An Overview of Recent Insights into the Response of TLR to SARS-CoV-2 Infection and the Potential of TLR Agonists as SARS-CoV-2 Vaccine Adjuvants
Abstract
:1. Introduction
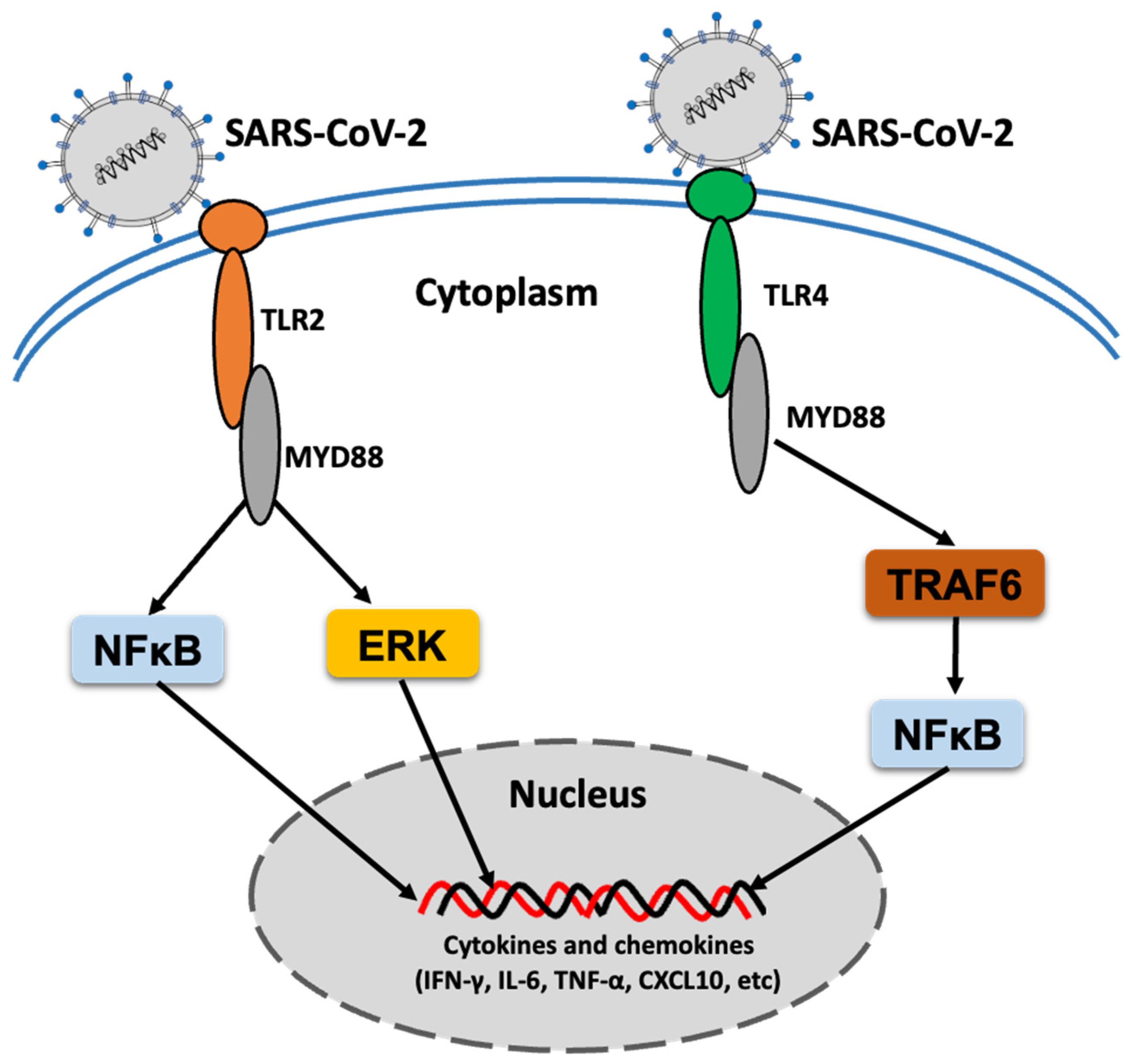
2. TLR Response to SARS-CoV-2 Infection
3. TLR Agonists as COVID-19 Vaccine Adjuvants
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Cheng, V.C.C.; Lau, S.K.P.; Woo, P.C.Y.; Yuen, K.-Y. Severe Acute Respiratory Syndrome Coronavirus as an Agent of Emerging and Reemerging Infection. Clin. Microbiol. Rev. 2007, 20, 660–694. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Su, S.; Wong, G.; Shi, W.; Liu, J.; Lai, A.C.K.; Zhou, J.; Liu, W.; Bi, Y.; Gao, G.F. Epidemiology, Genetic Recombination, and Pathogenesis of Coronaviruses. Trends Microbiol. 2016, 24, 490–502. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cavanagh, D. Coronaviruses in poultry and other birds. Avian Pathol. 2005, 34, 439–448. [Google Scholar] [CrossRef] [PubMed]
- Cui, J.; Li, F.; Shi, Z.-L. Origin and evolution of pathogenic coronaviruses. Nat. Rev. Microbiol. 2019, 17, 181–192. [Google Scholar] [CrossRef] [Green Version]
- Kapikian, A.Z. The coronaviruses. Dev. Biol. Stand. 1975, 28, 42–64. [Google Scholar]
- Lu, R.; Zhao, X.; Li, J.; Niu, P.; Yang, B.; Wu, H.; Wang, W.; Song, H.; Huang, B.; Zhu, N.; et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: Implications for virus origins and receptor binding. Lancet 2020, 395, 565–574. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.; Liu, Q.; Guo, D. Emerging coronaviruses: Genome structure, replication, and pathogenesis. J. Med. Virol. 2020, 92, 418–423. [Google Scholar] [CrossRef]
- Yadav, R.; Chaudhary, J.; Jain, N.; Chaudhary, P.; Khanra, S.; Dhamija, P.; Sharma, A.; Kumar, A.; Handu, S. Role of Structural and Non-Structural Proteins and Therapeutic Targets of SARS-CoV-2 for COVID-19. Cells 2021, 10, 821. [Google Scholar] [CrossRef]
- Zhou, P.; Yang, X.-L.; Wang, X.-G.; Hu, B.; Zhang, L.; Zhang, W.; Si, H.-R.; Zhu, Y.; Li, B.; Huang, C.-L.; et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 2020, 579, 270–273. [Google Scholar] [CrossRef] [Green Version]
- Peiris, J.S.M.; Chu, C.M.; Cheng, V.; Chan, K.; Hung, I.F.N.; Poon, L.; Law, K.; Tang, B.; Hon, T.; Chan, C.; et al. Clinical progression and viral load in a community outbreak of coronavirus-associated SARS pneumonia: A prospective study. Lancet 2003, 361, 1767–1772. [Google Scholar] [CrossRef] [Green Version]
- Zaki, A.M.; Van Boheemen, S.; Bestebroer, T.M.; Osterhaus, A.D.M.E.; Fouchier, R.A.M. Isolation of a Novel Coronavirus from a Man with Pneumonia in Saudi Arabia. N. Engl. J. Med. 2012, 367, 1814–1820. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization (WHO). WHO Coronavirus (COVID-19) Dashboard. Available online: https://covid19.who.int/ (accessed on 1 October 2021).
- Cucinotta, D.; Vanelli, M. WHO declares COVID-19 a pandemic. Acta Biomed. 2020, 91, 157–160. [Google Scholar] [CrossRef] [PubMed]
- Harvey, W.T.; Carabelli, A.M.; Jackson, B.; Gupta, R.K.; Thomson, E.C.; Harrison, E.M.; Ludden, C.; Reeve, R.; Rambaut, A.; Peacock, S.J.; et al. SARS-CoV-2 variants, spike mutations and immune escape. Nat. Rev. Genet. 2021, 19, 409–424. [Google Scholar] [CrossRef] [PubMed]
- Medzhitov, R.; Janeway, C. Innate Immunity. N. Engl. J. Med. 2000, 343, 338–344. [Google Scholar] [CrossRef]
- Mogensen, T.H. Pathogen Recognition and Inflammatory Signaling in Innate Immune Defenses. Clin. Microbiol. Rev. 2009, 22, 240–273. [Google Scholar] [CrossRef] [Green Version]
- Takeuchi, O.; Akira, S. Pattern Recognition Receptors and Inflammation. Cell 2010, 140, 805–820. [Google Scholar] [CrossRef] [Green Version]
- Akira, S.; Uematsu, S.; Takeuchi, O. Pathogen recognition and innate immunity. Cell 2006, 124, 783–801. [Google Scholar] [CrossRef] [Green Version]
