scarano, F.; Strickler, J.; Dietzschold, B.; Karush, F.; Nathanson, N. Organization of the middle RNA segment of snowshoe hare Bunyavirus. Virology 1988, 167, 422–432. [Google Scholar] [CrossRef]
- Shi, X.; Botting, C.H.; Li, P.; Niglas, M.; Brennan, B.; Shirran, S.L.; Szemiel, A.M.; Elliott, R.M. Bunyamwera orthobunyavirus glycoprotein precursor is processed by cellular signal peptidase and signal peptide peptidase. Proc. Natl. Acad. Sci. USA 2016, 113, 8825–8830. [Google Scholar] [CrossRef]
- Shi, X.; Kohl, A.; Leonard, V.H.; Li, P.; McLees, A.; Elliott, R.M. Requirement of the N-terminal region of orthobunyavirus nonstructural protein NSm for virus assembly and morphogenesis. J. Virol. 2006, 80, 8089–8099. [Google Scholar] [CrossRef]
- Fontana, J.; Lopez-Montero, N.; Elliott, R.M.; Fernandez, J.J.; Risco, C. The unique architecture of Bunyamwera virus factories around the Golgi complex. Cell Microbiol. 2008, 10, 2012–2028. [Google Scholar] [CrossRef] [PubMed]
- Ishihara, Y.; Shioda, C.; Bangphoomi, N.; Sugiura, K.; Saeki, K.; Tsuda, S.; Iwanaga, T.; Takenaka-Uema, A.; Kato, K.; Murakami, S.; et al. Akabane virus nonstructural protein NSm regulates viral growth and pathogenicity in a mouse model. J. Vet. Med. Sci. 2016, 78, 1391–1397. [Google Scholar] [CrossRef] [PubMed]
- Bergeron, E.; Zivcec, M.; Chakrabarti, A.K.; Nichol, S.T.; Albarino, C.G.; Spiropoulou, C.F. Recovery of Recombinant Crimean Congo Hemorrhagic Fever Virus Reveals a Function for Non-structural Glycoproteins Cleavage by Furin. PLoS Pathog. 2015, 11, e1004879. [Google Scholar] [CrossRef] [PubMed]
- Burt, F.J.; Spencer, D.C.; Leman, P.A.; Patterson, B.; Swanepoel, R. Investigation of tick-borne viruses as pathogens of humans in South Africa and evidence of Dugbe virus infection in a patient with prolonged thrombocytopenia. Epidemiol. Infect. 1996, 116, 353–361. [Google Scholar] [CrossRef] [PubMed]
- Marczinke, B.I.; Nichol, S.T. Nairobi sheep disease virus, an important tick-borne pathogen of sheep and goats in Africa, is also present in Asia. Virology 2002, 303, 146–151. [Google Scholar] [CrossRef]
- Walker, P.J.; Widen, S.G.; Firth, C.; Blasdell, K.R.; Wood, T.G.; Travassos da Rosa, A.P.; Guzman, H.; Tesh, R.B.; Vasilakis, N. Genomic Characterization of Yogue, Kasokero, Issyk-Kul, Keterah, Gossas, and Thiafora Viruses: Nairoviruses Naturally Infecting Bats, Shrews, and Ticks. Am. J. Trop. Med. Hyg. 2015, 93, 1041–1051. [Google Scholar] [CrossRef]
- Goedhals, D.; Bester, P.A.; Paweska, J.T.; Swanepoel, R.; Burt, F.J. Comparative analysis of the L, M, and S RNA segments of Crimean-Congo haemorrhagic fever virus isolates from southern Africa. J. Med. Virol. 2015, 87, 717–724. [Google Scholar] [CrossRef] [PubMed]
- Spengler, J.R.; Bente, D.A. Therapeutic intervention in Crimean-Congo hemorrhagic fever: Where are we now? Future Virol. 2015, 10, 203–206. [Google Scholar] [CrossRef]
- Barnwal, B.; Karlberg, H.; Mirazimi, A.; Tan, Y.J. The Non-structural Protein of Crimean-Congo Hemorrhagic Fever Virus Disrupts the Mitochondrial Membrane Potential and Induces Apoptosis. J. Biol. Chem. 2016, 291, 582–592. [Google Scholar] [CrossRef] [PubMed]
- Hewson, R.; Chamberlain, J.; Mioulet, V.; Lloyd, G.; Jamil, B.; Hasan, R.; Gmyl, A.; Gmyl, L.; Smirnova, S.E.; Lukashev, A.; et al. Crimean-Congo haemorrhagic fever virus: Sequence analysis of the small RNA segments from a collection of viruses world wide. Virus Res. 2004, 102, 185–189. [Google Scholar] [CrossRef]
- Fuller, J.; Surtees, R.A.; Shaw, A.B.; Alvarez-Rodriguez, B.; Slack, G.S.; Bell-Sakyi, L.; Mankouri, J.; Edwards, T.A.; Hewson, R.; Barr, J.N. Hazara nairovirus elicits differential induction of apoptosis and nucleocapsid protein cleavage in mammalian and tick cells. J. Gen. Virol. 2019, 100, 392–402. [Google Scholar] [CrossRef]
- Karlberg, H.; Tan, Y.J.; Mirazimi, A. Induction of caspase activation and cleavage of the viral nucleocapsid protein in different cell types during Crimean-Congo hemorrhagic fever virus infection. J. Biol. 2011, 286, 3227–3234. [Google Scholar] [CrossRef]
- Karlberg, H.; Tan, Y.J.; Mirazimi, A. Crimean-Congo haemorrhagic fever replication interplays with regulation mechanisms of apoptosis. J. Gen. Virol. 2015, 96, 538–546. [Google Scholar] [CrossRef] [PubMed]
- Hawman, D.W.; Meade-White, K.; Leventhal, S.; Feldmann, F.; Okumura, A.; Smith, B.; Scott, D.; Feldmann, H. Immunocompetent mouse model for Crimean-Congo hemorrhagic fever virus. Elife 2021, 10, e63906. [Google Scholar] [CrossRef]
- Mishra, A.K.; Moyer, C.L.; Abelson, D.M.; Deer, D.J.; El Omari, K.; Duman, R.; Lobel, L.; Lutwama, J.J.; Dye, J.M.; Wagner, A.; et al. Structure and Characterization of Crimean-Congo Hemorrhagic Fever Virus GP38. J. Virol. 2020, 94. [Google Scholar] [CrossRef]
