Abstract
Severe fever with thrombocytopenia syndrome (SFTS) is a novel tick-borne infectious disease, therefore, the information on the whole genome of the SFTS virus (SFTSV) is still limited. This study demonstrates a nearly whole genome of the SFTSV identified in Osaka in 2017 and 2018 by next-generation sequencing (NGS). The evolutionary lineage of two genotypes, C5 and J1, was identified in Osaka. The first case in Osaka belongs to suspect reassortment (L:C5, M:C5, S:C4), the other is genotype J1 (L: J1, M: J1, S: J1) according to the classification by a Japanese group. C5 was identified in China, indicating that C5 identified in this study may be transmitted by birds between China and Japan. This study revealed that different SFTSV genotypes were distributed in two local areas, suggesting the separate or focal transmission patterns in Osaka.
1. Introduction
Severe fever with thrombocytopenia syndrome (SFTS) is a novel tick-borne infectious disease with high mortality. The first reported cases of SFTS were reported in China in 2011 [1], and later the virus was identified in other countries in Asia such as Korea, Japan, Vietnam, Taiwan, and Myanmar [2,3,4,5,6].
SFTS virus (SFTSV), recently renamed Dabie bandavirus, belongs to the family Phenuiviridae, being an enveloped virus containing a three-segment negative RNA genome [1,7]. SFTSV is most closely related to Guertu virus and Heartland virus, which are tick-borne viruses like SFTSV, in the family Phenuiviridae [8]. The three segments are referred to as large (L), medium (M), and small (S) in order from the longest. The L segment encodes RNA-dependent RNA polymerase (RdRp), the M segment encodes two glycoproteins (GP), Gn and Gc, and the S segment encodes nucleoprotein (Np) and nonstructural protein (NS).
SFTS was identified for the first time in 2011; therefore, the genotyping has not been established yet in the world. Yoshikawa et al. classified SFTSV into eight genotypes: Chinese clades 1 to 5 (C1 to C5 clades) and Japanese clades 1 to 3 (J1 to J3 clades) [9]. These previous studies revealed that the distribution of SFTSV genotypes has regional characteristics. J1–J3 clades were mainly detected in Japan and Korea, whereas C1–C5 clades were found in China [9,10]. In Korea, it is estimated that 70% of the SFTSV belong to the J clade; in addition, recently, the C clade has also been identified there [9,11]. The C clade has been detected in Japan, but there are fewer reports than in Korea [9]. There are only few whole genome analyses limited to classifying genotypes. Some previous studies analyzed only a partial region or only the M or S segments; therefore, there are few complete SFTSV sequences in the public database. SFTSV showed different genotypes in each segment, indicating that SFTSV undergoes reassortment like the influenza virus and the rotavirus for genome segments [10,12].
We confirmed two cases in Osaka. The first case was found in 2017 and the other was found in 2018. Two confirmed cases were determined by RT-PCR. To investigate the genetic background of the SFTSV circulating in Osaka, we performed viral isolation, genome analysis using next-generation sequencing (NGS), and phylogenetic analysis.
2. Materials and Methods
2.1. Virus Isolation
The patient serum of the first case was collected in August 2017 and the second case’s serum was collected in July 2018. The specimen was inoculated with Vero cells in an incubator during one week for the isolation of the SFTSV. Vero cells were had been observed to detect the cytopathic effect (CPE) every day for one week. The supernatant was collected at −80 °C and used for extraction of viral RNA.
2.2. Whole Genome Sequencing by NGS
The viral RNA was extracted from the supernatant by using a QIAamp Viral RNA kit (Qiagen, Hilden, Germany) and stored at −80 °C. The cDNA was used for high-throughput sequencing using a Nextera XT DNA Sample Prep Kit (Illumina, San Diego, CA, USA). Sequence-specific adapters were ligated to the amplicons and then the fragments were amplified using seven cycles of the polymerase chain reaction. The sequencing was performed on an automated MiSeq (Illumina) after the preparation of DNA libraries for MiSeq. We used CLC Workbench (Filgen, Nagoya, Japan) to construct a nearly whole genome and assemble the short reads.
2.3. Phylogenetic Analysis
The maximum likelihood tree of the nucleotide sequences was constructed. Briefly, the sequences were aligned using CLUSTAL W (version 1.4) and a distance matrix of nucleotide substitutions per site was estimated from the alignment according to Kimura’s two-parameter method. The trees were generated with 100 bootstrap replicates from the matrix numbers using MEGA version 7.0 [13] (https://www.megasoftware.net/).