- Amarante-Mendes, G.P.; Adjemian, S.; Branco, L.M.; Zanetti, L.; Weinlich, R.; Bortoluci, K.R. Pattern Recognition Receptors and the Host Cell Death Molecular Machinery. Front. Immunol. 2018, 9, 2379. [Google Scholar] [CrossRef] [Green Version]
- Akira, S.; Takeda, K.; Kaisho, T. Toll-like receptors: Critical proteins linking innate and acquired immunity. Nat. Immunol. 2001, 2, 675–680. [Google Scholar] [CrossRef]
- Iwasaki, A.; Medzhitov, R. Toll-like receptor control of the adaptive immune responses. Nat. Immunol. 2004, 5, 987–995. [Google Scholar] [CrossRef]
- Takeda, K.; Akira, S. Toll-like receptors in innate immunity. Int. Immunol. 2005, 17, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Kawasaki, T.; Kawai, T. Toll-Like Receptor Signaling Pathways. Front. Immunol. 2014, 5, 461. [Google Scholar] [CrossRef] [Green Version]
- O’Neill, L.A.J.; Golenbock, D.T.; Bowie, A. The history of Toll-like receptors—Redefining innate immunity. Nat. Rev. Immunol. 2013, 13, 453–460. [Google Scholar] [CrossRef] [PubMed]
- Chaturvedi, A.; Pierce, S.K. How Location Governs Toll-Like Receptor Signaling. Traffic 2009, 10, 621–628. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kawai, T.; Akira, S. Toll-like Receptors and Their Crosstalk with Other Innate Receptors in Infection and Immunity. Immunity 2011, 34, 637–650. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yokota, S.-I.; Okabayashi, T.; Fujii, N. The Battle between Virus and Host: Modulation of Toll-Like Receptor Signaling Pathways by Virus Infection. Mediat. Inflamm. 2010, 2010, 184328. [Google Scholar] [CrossRef] [Green Version]
- Huang, B.; Zhao, J.; Unkeless, J.C.; Feng, Z.H.; Xiong, H. TLR signaling by tumor and immune cells: A double-edged sword. Oncogene 2008, 27, 218–224. [Google Scholar] [CrossRef] [Green Version]
- Ebermeyer, T.; Cognasse, F.; Berthelot, P.; Mismetti, P.; Garraud, O.; Hamzeh-Cognasse, H. Platelet Innate Immune Receptors and TLRs: A Double-Edged Sword. Int. J. Mol. Sci. 2021, 22, 7894. [Google Scholar] [CrossRef]
- Zheng, M.; Karki, R.; Williams, E.P.; Yang, D.; Fitzpatrick, E.; Vogel, P.; Jonsson, C.B.; Kanneganti, T.-D. TLR2 senses the SARS-CoV-2 envelope protein to produce inflammatory cytokines. Nat. Immunol. 2021, 22, 829–838. [Google Scholar] [CrossRef]
- Zuniga, E.I.; Macal, M.; Lewis, G.M.; Harker, J.A. Innate and Adaptive Immune Regulation during Chronic Viral Infections. Annu. Rev. Virol. 2015, 2, 573–597. [Google Scholar] [CrossRef] [Green Version]
- Modhiran, N.; Watterson, D.; Muller, D.A.; Panetta, A.K.; Sester, D.P.; Liu, L.; Hume, D.A.; Stacey, K.J.; Young, P.R. Dengue virus NS1 protein activates cells via Toll-like receptor 4 and disrupts endothelial cell monolayer integrity. Sci. Transl. Med. 2015, 7, 304ra142. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ozato, K.; Tsujimura, H.; Tamura, T. Toll-Like Receptor Signaling and Regulation of Cytokine Gene Expression in the Immune System. Biotechniques 2002, 33, S66–S75. [Google Scholar] [CrossRef] [Green Version]
- Salaun, B.; Romero, P.; Lebecque, S. Toll-like receptors’ two-edged sword: When immunity meets apoptosis. Eur. J. Immunol. 2007, 37, 3311–3318. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.M.; Hill, H.R. Role of Host Immune and Inflammatory Responses in COVID-19 Cases with Underlying Primary Immunodeficiency: A Review. J. Interf. Cytokine Res. 2020, 40, 549–554. [Google Scholar] [CrossRef] [PubMed]
- Ye, Q.; Wang, B.; Mao, J. The pathogenesis and treatment of the ‘Cytokine Storm’ in COVID-19. J. Infect. 2020, 80, 607–613. [Google Scholar] [CrossRef] [PubMed]
- Sheahan, T.; Morrison, T.E.; Funkhouser, W.; Uematsu, S.; Akira, S.; Baric, R.S.; Heise, M.T. MyD88 Is Required for Protection from Lethal Infection with a Mouse-Adapted SARS-CoV. PLoS Pathog. 2008, 4, e1000240. [Google Scholar] [CrossRef] [Green Version]