- Kuhn, J.H.; Amarasinghe, G.K.; Basler, C.F.; Bavari, S.; Bukreyev, A.; Chandran, K.; Crozier, I.; Dolnik, O.; Dye, J.M.; Formenty, P.B.H.; et al. ICTV Virus Taxonomy Profile: Filoviridae. J. Gen. Virol. 2019, 100, 911–912. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.Y.; Duckers, H.J.; Sullivan, N.J.; Sanchez, A.; Nabel, E.G.; Nabel, G.J. Identification of the Ebola virus glycoprotein as the main viral determinant of vascular cell cytotoxicity and injury. Nat. Med. 2000, 6, 886–889. [Google Scholar] [CrossRef] [PubMed]
- Freitas, N.; Enguehard, M.; Denolly, S.; Levy, C.; Neveu, G.; Lerolle, S.; Devignot, S.; Weber, F.; Bergeron, E.; Legros, V.; et al. The interplays between Crimean-Congo hemorrhagic fever virus (CCHFV) M segment-encoded accessory proteins and structural proteins promote virus assembly and infectivity. PLoS Pathog. 2020, 16, e1008850. [Google Scholar] [CrossRef] [PubMed]
- Golden, J.W.; Shoemaker, C.J.; Lindquist, M.E.; Zeng, X.; Daye, S.P.; Williams, J.A.; Liu, J.; Coffin, K.M.; Olschner, S.; Flusin, O.; et al. GP38-targeting monoclonal antibodies protect adult mice against lethal Crimean-Congo hemorrhagic fever virus infection. Sci. Adv. 2019, 5, eaaw9535. [Google Scholar] [CrossRef]
- Altamura, L.A.; Bertolotti-Ciarlet, A.; Paragas, J.; Schmaljohn, C.S.; Doms, R.W. Identification of a novel C-terminal cleavage of Crimean-Congo hemorrhagic fever virus PreGN that leads to generation of an NSM protein. J. Virol. 2007, 81, 6632–6642. [Google Scholar] [CrossRef] [PubMed]
- Sanchez, A.J.; Vincent, M.J.; Nichol, S.T. Characterization of the glycoproteins of Crimean-Congo hemorrhagic fever virus. J. Virol. 2002, 76, 7263–7275. [Google Scholar] [CrossRef] [PubMed]
- Dilcher, M.; Koch, A.; Hasib, L.; Dobler, G.; Hufert, F.T.; Weidmann, M. Genetic characterization of Erve virus, a European Nairovirus distantly related to Crimean-Congo hemorrhagic fever virus. Virus Genes 2012, 45, 426–432. [Google Scholar] [CrossRef]
- Ramanathan, H.N.; Jonsson, C.B. New and Old World hantaviruses differentially utilize host cytoskeletal components during their life cycles. Virology 2008, 374, 138–150. [Google Scholar] [CrossRef]
- Binder, F.; Ryll, R.; Drewes, S.; Jagdmann, S.; Reil, D.; Hiltbrunner, M.; Rosenfeld, U.M.; Imholt, C.; Jacob, J.; Heckel, G.; et al. Spatial and Temporal Evolutionary Patterns in Puumala Orthohantavirus (PUUV) S Segment. Pathogens 2020, 9, 548. [Google Scholar] [CrossRef]
- Mittler, E.; Dieterle, M.E.; Kleinfelter, L.M.; Slough, M.M.; Chandran, K.; Jangra, R.K. Hantavirus entry: Perspectives and recent advances. Adv. Virus Res. 2019, 104, 185–224. [Google Scholar] [CrossRef] [PubMed]
- Vera-Otarola, J.; Solis, L.; Soto-Rifo, R.; Ricci, E.P.; Pino, K.; Tischler, N.D.; Ohlmann, T.; Darlix, J.L.; Lopez-Lastra, M. The Andes hantavirus NSs protein is expressed from the viral small mRNA by a leaky scanning mechanism. J. Virol. 2012, 86, 2176–2187. [Google Scholar] [CrossRef]
- Virtanen, J.O.; Jaaskelainen, K.M.; Djupsjobacka, J.; Vaheri, A.; Plyusnin, A. Tula hantavirus NSs protein accumulates in the perinuclear area in infected and transfected cells. Arch. Virol. 2010, 155, 117–121. [Google Scholar] [CrossRef]
- Jaaskelainen, K.M.; Kaukinen, P.; Minskaya, E.S.; Plyusnina, A.; Vapalahti, O.; Elliott, R.M.; Weber, F.; Vaheri, A.; Plyusnin, A. Tula and Puumala hantavirus NSs ORFs are functional and the products inhibit activation of the interferon-beta promoter. J. Med. Virol. 2007, 79, 1527–1536. [Google Scholar] [CrossRef] [PubMed]
- Ronnberg, T.; Jaaskelainen, K.; Blot, G.; Parviainen, V.; Vaheri, A.; Renkonen, R.; Bouloy, M.; Plyusnin, A. Searching for cellular partners of hantaviral nonstructural protein NSs: Y2H screening of mouse cDNA library and analysis of cellular interactome. PLoS ONE 2012, 7, e34307. [Google Scholar] [CrossRef] [PubMed]
- Boshra, H.; Lorenzo, G.; Busquets, N.; Brun, A. Rift valley fever: Recent insights into pathogenesis and prevention. J. Virol. 2011, 85, 6098–6105. [Google Scholar] [CrossRef] [PubMed]
- Ly, H.J.; Ikegami, T. Rift Valley fever virus NSs protein functions and the similarity to other bunyavirus NSs proteins. Virol. J. 2016, 13, 118. [Google Scholar] [CrossRef] [PubMed]
- Lihoradova, O.; Ikegami, T. Countermeasure development for Rift Valley fever: Deletion, modification or targeting of major virulence factor NSs. Future Virol. 2014, 9, 27–39. [Google Scholar] [CrossRef] [PubMed]
- Copeland, A.M.; Van Deusen, N.M.; Schmaljohn, C.S. Rift Valley fever virus NSS gene expression correlates with a defect in nuclear mRNA export. Virology 2015, 486, 88–93. [Google Scholar] [CrossRef] [PubMed]
- Narayanan, A.; Amaya, M.; Voss, K.; Chung, M.; Benedict, A.; Sampey, G.; Kehn-Hall, K.; Luchini, A.; Liotta, L.; Bailey, C.; et al. Reactive oxygen species activate NFkappaB (p65) and p53 and induce apoptosis in RVFV infected liver cells. Virology 2014, 449, 270–286. [Google Scholar] [CrossRef]