3. Results and Discussion
Firstly, we investigated the phylogeny of the three fragments of the SFTSV’s near-full-length genome sequences including segment L, segment M, and segment S (about 10.0 kb). For this study, we used the sequences obtained in Osaka (n = 2), 78 reference sequences of a past global or Japanese epidemic reported by various studies, and various reported sequences of other genotypes (Supplementary Table S1). Case A (strain name: JaOF2017-7 ) was found in the south of Osaka, the putative exposure area was the south of Osaka or Wakayama in 2017. Case B (strain name: JaOT2018-6 ) was found in the north of Osaka, the putative exposure area was Shiga in 2018. Figure 1 shows a representative maximum likelihood tree constructed by using 78 reference sequences. The tree show that that the two newly obtained sequences are divisible into two distinct lineage groups with the high bootstrap value of 100/100 according to the classification by a Japanese group (Figure 1) [9]. The segment L of case A belonged to C5 and that of case B belonged to J1 (Figure 1A). The phylogenetic tree of segment M revealed that case A belonged to C5 and case B belonged to J1 (Figure 1B). The phylogenetic tree of segment S revealed that case A belonged to C4 and case B belonged to J1 (Figure 1C). The monophyletic relationship of the sequences was reproducible when the trees were constructed with different algorithms, such as neighbor joining and Unweighted Pair Group Method with Arithmetic mean (UPMGA), or with the segment L, segment M, or segment S sequences (data not shown). According to the phylogenetic tree analysis, case B formed a monophyletic group with the J1 references, which was the major dominant genotype in Japan. On the other hand, segments of case A formed a monophyletic group with the C5 references which were reported in China [9] and were closely related to the SPL161A isolated in Wakayama. In Japan, mainly, J1–J3 strains were detected, and C1–C5 genotypes were barely reported.
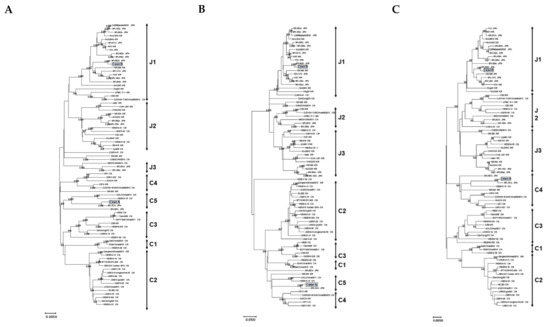
Figure 1.
Phylogenetic classification of the SFTSV. The maximum likelihood tree was constructed with the near-full-length genome sequences (about 10.0 kb) obtained from the specimens collected in Osaka in 2017 and 2018 in this study (n = 2), SFTSV reference sequences from past epidemics in Japan and other countries (n = 78). The phylogenetic trees are shown as segment L (A), segment M (B), and segment S (C). The bootstrap value is shown to be above 90.
In this study, we examined a nearly whole genome of the SFTSV identified in Osaka in 2017 and 2018 by NGS. We analyzed the evolutionary lineage of two genotypes, C5 and J1, in the north and the south of Osaka located at the distance of about 60 km. We first showed the nearly whole genome of the C5 identified in Osaka. Case A was found in the south of Osaka, the putative exposure area was Wakayama. The sequence of segments L, M, and S in case A shows high homology (99%) with that of the SPL161A isolated in Wakayama, indicating that C5 was cocirculating in Osaka and the Wakayama area where it had a possibility of spread and persisted. The presence of C5 may be related to the area near the border in Wakayama where C5 was reported recently, suggesting that C5 is circulating and spread in the Wakayama area. C5 is found in China and is less often reported in Japan. It is hard to think that humans (e.g., tourists) are able to carry SFTSV, because SFTS induces severe symptoms in humans. The birds are able to carry SFTS virus-bearing ticks for a long distance, indicating that birds between China and Japan may transmit C5 [10,14,15].
Interestingly, segments L and M were clustered in C5, but segment S was clustered in C4, indicating that this case A would be a possible reassortment strain, because it has already been reported that SPL161A which shows high homology (99%) with case A would be a possible reassortment strain [9]. There are few sequences of C5 genotypes in the public database; therefore, we are not able to confirm the conclusion that case A is a reassortment strain. According to the classification by a Japanese group, segments L and M were defined for C1–C5, but segment S was classified only for C1–C4 [9]. In addition, the classification of genotypes is still controversial depending on the country [11,16]. It is reported that bunyavirus sometimes occurred as a reassortment virus by changing segment M, it may emerge as a new reassortment strain of SFTSV [17]. However, the genotype pattern of case A revealed that only segment S was changed, not segment M. It may have been the result of accumulation of nucleotide variability, and case A may apparently show a reassortment strain between the genotypes.