- Totura, A.; Whitmore, A.; Agnihothram, S.; Schäfer, A.; Katze, M.G.; Heise, M.T.; Baric, R.S. Toll-Like Receptor 3 Signaling via TRIF Contributes to a Protective Innate Immune Response to Severe Acute Respiratory Syndrome Coronavirus Infection. MBio 2015, 6, e00638-15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Liu, L. The Membrane Protein of Severe Acute Respiratory Syndrome Coronavirus Functions as a Novel Cytosolic Pathogen-Associated Molecular Pattern to Promote Beta Interferon Induction via a Toll-Like-Receptor-Related TRAF3-Independent Mechanism. MBio 2016, 7, e01872-15. [Google Scholar] [CrossRef] [Green Version]
- Spiegel, M.; Weber, F. Inhibition of cytokine gene expression and induction of chemokine genes in non-lymphatic cells infected with SARS coronavirus. Virol. J. 2006, 3, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Bhattacharya, M.; Sharma, A.R.; Mallick, B.; Sharma, G.; Lee, S.-S.; Chakraborty, C. Immunoinformatics approach to understand molecular interaction between multi-epitopic regions of SARS-CoV-2 spike-protein with TLR4/MD-2 complex. Infect. Genet. Evol. 2020, 85, 104587. [Google Scholar] [CrossRef]
- Choudhury, A.; Mukherjee, S. In silico studies on the comparative characterization of the interactions of SARS-CoV-2 spike glycoprotein with ACE-2 receptor homologs and human TLRs. J. Med. Virol. 2020, 92, 2105–2113. [Google Scholar] [CrossRef] [PubMed]
- Shirato, K.; Kizaki, T. SARS-CoV-2 spike protein S1 subunit induces pro-inflammatory responses via toll-like receptor 4 signaling in murine and human macrophages. Heliyon 2021, 7, e06187. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Kuang, M.; Li, J.; Zhu, L.; Jia, Z.; Guo, X.; Hu, Y.; Kong, J.; Yin, H.; Wang, X.; et al. SARS-CoV-2 spike protein interacts with and activates TLR41. Cell Res. 2021, 31, 1–3. [Google Scholar] [CrossRef] [PubMed]
- Petruk, G.; Puthia, M.; Petrlova, J.; Samsudin, F.; Strömdahl, A.-C.; Cerps, S.; Uller, L.; Kjellström, S.; Bond, P.J.; Schmidtchen, A.A. SARS-CoV-2 spike protein binds to bacterial lipopolysaccharide and boosts proinflammatory activity. J. Mol. Cell Biol. 2021, 12, 916–932. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Nie, Y.; Wang, W.; Ma, N.; Lendlein, A. Periodic thermomechanical modulation of toll-like receptor expression and distribution in mesenchymal stromal cells. MRS Commun. 2021, 11, 425–431. [Google Scholar] [CrossRef]
- Kobayashi, K.; Hernandez, L.D.; Galán, J.E.; Janeway, C.A.; Medzhitov, R.; Flavell, R.A. IRAK-M Is a Negative Regulator of Toll-like Receptor Signaling. Cell 2002, 110, 191–202. [Google Scholar] [CrossRef] [Green Version]
- Pantazi, I.; Al-Qahtani, A.A.; Alhamlan, F.S.; Alothaid, H.; Matou-Nasri, S.; Sourvinos, G.; Vergadi, E.; Tsatsanis, C. SARS-CoV-2/ACE2 Interaction Suppresses IRAK-M Expression and Promotes Pro-Inflammatory Cytokine Production in Macrophages. Front. Immunol. 2021, 12, 683800. [Google Scholar] [CrossRef]
- Khan, S.; Shafiei, M.S.; Longoria, C.; Schoggins, J.; Savani, R.C.; Zaki, H. SARS-CoV-2 spike protein induces inflammation via TLR2-dependent activation of the NF-κB pathway. bioRxiv 2021. [Google Scholar] [CrossRef]
- Sariol, A.; Perlman, S. SARS-CoV-2 takes its Toll. Nat. Immunol. 2021, 22, 801–802. [Google Scholar] [CrossRef]
- Conti, P.; Caraffa, A.; Tete, G.; Gallenga, C.E.; Ross, R.; Kritas, S.K.; Frydas, I.; Younes, A.; Di Emidio, P.; Ronconi, G. Mast cells activated by SARS-CoV-2 release histamine which increases IL-1 levels causing cytokine storm and inflammatory reaction in COVID-19. J. Biol. Regul. Homeost. Agents 2020, 34, 1629–1632. [Google Scholar]