- Habjan, M.; Pichlmair, A.; Elliott, R.M.; Overby, A.K.; Glatter, T.; Gstaiger, M.; Superti-Furga, G.; Unger, H.; Weber, F. NSs protein of rift valley fever virus induces the specific degradation of the double-stranded RNA-dependent protein kinase. J. Virol. 2009, 83, 4365–4375. [Google Scholar] [CrossRef] [PubMed]
- de la Fuente, C.; Pinkham, C.; Dabbagh, D.; Beitzel, B.; Garrison, A.; Palacios, G.; Hodge, K.A.; Petricoin, E.F.; Schmaljohn, C.; Campbell, C.E.; et al. Phosphoproteomic analysis reveals Smad protein family activation following Rift Valley fever virus infection. PLoS ONE 2018, 13, e0191983. [Google Scholar] [CrossRef]
- Lau, S.; Weber, F. Nuclear pore protein Nup98 is involved in replication of Rift Valley fever virus and nuclear import of virulence factor NSs. J. Gen. Virol. 2020, 101, 712–716. [Google Scholar] [CrossRef] [PubMed]
- Kohl, A.; di Bartolo, V.; Bouloy, M. The Rift Valley fever virus nonstructural protein NSs is phosphorylated at serine residues located in casein kinase II consensus motifs in the carboxy-terminus. Virology 1999, 263, 517–525. [Google Scholar] [CrossRef]
- Terasaki, K.; Ramirez, S.I.; Makino, S. Mechanistic Insight into the Host Transcription Inhibition Function of Rift Valley Fever Virus NSs and Its Importance in Virulence. PLoS Negl. Trop. Dis. 2016, 10, e0005047. [Google Scholar] [CrossRef]
- Austin, D.; Baer, A.; Lundberg, L.; Shafagati, N.; Schoonmaker, A.; Narayanan, A.; Popova, T.; Panthier, J.J.; Kashanchi, F.; Bailey, C.; et al. p53 Activation following Rift Valley fever virus infection contributes to cell death and viral production. PLoS ONE 2012, 7, e36327. [Google Scholar] [CrossRef]
- Bamia, A.; Marcato, V.; Boissiere, M.; Mansuroglu, Z.; Tamietti, C.; Romani, M.; Simon, D.; Tian, G.; Niedergang, F.; Panthier, J.J.; et al. The NSs protein encoded by the virulent strain of Rift Valley fever virus targets the expression of Abl2 and the actin cytoskeleton of the host affecting cell mobility, cell shape and cell-cell adhesion. J. Virol. 2020, 95. [Google Scholar] [CrossRef]
- Vialat, P.; Billecocq, A.; Kohl, A.; Bouloy, M. The S segment of rift valley fever phlebovirus (Bunyaviridae) carries determinants for attenuation and virulence in mice. J. Virol. 2000, 74, 1538–1543. [Google Scholar] [CrossRef] [PubMed]
- Monteiro, G.E.R.; Jansen van Vuren, P.; Wichgers Schreur, P.J.; Odendaal, L.; Clift, S.J.; Kortekaas, J.; Paweska, J.T. Mutation of adjacent cysteine residues in the NSs protein of Rift Valley fever virus results in loss of virulence in mice. Virus Res. 2018, 249, 31–44. [Google Scholar] [CrossRef] [PubMed]
- Roberts, K.K.; Hill, T.E.; Davis, M.N.; Holbrook, M.R.; Freiberg, A.N. Cytokine response in mouse bone marrow derived macrophages after infection with pathogenic and non-pathogenic Rift Valley fever virus. J. Gen. Virol. 2015, 96, 1651–1663. [Google Scholar] [CrossRef] [PubMed]
- Pinkham, C.; An, S.; Lundberg, L.; Bansal, N.; Benedict, A.; Narayanan, A.; Kehn-Hall, K. The role of signal transducer and activator of transcription 3 in Rift Valley fever virus infection. Virology 2016, 496, 175–185. [Google Scholar] [CrossRef] [PubMed]
- Vaughn, V.M.; Streeter, C.C.; Miller, D.J.; Gerrard, S.R. Restriction of rift valley Fever virus virulence in mosquito cells. Viruses 2010, 2, 655–675. [Google Scholar] [CrossRef]
- Mroz, C.; Schmidt, K.M.; Reiche, S.; Groschup, M.H.; Eiden, M. Development of monoclonal antibodies to Rift Valley Fever Virus and their application in antigen detection and indirect immunofluorescence. J. Immunol. Methods 2018, 460, 36–44. [Google Scholar] [CrossRef]
- Leger, P.; Lara, E.; Jagla, B.; Sismeiro, O.; Mansuroglu, Z.; Coppee, J.Y.; Bonnefoy, E.; Bouloy, M. Dicer-2- and Piwi-mediated RNA interference in Rift Valley fever virus-infected mosquito cells. J. Virol. 2013, 87, 1631–1648. [Google Scholar] [CrossRef]
- Fernandez, J.C.; Billecocq, A.; Durand, J.P.; Cetre-Sossah, C.; Cardinale, E.; Marianneau, P.; Pepin, M.; Tordo, N.; Bouloy, M. The nonstructural protein NSs induces a variable antibody response in domestic ruminants naturally infected with Rift Valley fever virus. Clin. Vaccine Immunol. 2012, 19, 5–10. [Google Scholar] [CrossRef]
- Moutailler, S.; Krida, G.; Madec, Y.; Bouloy, M.; Failloux, A.B. Replication of Clone 13, a naturally attenuated avirulent isolate of Rift Valley fever virus, in Aedes and Culex mosquitoes. Vector Borne Zoonotic Dis. 2010, 10, 681–688. [Google Scholar] [CrossRef]
- Muller, R.; Saluzzo, J.F.; Lopez, N.; Dreier, T.; Turell, M.; Smith, J.; Bouloy, M. Characterization of clone 13, a naturally attenuated avirulent isolate of Rift Valley fever virus, which is altered in the small segment. Am. J. Trop. Med. Hyg. 1995, 53, 405–411. [Google Scholar] [CrossRef]
- Kortekaas, J.; Oreshkova, N.; van Keulen, L.; Kant, J.; Bosch, B.J.; Bouloy, M.; Moulin, V.; Goovaerts, D.; Moormann, R.J. Comparative efficacy of two next-generation Rift Valley fever vaccines. Vaccine 2014, 32, 4901–4908. [Google Scholar] [CrossRef] [PubMed]