In summary, we demonstrated distribution of different SFTSV genotypes in two local areas in Osaka, suggesting separate or focal transmission patterns in Osaka.
Supplementary Materials
The following are available online at https://www.mdpi.com/1999-4915/13/2/177/s1. Supplementary Table S1. The SFTSV strains and accession numbers used in the phylogenetic analyses.
Author Contributions
Conceptualization, R.I., I.A., and T.S.; methodology, R.I., I.A., T.S., and T.Y.; software, R.I., I.A., and K.M. (Kazushi Motomura); validation, R.I., I.A., T.S., and T.Y.; investigation, R.I., I.A., T.S., and T.Y.; resources, H.T., K.M. (Kazutoshi Morisada), and M.K.; writing—original draft preparation, R.I., I.A.; writing—review and editing, K.M. (Kazushi Motomura); supervision, K.M. (Kazushi Motomura); project administration, K.M. (Kazushi Motomura); funding acquisition, K.I. All authors have read and agreed to the published version of the manuscript.
Funding
This study was supported by Kakenhi (18KK0271) from the Ministry of Education, Culture, Sports, Science, and Technology of Japan. We would like to thank our enthusiastic laboratory participants who always provide useful suggestions for continuous quality improvement.
Institutional Review Board Statement
The study was conducted according to the guidelines of the Declaration of Helsinki and approved by the Ethics Committee of Osaka Institute of Public Health (protocol code 1003-08-04 and 2018-Sep.-26 as date of approval).
Informed Consent Statement
Not applicable.
Data Availability Statement
Data available in a publicly accessible repository; The data presented in this study are openly available in GenBank, accession number [LC590890-LC590895].
Conflicts of Interest
The authors declare that they have no competing interests of either a financial or nonfinancial nature regarding the work described in the present manuscript and its publication.
References
- Yu, X.-J.; Liang, M.-F.; Zhang, S.-Y.; Liu, Y.; Li, J.-D.; Sun, Y.-L.; Zhang, L.; Zhang, Q.-F.; Popov, V.L.; Li, C.; et al. Fever with Thrombocytopenia Associated with a Novel Bunyavirus in China. N. Engl. J. Med. 2011, 364, 1523–1532. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.-H.; Yi, J.; Kim, G.; Choi, S.J.; Jun, K., II; Kim, N.-H.; Choe, P.G.; Kim, N.-J.; Lee, J.-K.; Oh, M. Severe Fever with Thrombocytopenia Syndrome, South Korea, 2012. Emerg. Infect. Dis. 2013, 19, 1892. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, T.; Maeda, K.; Suzuki, T.; Ishido, A.; Shigeoka, T.; Tominaga, T.; Kamei, T.; Honda, M.; Ninomiya, D.; Sakai, T.; et al. The First Identification and Retrospective Study of Severe Fever With Thrombocytopenia Syndrome in Japan. J. Infect. Dis. 2014, 209, 816–827. [Google Scholar] [CrossRef] [PubMed]
- Tran, X.C.; Yun, Y.; Van An, L.; Kim, S.-H.; Thao, N.P.; Man, P.K.; Yoo, J.R.; Heo, S.T.; Cho, N.-H.; Lee, K.H. Endemic Severe Fever with Thrombocytopenia Syndrome, Vietnam. Emerg. Infect. Dis. 2019, 25, 1029–1031. [Google Scholar] [CrossRef] [PubMed]
- Peng, S.-H.; Yang, S.-L.; Tang, S.-E.; Wang, T.-C.; Hsu, T.-C.; Su, C.-L.; Chen, M.-Y.; Shimojima, M.; Yoshikawa, T.; Shu, P.-Y. Human Case of Severe Fever with Thrombocytopenia Syndrome Virus Infection, Taiwan, 2019. Emerg. Infect. Dis. 2020, 26, 1612–1614. [Google Scholar] [CrossRef] [PubMed]
- Win, A.M.; Nguyen, Y.T.H.; Kim, Y.; Ha, N.-Y.; Kang, J.-G.; Kim, H.; San, B.; Kyaw, O.; Htike, W.W.; Choi, D.-O.; et al. Genotypic Heterogeneity of Orientia tsutsugamushi in Scrub Typhus Patients and Thrombocytopenia Syndrome Co-infection, Myanmar. Emerg. Infect. Dis. 2020, 26, 1878–1881. [Google Scholar] [CrossRef] [PubMed]