- Gebremeskel, S.; Schanin, J.; Coyle, K.M.; Butuci, M.; Luu, T.; Brock, E.C.; Xu, A.; Wong, A.; Leung, J.; Korver, W.; et al. Mast Cell and Eosinophil Activation Are Associated With COVID-19 and TLR-Mediated Viral Inflammation: Implications for an Anti-Siglec-8 Antibody. Front. Immunol. 2021, 12, 641. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, R.; Al-Mass, A.; Atizado, V.; Al-Hubail, A.; Al-Ghimlas, F.; Al-Arouj, M.; Bennakhi, A.; Dermime, S.; Behbehani, K. Elevated expression of the toll like receptors 2 and 4 in obese individuals: Its significance for obesity-induced inflammation. J. Inflamm. 2012, 9, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Cuevas, A.M.; Clark, J.M.; Potter, J.J. Increased TLR/MyD88 signaling in patients with obesity: Is there a link to COVID-19 disease severity? Int. J. Obes. 2021, 45, 1152–1154. [Google Scholar] [CrossRef]
- Englmeier, L. A theory on SARS-COV-2 susceptibility: Reduced TLR7-activity as a mechanistic link between men, obese and elderly. J. Biol. Regul. Homeost. Agents 2020, 34, 1125–1129. [Google Scholar]
- Van Der Made, C.I.; Simons, A.; Schuurs-Hoeijmakers, J.; Heuvel, G.V.D.; Mantere, T.; Kersten, S.; Van Deuren, R.C.; Steehouwer, M.; Van Reijmersdal, S.V.; Jaeger, M.; et al. Presence of Genetic Variants Among Young Men With Severe COVID-19. JAMA 2020, 324, 663–673. [Google Scholar] [CrossRef] [PubMed]
- Fallerini, C.; Daga, S.; Mantovani, S.; Benetti, E.; Picchiotti, N.; Francisci, D.; Paciosi, F.; Schiaroli, E.; Baldassarri, M.; Fava, F.; et al. Association of Toll-like receptor 7 variants with life-threatening COVID-19 disease in males: Findings from a nested case-control study. eLife 2021, 10, e67569. [Google Scholar] [CrossRef] [PubMed]
- Severa, M.; Diotti, R.A.; Etna, M.P.; Rizzo, F.; Fiore, S.; Ricci, D.; Iannetta, M.; Sinigaglia, A.; Lodi, A.; Mancini, N.; et al. Differential plasmacytoid dendritic cell phenotype and type I Interferon response in asymptomatic and severe COVID-19 infection. PLoS Pathog. 2021, 17, e1009878. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Q.; Chen, V.; Shannon, C.P.; Wei, X.-S.; Xiang, X.; Wang, X.; Wang, Z.-H.; Tebbutt, S.J.; Kollmann, T.R.; Fish, E.N. Interferon-α2b Treatment for COVID-19. Front. Immunol. 2020, 11, 1061. [Google Scholar] [CrossRef]
- Sohn, K.M.; Lee, S.-G.; Kim, H.J.; Cheon, S.; Jeong, H.; Lee, J.; Kim, I.S.; Silwal, P.; Kim, Y.J.; Paik, S.; et al. COVID-19 Patients Upregulate Toll-like Receptor 4-mediated Inflammatory Signaling That Mimics Bacterial Sepsis. J. Korean Med. Sci. 2020, 35, e343. [Google Scholar] [CrossRef]
- Aguida, B.; Pooam, M.; Ahmad, M.; Jourdan, N. Infrared light therapy relieves TLR-4 dependent hyper-inflammation of the type induced by COVID-19. Commun. Integr. Biol. 2021, 14, 200–211. [Google Scholar] [CrossRef]
- Pessoa, N.L.; Bentes, A.A.; de Carvalho, A.L.; Silva, T.B.D.S.; Alves, P.A.; Reis, E.V.D.S.; Rodrigues, T.A.; Kroon, E.G.; Campos, M.A. Case report: Hepatitis in a child infected with SARS-CoV-2 presenting toll-like receptor 7 Gln11Leu single nucleotide polymorphism. Virol. J. 2021, 18, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.; Chen, X.; Cai, Y.; Xia, J.; Zhou, X.; Xu, S.; Huang, H.; Zhang, L.; Zhou, X.; Du, C.; et al. Risk Factors Associated With Acute Respiratory Distress Syndrome and Death in Patients With Coronavirus Disease 2019 Pneumonia in Wuhan, China. JAMA Intern. Med. 2020, 180, 934–943. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Curatola, A.; Chiaretti, A.; Ferretti, S.; Bersani, G.; Lucchetti, D.; Capossela, L.; Sgambato, A.; Gatto, A. Cytokine Response to SARS-CoV-2 Infection in Children. Viruses 2021, 13, 1868. [Google Scholar] [CrossRef] [PubMed]
- Scully, E.P.; Haverfield, J.; Ursin, R.L.; Tannenbaum, C.; Klein, S.L. Considering how biological sex impacts immune responses and COVID-19 outcomes. Nat. Rev. Immunol. 2020, 20, 442–447. [Google Scholar] [CrossRef] [PubMed]