- Gowen, B.B.; Westover, J.B.; Sefing, E.J.; Bailey, K.W.; Nishiyama, S.; Wandersee, L.; Scharton, D.; Jung, K.H.; Ikegami, T. MP-12 virus containing the clone 13 deletion in the NSs gene prevents lethal disease when administered after Rift Valley fever virus infection in hamsters. Front. Microbiol. 2015, 6, 651. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Vialat, P.; Muller, R.; Vu, T.H.; Prehaud, C.; Bouloy, M. Mapping of the mutations present in the genome of the Rift Valley fever virus attenuated MP12 strain and their putative role in attenuation. Virus Res. 1997, 52, 43–50. [Google Scholar] [CrossRef]
- Gowen, B.B.; Bailey, K.W.; Scharton, D.; Vest, Z.; Westover, J.B.; Skirpstunas, R.; Ikegami, T. Post-exposure vaccination with MP-12 lacking NSs protects mice against lethal Rift Valley fever virus challenge. Antivir. Res. 2013, 98, 135–143. [Google Scholar] [CrossRef] [PubMed]
- Kalveram, B.; Lihoradova, O.; Indran, S.V.; Ikegami, T. Using reverse genetics to manipulate the NSs gene of the Rift Valley fever virus MP-12 strain to improve vaccine safety and efficacy. J. Vis. Exp. 2011, e3400. [Google Scholar] [CrossRef]
- Terasaki, K.; Juelich, T.L.; Smith, J.K.; Kalveram, B.; Perez, D.D.; Freiberg, A.N.; Makino, S. A single-cycle replicable Rift Valley fever phlebovirus vaccine carrying a mutated NSs confers full protection from lethal challenge in mice. Sci. Rep. 2018, 8, 17097. [Google Scholar] [CrossRef]
- Murakami, S.; Terasaki, K.; Makino, S. Generation of a Single-Cycle Replicable Rift Valley Fever Vaccine. Methods Mol. Biol. 2016, 1403, 187–206. [Google Scholar] [CrossRef]
- Ikegami, T.; Won, S.; Peters, C.J.; Makino, S. Rescue of infectious rift valley fever virus entirely from cDNA, analysis of virus lacking the NSs gene, and expression of a foreign gene. J. Virol. 2006, 80, 2933–2940. [Google Scholar] [CrossRef]
- Bird, B.H.; Maartens, L.H.; Campbell, S.; Erasmus, B.J.; Erickson, B.R.; Dodd, K.A.; Spiropoulou, C.F.; Cannon, D.; Drew, C.P.; Knust, B.; et al. Rift Valley fever virus vaccine lacking the NSs and NSm genes is safe, nonteratogenic, and confers protection from viremia, pyrexia, and abortion following challenge in adult and pregnant sheep. J. Virol. 2011, 85, 12901–12909. [Google Scholar] [CrossRef]
- Bird, B.H.; Albarino, C.G.; Hartman, A.L.; Erickson, B.R.; Ksiazek, T.G.; Nichol, S.T. Rift valley fever virus lacking the NSs and NSm genes is highly attenuated, confers protective immunity from virulent virus challenge, and allows for differential identification of infected and vaccinated animals. J. Virol. 2008, 82, 2681–2691. [Google Scholar] [CrossRef] [PubMed]
- Smith, D.R.; Johnston, S.C.; Piper, A.; Botto, M.; Donnelly, G.; Shamblin, J.; Albarino, C.G.; Hensley, L.E.; Schmaljohn, C.; Nichol, S.T.; et al. Attenuation and efficacy of live-attenuated Rift Valley fever virus vaccine candidates in non-human primates. PLoS Negl. Trop. Dis. 2018, 12, e0006474. [Google Scholar] [CrossRef] [PubMed]
- Wichgers Schreur, P.J.; Kant, J.; van Keulen, L.; Moormann, R.J.; Kortekaas, J. Four-segmented Rift Valley fever virus induces sterile immunity in sheep after a single vaccination. Vaccine 2015, 33, 1459–1464. [Google Scholar] [CrossRef]
- Keck, F.; Amaya, M.; Kehn-Hall, K.; Roberts, B.; Bailey, C.; Narayanan, A. Characterizing the effect of Bortezomib on Rift Valley Fever Virus multiplication. Antivir. Res. 2015, 120, 48–56. [Google Scholar] [CrossRef]
- Narayanan, A.; Kehn-Hall, K.; Senina, S.; Lundberg, L.; Van Duyne, R.; Guendel, I.; Das, R.; Baer, A.; Bethel, L.; Turell, M.; et al. Curcumin inhibits Rift Valley fever virus replication in human cells. J. Biol. Chem. 2012, 287, 33198–33214. [Google Scholar] [CrossRef]
- Sall, A.A.; de A Zanotto, P.M.; Zeller, H.G.; Digoutte, J.P.; Thiongane, Y.; Bouloy, M. Variability of the NS(S) protein among Rift Valley fever virus isolates. J. Gen. Virol. 1997, 78 Pt 11, 2853–2858. [Google Scholar] [CrossRef]
- Giorgi, C.; Accardi, L.; Nicoletti, L.; Gro, M.C.; Takehara, K.; Hilditch, C.; Morikawa, S.; Bishop, D.H. Sequences and coding strategies of the S RNAs of Toscana and Rift Valley fever viruses compared to those of Punta Toro, Sicilian Sandfly fever, and Uukuniemi viruses. Virology 1991, 180, 738–753. [Google Scholar] [CrossRef]
- Rezelj, V.V.; Overby, A.K.; Elliott, R.M. Generation of mutant Uukuniemi viruses lacking the nonstructural protein NSs by reverse genetics indicates that NSs is a weak interferon antagonist. J. Virol. 2015, 89, 4849–4856. [Google Scholar] [CrossRef]
- Simons, J.F.; Persson, R.; Pettersson, R.F. Association of the nonstructural protein NSs of Uukuniemi virus with the 40S ribosomal subunit. J. Virol. 1992, 66, 4233–4241. [Google Scholar] [CrossRef]
- Rezelj, V.V.; Li, P.; Chaudhary, V.; Elliott, R.M.; Jin, D.Y.; Brennan, B. Differential Antagonism of Human Innate Immune Responses by Tick-Borne Phlebovirus Nonstructural Proteins. mSphere 2017, 2. [Google Scholar] [CrossRef]