- Kuhn, J.H.; Adkins, S.; Alioto, D.; Alkhovsky, S.V.; Amarasinghe, G.K.; Anthony, S.J.; Avšič-Županc, T.; Ayllón, M.A.; Bahl, J.; Balkema-Buschmann, A.; et al. 2020 taxonomic update for phylum Negarnaviricota (Riboviria: Orthornavirae), including the large orders Bunyavirales and Mononegavirales. Arch. Virol. 2020, 165, 3023–3072. [Google Scholar] [CrossRef] [PubMed]
- Shen, S.; Duan, X.; Wang, B.; Zhu, L.; Zhang, Y.; Zhang, J.; Wang, J.; Luo, T.; Kou, C.; Liu, D.; et al. A novel tick-borne phlebovirus, closely related to severe fever with thrombocytopenia syndrome virus and Heartland virus, is a potential pathogen. Emerg. Microbes Infect. 2018, 7, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Yoshikawa, T.; Shimojima, M.; Fukushi, S.; Tani, H.; Fukuma, A.; Taniguchi, S.; Singh, H.; Suda, Y.; Shirabe, K.; Toda, S.; et al. Phylogenetic and Geographic Relationships of Severe Fever with Thrombocytopenia Syndrome Virus in China, South Korea, and Japan. J. Infect. Dis. 2015, 212, 889–898. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; Hu, S.; Liu, X.; Yang, J.; Liu, D.; Wu, L.; Wang, H.; Hu, Z.; Deng, F.; Shen, S. Migration, recombination, and reassortment are involved in the evolution of severe fever with thrombocytopenia syndrome bunyavirus. Infect. Genet. Evol. 2017, 47, 109–117. [Google Scholar] [CrossRef] [PubMed]
- Yun, S.-M.; Park, S.-J.; Kim, Y.-I.; Park, S.-W.; Yu, M.-A.; Kwon, H.-I.; Kim, E.-H.; Yu, K.-M.; Jeong, H.W.; Ryou, J.; et al. Genetic and pathogenic diversity of severe fever with thrombocytopenia syndrome virus (SFTSV) in South Korea. JCI Insight 2020, 5, 1–12. [Google Scholar] [CrossRef]
- Li, Z.; Hu, J.; Cui, L.; Hong, Y.; Liu, J.; Li, P.; Guo, X.; Liu, W.; Wang, X.; Qi, X.; et al. Increased Prevalence of Severe Fever with Thrombocytopenia Syndrome in Eastern China Clustered with Multiple Genotypes and Reasserted Virus during 2010–2015. Sci. Rep. 2017, 7, 6503. [Google Scholar] [CrossRef]
- Kumar, S.; Tamura, K.; Nei, M. MEGA: Molecular Evolutionary Genetics Analysis software for microcomputers. Bioinformatics 1994, 10, 189–191. [Google Scholar] [CrossRef]
- Yun, Y.; Heo, S.T.; Kim, G.; Hewson, R.; Kim, H.; Park, D.; Cho, N.-H.; Oh, W.S.; Ryu, S.Y.; Kwon, K.T.; et al. Phylogenetic Analysis of Severe Fever with Thrombocytopenia Syndrome Virus in South Korea and Migratory Bird Routes Between China, South Korea, and Japan. Am. J. Trop. Med. Hyg. 2015, 93, 468–474. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.; Li, S.; Zhang, Z.; Man, S.; Li, X.; Zhang, W.; Zhang, C.; Cheng, X. Phylogeographic analysis of severe fever with thrombocytopenia syndrome virus from Zhoushan Islands, China: Implication for transmission across the ocean. Sci. Rep. 2016, 6, 19563. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.-W.; Zhao, L.; Luo, L.-M.; Liu, M.-M.; Sun, Y.; Su, X.; Yu, X. Molecular Evolution and Spatial Transmission of Severe Fever with Thrombocytopenia Syndrome Virus Based on Complete Genome Sequences. PLoS ONE 2016, 11, e0151677. [Google Scholar] [CrossRef] [PubMed]
- Rezelj, V.V.; Mottram, T.J.; Hughes, J.; Elliott, R.M.; Kohl, A.; Brennan, B. M Segment-Based Minigenomes and Virus-Like Particle Assays as an Approach To Assess the Potential of Tick-Borne Phlebovirus Genome Reassortment. J. Virol. 2019, 93, e02068-18. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).