- Souyris, M.; Cenac, C.; Azar, P.; Daviaud, D.; Canivet, A.; Grunenwald, S.; Pienkowski, C.; Chaumeil, J.; Mejía, J.E.; Guéry, J.-C. TLR7 escapes X chromosome inactivation in immune cells. Sci. Immunol. 2018, 3. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leng, J.; Goldstein, D.R. Impact of aging on viral infections. Microbes Infect. 2010, 12, 1120–1124. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oh, S.-J.; Lee, J.K.; Shin, O.S. Aging and the Immune System: The Impact of Immunosenescence on Viral Infection, Immunity and Vaccine Immunogenicity. Immune Netw. 2019, 19, e37. [Google Scholar] [CrossRef]
- Bartleson, J.M.; Radenkovic, D.; Covarrubias, A.J.; Furman, D.; Winer, D.A.; Verdin, E. SARS-CoV-2, COVID-19 and the aging immune system. Nat. Aging 2021, 1, 769–782. [Google Scholar] [CrossRef]
- Williamson, E.J.; Walker, A.J.; Bhaskaran, K.; Bacon, S.; Bates, C.; Morton, C.E.; Curtis, H.J.; Mehrkar, A.; Evans, D.; Inglesby, P.; et al. Factors associated with COVID-19-related death using OpenSAFELY. Nature 2020, 584, 430–436. [Google Scholar] [CrossRef]
- Richardson, S.; Hirsch, J.S.; Narasimhan, M.; Crawford, J.M.; McGinn, T.; Davidson, K.W.; Northwell COVID-19 Research Consortium. Presenting Characteristics, Comorbidities, and Outcomes Among 5700 Patients Hospitalized with COVID-19 in the New York City Area. JAMA 2020, 323, 2052–2059. [Google Scholar] [CrossRef]
- Su, Y.; Chen, D.; Yuan, D.; Lausted, C.; Choi, J.; Dai, C.L.; Voillet, V.; Duvvuri, V.R.; Scherler, K.; Troisch, P.; et al. Multi-Omics Resolves a Sharp Disease-State Shift between Mild and Moderate COVID-19. Cell 2020, 183, 1479–1495. [Google Scholar] [CrossRef] [PubMed]
- Lucas, C.; Wong, P.; Klein, J.; Castro, T.B.R.; Silva, J.; Sundaram, M.; Ellingson, M.K.; Mao, T.; Oh, J.E.; Israelow, B.; et al. Longitudinal analyses reveal immunological misfiring in severe COVID-19. Nat. Cell Biol. 2020, 584, 463–469. [Google Scholar] [CrossRef]
- Peng, T.; Du, S.-Y.; Son, M.; Diamond, B. HIF-1α is a negative regulator of interferon regulatory factors: Implications for interferon production by hypoxic monocytes. Proc. Natl. Acad. Sci. USA 2021, 118, e2106017118. [Google Scholar] [CrossRef] [PubMed]
- Taniguchi-Ponciano, K.; Vadillo, E.; Mayani, H.; Gonzalez-Bonilla, C.R.; Torres, J.; Majluf, A.; Flores-Padilla, G.; Wacher-Rodarte, N.; Galan, J.C.; Ferat-Osorio, E.; et al. Increased expression of hypoxia-induced factor 1α mRNA and its related genes in myeloid blood cells from critically ill COVID-19 patients. Ann. Med. 2021, 53, 197–207. [Google Scholar] [CrossRef] [PubMed]
- Hadjadj, J.; Yatim, N.; Barnabei, L.; Corneau, A.; Boussier, J.; Smith, N.; Péré, H.; Charbit, B.; Bondet, V.; Chenevier-Gobeaux, C.; et al. Impaired type I interferon activity and inflammatory responses in severe COVID-19 patients. Science 2020, 369, 718–724. [Google Scholar] [CrossRef] [PubMed]
- Loffredo, M.; Lucero, H.; Chen, D.-Y.; O’Connell, A.; Bergqvist, S.; Munawar, A.; Bandara, A.; De Graef, S.; Weeks, S.D.; Douam, F.; et al. The in-vitro effect of famotidine on SARS-CoV-2 proteases and virus replication. Sci. Rep. 2021, 11, 1–9. [Google Scholar] [CrossRef]
- Freedberg, D.E.; Conigliaro, J.; Wang, T.C.; Tracey, K.J.; Callahan, M.V.; Abrams, J.A.; Sobieszczyk, M.E.; Markowitz, D.D.; Gupta, A.; O’Donnell, M.R.; et al. Famotidine Use Is Associated With Improved Clinical Outcomes in Hospitalized COVID-19 Patients: A Propensity Score Matched Retrospective Cohort Study. Gastroenterology 2020, 159, 1129–1131. [Google Scholar] [CrossRef]
- Mather, J.F.; Seip, R.L.; McKay, R.G. Impact of Famotidine Use on Clinical Outcomes of Hospitalized Patients with COVID-19. Am. J. Gastroenterol. 2020, 115, 1617–1623. [Google Scholar] [CrossRef]
- Mukherjee, R.; Bhattacharya, A.; Bojkova, D.; Mehdipour, A.R.; Shin, D.; Khan, K.S.; Cheung, H.H.-Y.; Wong, K.-B.; Ng, W.-L.; Cinatl, J.; et al. Famotidine inhibits toll-like receptor 3-mediated inflammatory signaling in SARS-CoV-2 infection. J. Biol. Chem. 2021, 297, 100925. [Google Scholar] [CrossRef]