- Hallam, H.J.; Lokugamage, N.; Ikegami, T. Rescue of infectious Arumowot virus from cloned cDNA: Posttranslational degradation of Arumowot virus NSs protein in human cells. PLoS Negl. Trop. Dis. 2019, 13, e0007904. [Google Scholar] [CrossRef]
- Simons, J.F.; Hellman, U.; Pettersson, R.F. Uukuniemi virus S RNA segment: Ambisense coding strategy, packaging of complementary strands into virions, and homology to members of the genus Phlebovirus. J. Virol. 1990, 64, 247–255. [Google Scholar] [CrossRef]
- Li, G.; Ren, J.; Xu, F.; Ferguson, M.R. Non-structural and nucleocapsid proteins of Punta Toro virus induce apoptosis of hepatocytes through both intrinsic and extrinsic pathways. Microbiol. Immunol. 2010, 54, 20–30. [Google Scholar] [CrossRef]
- Perrone, L.A.; Narayanan, K.; Worthy, M.; Peters, C.J. The S segment of Punta Toro virus (Bunyaviridae, Phlebovirus) is a major determinant of lethality in the Syrian hamster and codes for a type I interferon antagonist. J. Virol. 2007, 81, 884–892. [Google Scholar] [CrossRef] [PubMed]
- Alkan, C.; Bichaud, L.; de Lamballerie, X.; Alten, B.; Gould, E.A.; Charrel, R.N. Sandfly-borne phleboviruses of Eurasia and Africa: Epidemiology, genetic diversity, geographic range, control measures. Antivir. Res. 2013, 100, 54–74. [Google Scholar] [CrossRef] [PubMed]
- Wuerth, J.D.; Habjan, M.; Wulle, J.; Superti-Furga, G.; Pichlmair, A.; Weber, F. NSs Protein of Sandfly Fever Sicilian Phlebovirus Counteracts Interferon (IFN) Induction by Masking the DNA-Binding Domain of IFN Regulatory Factor 3. J. Virol. 2018, 92, e01202-18. [Google Scholar] [CrossRef]
- Charrel, R.N.; Bichaud, L.; de Lamballerie, X. Emergence of Toscana virus in the mediterranean area. World J. Virol. 2012, 1, 135–141. [Google Scholar] [CrossRef] [PubMed]
- Gori Savellini, G.; Gandolfo, C.; Cusi, M.G. Truncation of the C-terminal region of Toscana Virus NSs protein is critical for interferon-beta antagonism and protein stability. Virology 2015, 486, 255–262. [Google Scholar] [CrossRef] [PubMed]
- Gori-Savellini, G.; Valentini, M.; Cusi, M.G. Toscana virus NSs protein inhibits the induction of type I interferon by interacting with RIG-I. J. Virol. 2013, 87, 6660–6667. [Google Scholar] [CrossRef]
- Gori Savellini, G.; Weber, F.; Terrosi, C.; Habjan, M.; Martorelli, B.; Cusi, M.G. Toscana virus induces interferon although its NSs protein reveals antagonistic activity. J. Gen. Virol. 2011, 92, 71–79. [Google Scholar] [CrossRef]
- Di Bonito, P.; Nicoletti, L.; Mochi, S.; Accardi, L.; Marchi, A.; Giorgi, C. Immunological characterization of Toscana virus proteins. Arch. Virol. 1999, 144, 1947–1960. [Google Scholar] [CrossRef]
- Woelfl, F.; Leger, P.; Oreshkova, N.; Pahmeier, F.; Windhaber, S.; Koch, J.; Stanifer, M.; Roman Sosa, G.; Uckeley, Z.M.; Rey, F.A.; et al. Novel Toscana Virus Reverse Genetics System Establishes NSs as an Antagonist of Type I Interferon Responses. Viruses 2020, 12, 400. [Google Scholar] [CrossRef] [PubMed]
- Gori Savellini, G.; Anichini, G.; Gandolfo, C.; Prathyumnan, S.; Cusi, M.G. Toscana virus non-structural protein NSs acts as E3 ubiquitin ligase promoting RIG-I degradation. PLoS Pathog. 2019, 15, e1008186. [Google Scholar] [CrossRef] [PubMed]
- Indran, S.V.; Lihoradova, O.A.; Phoenix, I.; Lokugamage, N.; Kalveram, B.; Head, J.A.; Tigabu, B.; Smith, J.K.; Zhang, L.; Juelich, T.L.; et al. Rift Valley fever virus MP-12 vaccine encoding Toscana virus NSs retains neuroinvasiveness in mice. J. Gen. Virol. 2013, 94, 1441–1450. [Google Scholar] [CrossRef]
- Kalveram, B.; Ikegami, T. Toscana virus NSs protein promotes degradation of double-stranded RNA-dependent protein kinase. J. Virol. 2013, 87, 3710–3718. [Google Scholar] [CrossRef]
- Min, Y.Q.; Shi, C.; Yao, T.; Feng, K.; Mo, Q.; Deng, F.; Wang, H.; Ning, Y.J. The Nonstructural Protein of Guertu Virus Disrupts Host Defenses by Blocking Antiviral Interferon Induction and Action. ACS Infect. Dis. 2020, 6, 857–870. [Google Scholar] [CrossRef]
- Brennan, B.; Rezelj, V.V.; Elliott, R.M. Mapping of Transcription Termination within the S Segment of SFTS Phlebovirus Facilitated Generation of NSs Deletant Viruses. J. Virol. 2017, 91. [Google Scholar] [CrossRef]
- Savage, H.M.; Godsey, M.S.; Lambert, A.; Panella, N.A.; Burkhalter, K.L.; Harmon, J.R.; Lash, R.R.; Ashley, D.C.; Nicholson, W.L. First detection of heartland virus (Bunyaviridae: Phlebovirus) from field collected arthropods. Am. J. Trop. Med. Hyg. 2013, 89, 445–452. [Google Scholar] [CrossRef]
- Ning, Y.J.; Mo, Q.; Feng, K.; Min, Y.Q.; Li, M.; Hou, D.; Peng, C.; Zheng, X.; Deng, F.; Hu, Z.; et al. Interferon-gamma-Directed Inhibition of a Novel High-Pathogenic Phlebovirus and Viral Antagonism of the Antiviral Signaling by Targeting STAT1. Front. Immunol. 2019, 10, 1182. [Google Scholar] [CrossRef]
- Chaudhary, V.; Zhang, S.; Yuen, K.S.; Li, C.; Lui, P.Y.; Fung, S.Y.; Wang, P.H.; Chan, C.P.; Li, D.; Kok, K.H.; et al. Suppression of type I and type III IFN signalling by NSs protein of severe fever with thrombocytopenia syndrome virus through inhibition of STAT1 phosphorylation and activation. J. Gen. Virol. 2015, 96, 3204–3211. [Google Scholar] [CrossRef] [PubMed]
- Feng, K.; Deng, F.; Hu, Z.; Wang, H.; Ning, Y.J. Heartland virus antagonizes type I and III interferon antiviral signaling by inhibiting phosphorylation and nuclear translocation of STAT2 and STAT1. J. Biol. Chem. 2019, 294, 9503–9517. [Google Scholar] [CrossRef]
- Ning, Y.J.; Feng, K.; Min, Y.Q.; Deng, F.; Hu, Z.; Wang, H. Heartland virus NSs protein disrupts host defenses by blocking the TBK1 kinase-IRF3 transcription factor interaction and signaling required for interferon induction. J. Biol. Chem. 2017, 292, 16722–16733. [Google Scholar] [CrossRef]
- Zhang, S.; Zheng, B.; Wang, T.; Li, A.; Wan, J.; Qu, J.; Li, C.H.; Li, D.; Liang, M. NSs protein of severe fever with thrombocytopenia syndrome virus suppresses interferon production through different mechanism than Rift Valley fever virus. Acta Virol. 2017, 61, 289–298. [Google Scholar] [CrossRef] [PubMed]
- Moriyama, M.; Igarashi, M.; Koshiba, T.; Irie, T.; Takada, A.; Ichinohe, T. Two Conserved Amino Acids within the NSs of Severe Fever with Thrombocytopenia Syndrome Phlebovirus Are Essential for Anti-interferon Activity. J. Virol. 2018, 92. [Google Scholar] [CrossRef]
- Santiago, F.W.; Covaleda, L.M.; Sanchez-Aparicio, M.T.; Silvas, J.A.; Diaz-Vizarreta, A.C.; Patel, J.R.; Popov, V.; Yu, X.J.; Garcia-Sastre, A.; Aguilar, P.V. Hijacking of RIG-I signaling proteins into virus-induced cytoplasmic structures correlates with the inhibition of type I interferon responses. J. Virol. 2014, 88, 4572–4585. [Google Scholar] [CrossRef]
- Sun, Y.; Liu, M.M.; Lei, X.Y.; Yu, X.J. SFTS phlebovirus promotes LC3-II accumulation and nonstructural protein of SFTS phlebovirus co-localizes with autophagy proteins. Sci. Rep. 2018, 8, 5287. [Google Scholar] [CrossRef] [PubMed]
- Kitagawa, Y.; Sakai, M.; Shimojima, M.; Saijo, M.; Itoh, M.; Gotoh, B. Nonstructural protein of severe fever with thrombocytopenia syndrome phlebovirus targets STAT2 and not STAT1 to inhibit type I interferon-stimulated JAK-STAT signaling. Microbes Infect. 2018, 20, 360–368. [Google Scholar] [CrossRef]
- Ning, Y.J.; Feng, K.; Min, Y.Q.; Cao, W.C.; Wang, M.; Deng, F.; Hu, Z.; Wang, H. Disruption of type I interferon signaling by the nonstructural protein of severe fever with thrombocytopenia syndrome virus via the hijacking of STAT2 and STAT1 into inclusion bodies. J. Virol. 2015, 89, 4227–4236. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Ye, H.; Li, S.; Jiao, B.; Wu, J.; Zeng, P.; Chen, L. Severe fever with thrombocytopenia syndrome virus inhibits exogenous Type I IFN signaling pathway through its NSs invitro. PLoS ONE 2017, 12, e0172744. [Google Scholar] [CrossRef]
- Sun, Q.; Qi, X.; Zhang, Y.; Wu, X.; Liang, M.; Li, C.; Li, D.; Cardona, C.J.; Xing, Z. Synaptogyrin-2 Promotes Replication of a Novel Tick-borne Bunyavirus through Interacting with Viral Nonstructural Protein NSs. J. Biol. Chem. 2016, 291, 16138–16149. [Google Scholar] [CrossRef]
- Shen, S.; Duan, X.; Wang, B.; Zhu, L.; Zhang, Y.; Zhang, J.; Wang, J.; Luo, T.; Kou, C.; Liu, D.; et al. A novel tick-borne phlebovirus, closely related to severe fever with thrombocytopenia syndrome virus and Heartland virus, is a potential pathogen. Emerg. Microbes Infect. 2018, 7, 95. [Google Scholar] [CrossRef]
- Hong, Y.; Bai, M.; Qi, X.; Li, C.; Liang, M.; Li, D.; Cardona, C.J.; Xing, Z. Suppression of the IFN-alpha and -beta Induction through Sequestering IRF7 into Viral Inclusion Bodies by Nonstructural Protein NSs in Severe Fever with Thrombocytopenia Syndrome Bunyavirus Infection. J. Immunol. 2019, 202, 841–856. [Google Scholar] [CrossRef]
- Choi, Y.; Jiang, Z.; Shin, W.J.; Jung, J.U. Severe Fever with Thrombocytopenia Syndrome Virus NSs Interacts with TRIM21 To Activate the p62-Keap1-Nrf2 Pathway. J. Virol. 2020, 94. [Google Scholar] [CrossRef] [PubMed]
- Choi, Y.; Park, S.J.; Sun, Y.; Yoo, J.S.; Pudupakam, R.S.; Foo, S.S.; Shin, W.J.; Chen, S.B.; Tsichlis, P.N.; Lee, W.J.; et al. Severe fever with thrombocytopenia syndrome phlebovirus non-structural protein activates TPL2 signalling pathway for viral immunopathogenesis. Nat. Microbiol. 2019, 4, 429–437. [Google Scholar] [CrossRef]
- Qu, B.; Qi, X.; Wu, X.; Liang, M.; Li, C.; Cardona, C.J.; Xu, W.; Tang, F.; Li, Z.; Wu, B.; et al. Suppression of the interferon and NF-kappaB responses by severe fever with thrombocytopenia syndrome virus. J. Virol. 2012, 86, 8388–8401. [Google Scholar] [CrossRef]
- Min, Y.Q.; Ning, Y.J.; Wang, H.; Deng, F. A RIG-I-like receptor directs antiviral responses to a bunyavirus and is antagonized by virus-induced blockade of TRIM25-mediated ubiquitination. J. Biol. Chem 2020, 295, 9691–9711. [Google Scholar] [CrossRef]