- Tiwari, S.K.; Wang, S.; Smith, D.; Carlin, A.F.; Rana, T.M. Revealing Tissue-Specific SARS-CoV-2 Infection and Host Responses using Human Stem Cell-Derived Lung and Cerebral Organoids. Stem Cell Rep. 2021, 16, 437–445. [Google Scholar] [CrossRef]
- Salaris, C.; Scarpa, M.; Elli, M.; Bertolini, A.; Guglielmetti, S.; Pregliasco, F.; Blandizzi, C.; Brun, P.; Castagliuolo, I. Protective Effects of Lactoferrin against SARS-CoV-2 Infection In Vitro. Nutrients 2021, 13, 328. [Google Scholar] [CrossRef]
- Blanco-Melo, D.; Nilsson-Payant, B.E.; Liu, W.-C.; Uhl, S.; Hoagland, D.; Møller, R.; Jordan, T.X.; Oishi, K.; Panis, M.; Sachs, D.; et al. Imbalanced Host Response to SARS-CoV-2 Drives Development of COVID-19. Cell 2020, 181, 1036–1045. [Google Scholar] [CrossRef]
- Venet, M.; Ribeiro, M.S.; Décembre, E.; Bellomo, A.; Joshi, G.; Villard, M.; Cluet, D.; Perret, M.; Pescamona, R.; Paidassi, H.; et al. SARS-CoV-2 infected cells trigger an acute antiviral response mediated by Plasmacytoid dendritic cells in mild but not severe COVID-19 patients. medRxiv 2021. [Google Scholar] [CrossRef]
- Bastard, P.; Rosen, L.B.; Zhang, Q.; Michailidis, E.; Hoffmann, H.-H.; Zhang, Y.; Dorgham, K.; Philippot, Q.; Rosain, J.; Béziat, V.; et al. Auto-antibodies against type I IFNs in patients with life-threatening COVID-19. Science 2020, 370. [Google Scholar] [CrossRef] [PubMed]
- Park, A.; Iwasaki, A. Type I and Type III Interferons—Induction, Signaling, Evasion, and Application to Combat COVID-19. Cell Host Microbe 2020, 27, 870–878. [Google Scholar] [CrossRef]
- Lei, X.; Dong, X.; Ma, R.; Wang, W.; Xiao, X.; Tian, Z.; Wang, C.; Wang, Y.; Li, L.; Ren, L.; et al. Activation and evasion of type I interferon responses by SARS-CoV-2. Nat. Commun. 2020, 11, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Han, L.; Zhuang, M.; Deng, J.; Zheng, Y.; Zhang, J.; Nan, M.; Zhang, X.; Gao, C.; Wang, P. SARS-CoV-2 ORF9b antagonizes type I and III interferons by targeting multiple components of the RIG-I/MDA-5–MAVS, TLR3–TRIF, and cGAS–STING signaling pathways. J. Med. Virol. 2021, 93, 5376–5389. [Google Scholar] [CrossRef] [PubMed]
- Schreiber, G. The Role of Type I Interferons in the Pathogenesis and Treatment of COVID-19. Front. Immunol. 2020, 11, 595739. [Google Scholar] [CrossRef]
- Li, J.-Y.; Liao, C.-H.; Wang, Q.; Tan, Y.-J.; Luo, R.; Qiu, Y.; Ge, X.-Y. The ORF6, ORF8 and nucleocapsid proteins of SARS-CoV-2 inhibit type I interferon signaling pathway. Virus Res. 2020, 286, 198074. [Google Scholar] [CrossRef]
- Sui, L.; Zhao, Y.; Wang, W.; Wu, P.; Wang, Z.; Yu, Y.; Hou, Z.; Tan, G.; Liu, Q. SARS-CoV-2 Membrane Protein Inhibits Type I Interferon Production Through Ubiquitin-Mediated Degradation of TBK1. Front. Immunol. 2021, 12, 662989. [Google Scholar] [CrossRef]
- Ricci, D.; Etna, M.; Rizzo, F.; Sandini, S.; Severa, M.; Coccia, E. Innate Immune Response to SARS-CoV-2 Infection: From Cells to Soluble Mediators. Int. J. Mol. Sci. 2021, 22, 7017. [Google Scholar] [CrossRef]
- Mullins, S.R.; Vasilakos, J.P.; Deschler, K.; Grigsby, I.; Gillis, P.; John, J.; Elder, M.J.; Swales, J.; Timosenko, E.; Cooper, Z.; et al. Intratumoral immunotherapy with TLR7/8 agonist MEDI9197 modulates the tumor microenvironment leading to enhanced activity when combined with other immunotherapies. J. Immunother. Cancer 2019, 7, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Surendran, N.; Simmons, A.; Pichichero, M.E. TLR agonist combinations that stimulate Th type I polarizing responses from human neonates. Innate Immun. 2018, 24, 240–251. [Google Scholar] [CrossRef] [PubMed]
- Hennessy, E.J.; Parker, A.E.; O’Neill, L. Targeting Toll-like receptors: Emerging therapeutics? Nat. Rev. Drug Discov. 2010, 9, 293–307. [Google Scholar] [CrossRef]