- Liu, S.; Liu, H.; Kang, J.; Xu, L.; Zhang, K.; Li, X.; Hou, W.; Wang, Z.; Wang, T. The Severe Fever with Thrombocytopenia Syndrome Virus NSs Protein Interacts with CDK1 To Induce G2 Cell Cycle Arrest and Positively Regulate Viral Replication. J. Virol. 2020, 94. [Google Scholar] [CrossRef]
- Wu, X.; Qi, X.; Liang, M.; Li, C.; Cardona, C.J.; Li, D.; Xing, Z. Roles of viroplasm-like structures formed by nonstructural protein NSs in infection with severe fever with thrombocytopenia syndrome virus. FASEB J. 2014, 28, 2504–2516. [Google Scholar] [CrossRef] [PubMed]
- Kreher, F.; Tamietti, C.; Gommet, C.; Guillemot, L.; Ermonval, M.; Failloux, A.B.; Panthier, J.J.; Bouloy, M.; Flamand, M. The Rift Valley fever accessory proteins NSm and P78/NSm-GN are distinct determinants of virus propagation in vertebrate and invertebrate hosts. Emerg. Microbes Infect. 2014, 3, e71. [Google Scholar] [CrossRef] [PubMed]
- Crabtree, M.B.; Kent Crockett, R.J.; Bird, B.H.; Nichol, S.T.; Erickson, B.R.; Biggerstaff, B.J.; Horiuchi, K.; Miller, B.R. Infection and transmission of Rift Valley fever viruses lacking the NSs and/or NSm genes in mosquitoes: Potential role for NSm in mosquito infection. PLoS Negl. Trop. Dis. 2012, 6, e1639. [Google Scholar] [CrossRef]
- Won, S.; Ikegami, T.; Peters, C.J.; Makino, S. NSm protein of Rift Valley fever virus suppresses virus-induced apoptosis. J. Virol. 2007, 81, 13335–13345. [Google Scholar] [CrossRef] [PubMed]
- Gerrard, S.R.; Bird, B.H.; Albarino, C.G.; Nichol, S.T. The NSm proteins of Rift Valley fever virus are dispensable for maturation, replication and infection. Virology 2007, 359, 459–465. [Google Scholar] [CrossRef]
- Won, S.; Ikegami, T.; Peters, C.J.; Makino, S. NSm and 78-kilodalton proteins of Rift Valley fever virus are nonessential for viral replication in cell culture. J. Virol. 2006, 80, 8274–8278. [Google Scholar] [CrossRef]
- Terasaki, K.; Won, S.; Makino, S. The C-terminal region of Rift Valley fever virus NSm protein targets the protein to the mitochondrial outer membrane and exerts antiapoptotic function. J. Virol. 2013, 87, 676–682. [Google Scholar] [CrossRef]
- Narayanan, A.; Popova, T.; Turell, M.; Kidd, J.; Chertow, J.; Popov, S.G.; Bailey, C.; Kashanchi, F.; Kehn-Hall, K. Alteration in superoxide dismutase 1 causes oxidative stress and p38 MAPK activation following RVFV infection. PLoS ONE 2011, 6, e20354. [Google Scholar] [CrossRef]
- Engdahl, C.; Naslund, J.; Lindgren, L.; Ahlm, C.; Bucht, G. The Rift Valley Fever virus protein NSm and putative cellular protein interactions. Virol. J. 2012, 9, 139. [Google Scholar] [CrossRef] [PubMed]
- Ihara, T.; Smith, J.; Dalrymple, J.M.; Bishop, D.H. Complete sequences of the glycoproteins and M RNA of Punta Toro phlebovirus compared to those of Rift Valley fever virus. Virology 1985, 144, 246–259. [Google Scholar] [CrossRef]
- Chen, S.Y.; Matsuoka, Y.; Compans, R.W. Golgi complex localization of the Punta Toro virus G2 protein requires its association with the G1 protein. Virology 1991, 183, 351–365. [Google Scholar] [CrossRef]
- Di Bonito, P.; Mochi, S.; Gro, M.C.; Fortini, D.; Giorgi, C. Organization of the M genomic segment of Toscana phlebovirus. J. Gen. Virol. 1997, 78 Pt 1, 77–81. [Google Scholar] [CrossRef]
- Gro, M.C.; Di Bonito, P.; Fortini, D.; Mochi, S.; Giorgi, C. Completion of molecular characterization of Toscana phlebovirus genome: Nucleotide sequence, coding strategy of M genomic segment and its amino acid sequence comparison to other phleboviruses. Virus Res. 1997, 51, 81–91. [Google Scholar] [CrossRef]
- Hart, T.J.; Kohl, A.; Elliott, R.M. Role of the NSs protein in the zoonotic capacity of Orthobunyaviruses. Zoonoses Public Health 2009, 56, 285–296. [Google Scholar] [CrossRef] [PubMed]

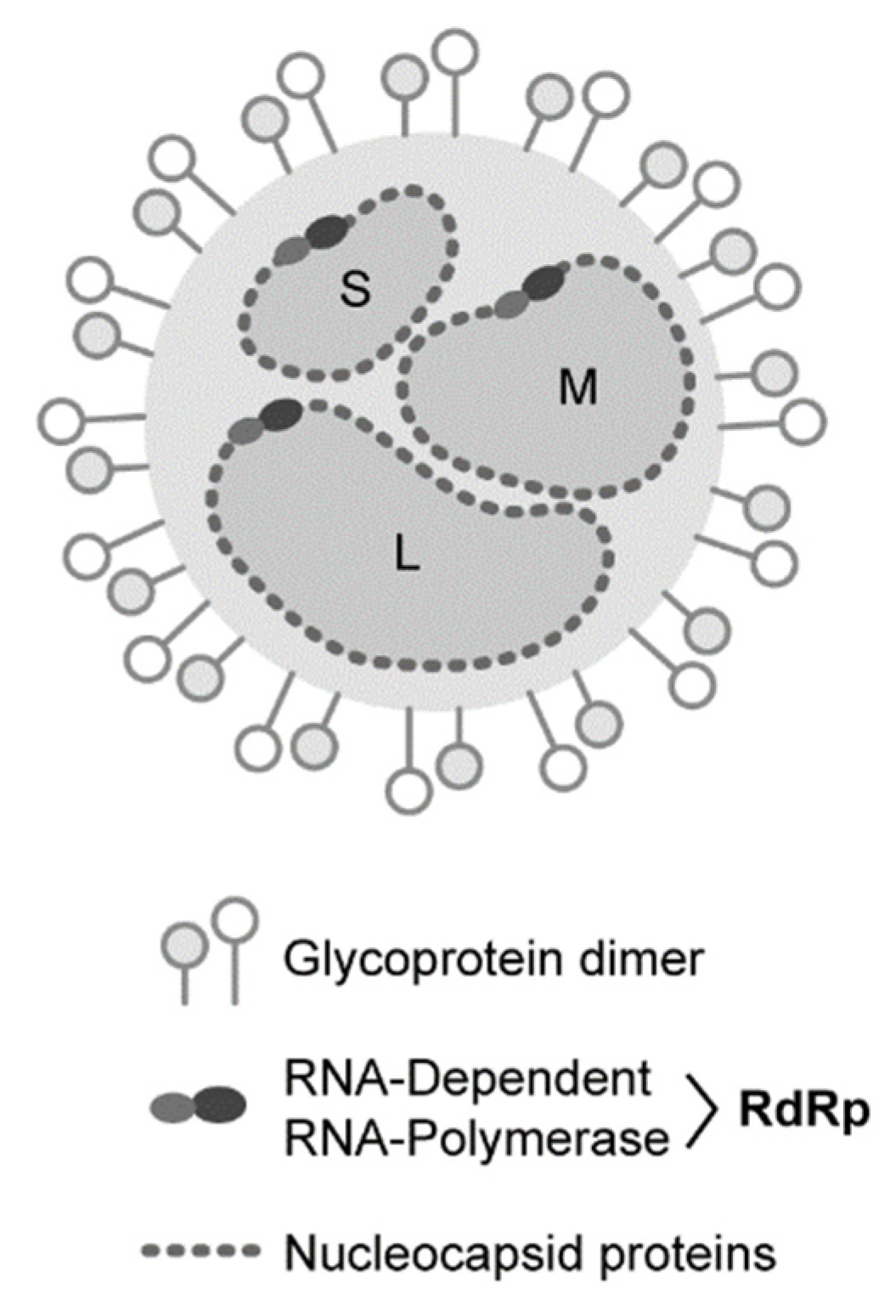
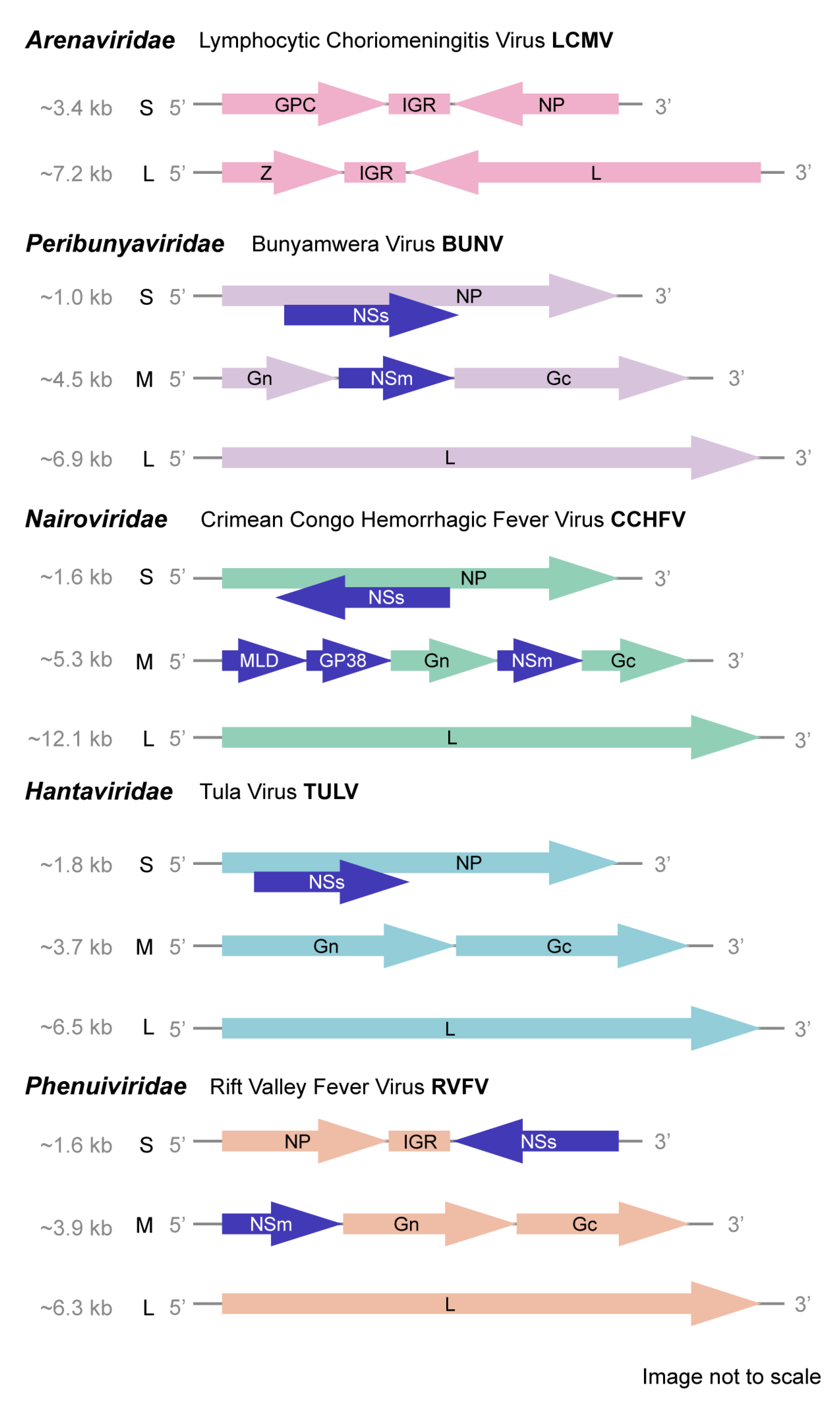
| Family | Genus | Species | Common Name |
|---|---|---|---|
| Mypoviridae | Hubavirus | * Myriapod hubavirus | Húběi Myriapoda Virus 5 (HbMV-5) |
| Wupedeviridae | Wumivirus | * Millipede wumivirus | Wǔhàn Millipede Virus 2 (WhMV-2) |
| Nairoviridae | Orthonairovirus | * Dugbe orthonairovirus | Dugbe Virus (DUGV) |
| Crimean-Congo hemorrhagic fever orthonairovirus | Crimean-Congo Hemorrhagic Fever Virus (CCHFV) | ||
| Hazara orthonairovirus | Hazara Virus (HAZV) | ||
| Nairobi sheep disease orthonairovirus | Nairobi Sheep Disease Virus (NSDV) | ||
| Thiafora orthonairovirus | Erve Virus (ERVEV) | ||
| Tospoviridae | Orthotospovirus | * Tomato spotted wilt tospovirus | Tomato Spotted Wilt Virus (TSWV) |
| Peribunyaviridae | Orthobunyavirus | * Bunyamwera orthobunyavirus | Bunyamwera Virus (BUNV) |
| Ngari Virus (NRIV) | |||
| Akabane orthobunyavirus | Akabane virus (AKAV) | ||
| Batai orthobunyavirus | Batai Virus (BATV) | ||
| California encephalitis orthobunyavirus | California Encephalitis Virus (CEV) | ||
| La Crosse orthobunyavirus | La Crosse Virus (LACV) | ||