- Kayesh, M.E.H.; Kohara, M.; Tsukiyama-Kohara, K. Toll-Like Receptor Response to Hepatitis B Virus Infection and Potential of TLR Agonists as Immunomodulators for Treating Chronic Hepatitis B: An Overview. Int. J. Mol. Sci. 2021, 22, 10462. [Google Scholar] [CrossRef] [PubMed]
- Alderson, M.R.; McGowan, P.; Baldridge, J.R.; Probst, P. TLR4 agonists as immunomodulatory agents. J. Endotoxin Res. 2006, 12, 313–319. [Google Scholar] [CrossRef]
- Baldridge, J.R.; McGowan, P.; Evans, J.T.; Cluff, C.; Mossman, S.; Johnson, D.; Persing, D. Taking a Toll on human disease: Toll-like receptor 4 agonists as vaccine adjuvants and monotherapeutic agents. Expert Opin. Biol. Ther. 2004, 4, 1129–1138. [Google Scholar] [CrossRef]
- Evans, J.T.; Cluff, C.W.; Johnson, D.A.; Lacy, M.J.; Persing, D.H.; Baldridge, J.R. Enhancement of antigen-specific immunity via the TLR4 ligands MPL™ adjuvant and Ribi.529. Expert Rev. Vaccines 2003, 2, 219–229. [Google Scholar] [CrossRef]
- Sánchez-Ramón, S.; Conejero, L.; Netea, M.G.; Sancho, D.; Palomares, Ó.; Subiza, J.L. Trained Immunity-Based Vaccines: A New Paradigm for the Development of Broad-Spectrum Anti-infectious Formulations. Front. Immunol. 2018, 9, 2936. [Google Scholar] [CrossRef] [PubMed]
- Reed, S.G.; Orr, M.; Fox, C. Key roles of adjuvants in modern vaccines. Nat. Med. 2013, 19, 1597–1608. [Google Scholar] [CrossRef]
- Roßmann, L.; Bagola, K.; Stephen, T.; Gerards, A.-L.; Walber, B.; Ullrich, A.; Schülke, S.; Kamp, C.; Spreitzer, I.; Hasan, M.; et al. Distinct single-component adjuvants steer human DC-mediated T-cell polarization via Toll-like receptor signaling toward a potent antiviral immune response. Proc. Natl. Acad. Sci. USA 2021, 118, e2103651118. [Google Scholar] [CrossRef]
- Jeong, S.-K.; Heo, Y.-K.; Jeong, J.-H.; Ham, S.-J.; Yum, J.-S.; Ahn, B.-C.; Song, C.-S.; Chun, E.-Y. COVID-19 Subunit Vaccine with a Combination of TLR1/2 and TLR3 Agonists Induces Robust and Protective Immunity. Vaccines 2021, 9, 957. [Google Scholar] [CrossRef] [PubMed]
- Proud, P.C.; Tsitoura, D.; Watson, R.J.; Chua, B.Y.; Aram, M.J.; Bewley, K.R.; Cavell, B.E.; Cobb, R.; Dowall, S.; Fotheringham, S.A.; et al. Prophylactic intranasal administration of a TLR2/6 agonist reduces upper respiratory tract viral shedding in a SARS-CoV-2 challenge ferret model. EBioMedicine 2021, 63, 103153. [Google Scholar] [CrossRef]
- Kumar, S.; Kesharwani, S.S.; Kuppast, B.; Rajput, M.; Bakkari, M.A.; Tummala, H. Discovery of inulin acetate as a novel immune-active polymer and vaccine adjuvant: Synthesis, material characterization, and biological evaluation as a toll-like receptor-4 agonist. J. Mater. Chem. B 2016, 4, 7950–7960. [Google Scholar] [CrossRef] [PubMed]
- Bakkari, M.A.; Valiveti, C.K.; Kaushik, R.S.; Tummala, H. Toll-like Receptor-4 (TLR4) Agonist-Based Intranasal Nanovaccine Delivery System for Inducing Systemic and Mucosal Immunity. Mol. Pharm. 2021, 18, 2233–2241. [Google Scholar] [CrossRef] [PubMed]
- Zhao, K.; Wang, H.; Wu, C. The immune responses of HLA-A*0201 restricted SARS-CoV S peptide-specific CD8+ T cells are augmented in varying degrees by CpG ODN, PolyI:C and R848. Vaccine 2011, 29, 6670–6678. [Google Scholar] [CrossRef] [PubMed]
- Kared, H.; Redd, A.D.; Bloch, E.M.; Bonny, T.S.; Sumatoh, H.R.; Kairi, F.; Carbajo, D.; Abel, B.; Newell, E.W.; Bettinotti, M.P.; et al. SARS-CoV-2–specific CD8+ T cell responses in convalescent COVID-19 individuals. J. Clin. Investig. 2021, 131. [Google Scholar] [CrossRef]
- Pan, Y.; Jiang, X.; Yang, L.; Chen, L.; Zeng, X.; Liu, G.; Tang, Y.; Qian, C.; Wang, X.; Cheng, F.; et al. SARS-CoV-2-specific immune response in COVID-19 convalescent individuals. Signal. Transduct. Target. Ther. 2021, 6, 1–10. [Google Scholar] [CrossRef]