| Tacaiuma orthobunyavirus | Tacaiuma Virus (TCMV) | ||
| Turlock orthobunyavirus | Umbre Virus (UMBV) | ||
| Witwatersrand orthobunyavirus | Witwatersrand Virus (WITV) | ||
| Arenaviridae | Mammarenavirus | * Lymphocytic choriomeningitis mammarenavirus | Lymphocytic Choriomeningitis Virus (LCMV) |
| Lassa mammarenavirus | Lassa Virus (LASV) | ||
| Argentinian mammarenavirus | Junin Virus (JUNV) | ||
| Hantaviridae | Orthohantavirus | * Hantaan orthohantavirus | Hantaan Virus (HTNV) |
| Dobrava-Belgrade orthohantavirus | Dobrava Virus (DOBV) | ||
| Andes orthohantavirus | Andes Virus (ANDV) | ||
| Puumala orthohantavirus | Puumala Virus (PUUV) | ||
| Seoul orthohantavirus | Seoul Virus (SEOV) | ||
| Sin Nombre ortohantavirus | Sin Nombre Virus (SNV) | ||
| Tula orthohantavirus | Tula Virus (TULV) | ||
| Leishbuviridae | Shilevirus | * Leptomonas shilevirus | Leptomonas Moramango Virus (LEPMV) |
| Phenuiviridae | Banyangvirus | * Huaiyangshan banyangvirus | Severe Fever with Thrombocytopenia Syndrome Virus (SFTSV) |
| Heartland banyangvirus | Heartland Virus (HRTV) | ||
| Guertu banyangvirus | Guertu Virus (GTV) | ||
| Phlebovirus | * Rift Valley fever phlebovirus | Rift Valley Fever Virus (RVFV) | |
| Punta Toro phlebovirus | Punta Toro Virus (PTV) | ||
| Salehabad phlebovirus | Arumowot Virus (AMTV) | ||
| Sandfly fever Naples phlebovirus | Sandfly Fever Sicilian Virus (SFSV) | ||
| Toscana Virus (TOSV) | |||
| Uukuniemi phlebovirus | Uukuniemi Virus (UUKV) | ||
| Unassigned | Coguvirus | * Citrus coguvirus | Citrus Concave Gum-Associated Virus (CCGaV) |
| Cruliviridae | Lincruvirus | * Crustacean lincruvirus | Wēnlǐng Crustacean Virus 9 (WICV-9) |
| Fimoviridae | Emaravirus | * European mountain ash ringspotassociated emaravirus | European Mountain Ash Ringspot-Associated Virus (EMARaV) |
| Phasmaviridae | Feravirus | * Ferak feravirus | Ferak Virus (FRKV) |
| Family | NS Protein | Functions |
|---|---|---|
| Peribunyaviridae | NSs | Blocking production of type I IFN [27,28,29,30,31,38] |
| Blocking transcription and translation [28,29,162] | ||
| Inducing apoptosis [24,162] Inhibiting apoptosis [38] | ||
| NSm | Potential role in infection [42,43] | |
| Nairoviridae | NSs | Inducing apoptosis [50,52] |
| MLD | Potential role in Gn/Gc incorporation into viral particles [59] | |
| Potential impact on GP38 conformation [59] | ||
| Potential role in regulating Gc accumulation in the Golgi [59] | ||
| GP38 | Potential role in Gn/Gc maturation [44,59] | |
| Potential role in viral replication [56] | ||
| NSm | Role in viral replication and particle formation [48,59] | |
| Potential role in virulence [47] | ||
| Hantaviridae | NSs | Blocking IFN signaling [68] |
| Blocking NF-kB signaling [69] | ||
| Limiting dsRNA production [69] | ||
| Potential role in chromatin remodeling [70] | ||
| Potential role in viral replication [70] | ||
| Phenuiviridae | NSs | Aiding in viral evasion of host immunity [72,73,74] |
| Inhibiting general transcription [72] | ||
| Downregulating or Degrading PKR [72,126] | ||
| Segregating chromatin DNA [72] | ||
| Role in filament formation [72] | ||
| Inducing apoptosis [72] | ||
| Antagonizing of type I IFN system [72,111,115,117,119,120,127,132,133,134,135,136,137,138,139,140,143] | ||
| Antagonizing of type II IFN system (SFTSV only) [130] | ||
| Antagonizing of type III IFN system (SFTSV/HRTV only) [131,132] | ||
| Inducing cellular damage [75,76,77,78,79,80,81,82,148] | ||
| Inducing apoptosis [81,114] | ||
| Promoting viral replication [122,149] | ||
| Upregulating the p62-Keap1-Nrf2 antioxidant pathway [144] | ||
| Inducing IL-10 expression [145] | ||
| Blocking NF-kB signaling [146] | ||
| Mediating antiviral signaling [147] | ||
| Promoting cell cycle arrest [81,148] | ||
| NSm | Maintaining mosquito vector infection [88,150,151] | |
| Role in antiapoptotic function [152,155,156] | ||
| Potential role in neuro-invasion [157] | ||
| Potential role in protein trafficking [157] | ||
| Potential role in mRNA nuclear transport [157] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Leventhal, S.S.; Wilson, D.; Feldmann, H.; Hawman, D.W. A Look into Bunyavirales Genomes: Functions of Non-Structural (NS) Proteins. Viruses 2021, 13, 314. https://doi.org/10.3390/v13020314
Leventhal SS, Wilson D, Feldmann H, Hawman DW. A Look into Bunyavirales Genomes: Functions of Non-Structural (NS) Proteins. Viruses. 2021; 13(2):314. https://doi.org/10.3390/v13020314
Chicago/Turabian StyleLeventhal, Shanna S., Drew Wilson, Heinz Feldmann, and David W. Hawman. 2021. "A Look into Bunyavirales Genomes: Functions of Non-Structural (NS) Proteins" Viruses 13, no. 2: 314. https://doi.org/10.3390/v13020314
APA StyleLeventhal, S. S., Wilson, D., Feldmann, H., & Hawman, D. W. (2021). A Look into Bunyavirales Genomes: Functions of Non-Structural (NS) Proteins. Viruses, 13(2), 314. https://doi.org/10.3390/v13020314