- Moderbacher, C.R.; Ramirez, S.I.; Dan, J.M.; Grifoni, A.; Hastie, K.M.; Weiskopf, D.; Belanger, S.; Abbott, R.K.; Kim, C.; Choi, J.; et al. Antigen-Specific Adaptive Immunity to SARS-CoV-2 in Acute COVID-19 and Associations with Age and Disease Severity. Cell 2020, 183, 996–1012. [Google Scholar] [CrossRef]
- Arunachalam, P.S.; Walls, A.C.; Golden, N.; Atyeo, C.; Fischinger, S.; Li, C.; Aye, P.; Navarro, M.J.; Lai, L.; Edara, V.V.; et al. Adjuvanting a subunit COVID-19 vaccine to induce protective immunity. Nat. Cell Biol. 2021, 594, 253–258. [Google Scholar] [CrossRef] [PubMed]
- Richmond, P.; Hatchuel, L.; Dong, M.; Ma, B.; Hu, B.; Smolenov, I.; Li, P.; Liang, P.; Han, H.H.; Liang, J.; et al. Safety and immunogenicity of S-Trimer (SCB-2019), a protein subunit vaccine candidate for COVID-19 in healthy adults: A phase 1, randomised, double-blind, placebo-controlled trial. Lancet 2021, 397, 682–694. [Google Scholar] [CrossRef]
- Angelopoulou, A.; Alexandris, N.; Konstantinou, E.; Mesiakaris, K.; Zanidis, C.; Farsalinos, K.; Poulas, K. Imiquimod-A toll like receptor 7 agonist-Is an ideal option for management of COVID 19. Environ. Res. 2020, 188, 109858. [Google Scholar] [CrossRef] [PubMed]
- Dyavar, S.R.; Singh, R.; Emani, R.; Pawar, G.P.; Chaudhari, V.D.; Podany, A.T.; Avedissian, S.N.; Fletcher, C.V.; Salunke, D.B. Role of toll-like receptor 7/8 pathways in regulation of interferon response and inflammatory mediators during SARS-CoV2 infection and potential therapeutic options. Biomed. Pharmacother. 2021, 141, 111794. [Google Scholar] [CrossRef] [PubMed]
- Khanmohammadi, S.; Rezaei, N. Role of Toll-like receptors in the pathogenesis of COVID-19. J. Med. Virol. 2021, 93, 2735–2739. [Google Scholar] [CrossRef] [PubMed]
| Vaccine | Sponsor | TLR Agonist Adjuvant | Target TLR | Clinical Phase of Development | Effects on Host Immunity | Clinical Trials. Gov Identifier/Reference |
|---|---|---|---|---|---|---|
| IMP CoVac-1 (SARS-CoV-2-derived multi-peptide vaccine) | University Hospital Tuebingen | TLR1/2 ligand XS15 | TLR1/2 | Phase I | Results not published yet | NCT04546841 |
| VXA-CoV2-1 | Vaxart | dsRNA | TLR3 | Phase I | Results not published yet | NCT04563702 |
| VXA-CoV2-1.1-S | Vaxart | dsRNA | TLR3 | Phase II | Results not published yet | NCT05067933 |
| SCB-2019 vaccine | Clover Biopharmaceuticals AUS Pty Ltd. | CpG 1018 plus alum | TLR9 | Phase I | Well-tolerated in healthy volunteers; elicited T-helper-1-biased CD4+ T-cell responses | [112] |
| SCB-2019 vaccine | Zhejiang Clover Biopharmaceuticals Inc. | CpG 1018 and alhydrogel | TLR9 | Phase II | Results not published yet | NCT04954131 |
| SCB-2019 vaccine | Clover Biopharmaceuticals AUS Pty Ltd. | CpG 1018 plus alum | TLR9 | Phase III | Results not published yet | NCT05012787 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kayesh, M.E.H.; Kohara, M.; Tsukiyama-Kohara, K. An Overview of Recent Insights into the Response of TLR to SARS-CoV-2 Infection and the Potential of TLR Agonists as SARS-CoV-2 Vaccine Adjuvants. Viruses 2021, 13, 2302. https://doi.org/10.3390/v13112302
Kayesh MEH, Kohara M, Tsukiyama-Kohara K. An Overview of Recent Insights into the Response of TLR to SARS-CoV-2 Infection and the Potential of TLR Agonists as SARS-CoV-2 Vaccine Adjuvants. Viruses. 2021; 13(11):2302. https://doi.org/10.3390/v13112302
Chicago/Turabian StyleKayesh, Mohammad Enamul Hoque, Michinori Kohara, and Kyoko Tsukiyama-Kohara. 2021. "An Overview of Recent Insights into the Response of TLR to SARS-CoV-2 Infection and the Potential of TLR Agonists as SARS-CoV-2 Vaccine Adjuvants" Viruses 13, no. 11: 2302. https://doi.org/10.3390/v13112302
APA StyleKayesh, M. E. H., Kohara, M., & Tsukiyama-Kohara, K. (2021). An Overview of Recent Insights into the Response of TLR to SARS-CoV-2 Infection and the Potential of TLR Agonists as SARS-CoV-2 Vaccine Adjuvants. Viruses, 13(11), 2302. https://doi.org/10.3390/v13112302






