Autophagy Inhibits Intercellular Transport of Citrus Leaf Blotch Virus by Targeting Viral Movement Protein
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant and Virus Materials
2.2. Plasmid Construction
2.3. Agroinfiltration
2.4. Fluorescence Protein Observation
2.5. Maltose-Binding Protein (MBP) Pull-Down Assay
2.6. Chemical Treatments
2.7. Transmission Electron Microscopy (TEM) Observation
2.8. Immunoblot Analysis
2.9. RT-PCR, Quantitative RT-PCR (qRT-PCR) and Northern Blot Analyses
3. Results
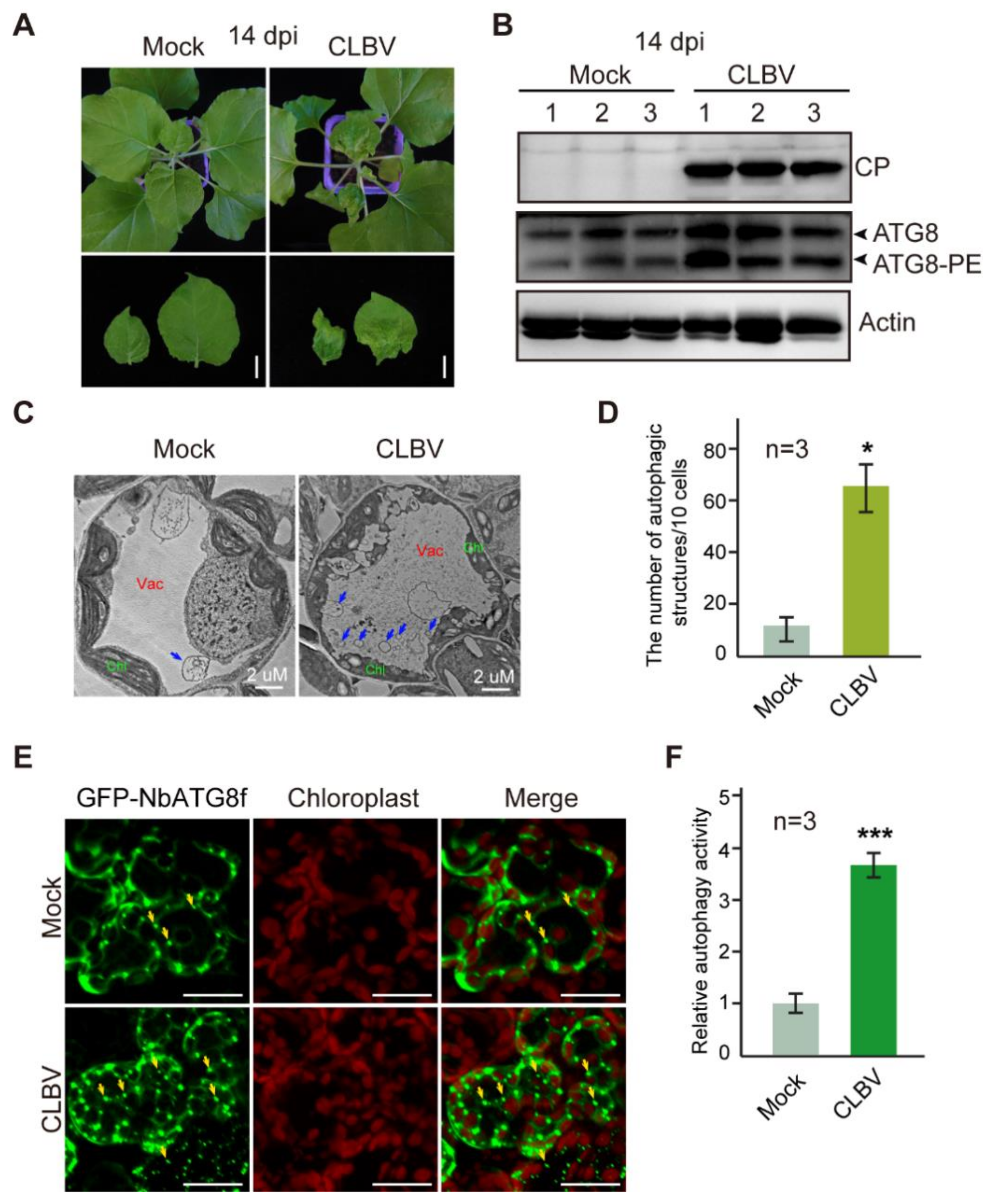
3.1. CLBV Infection Activates Autophagy
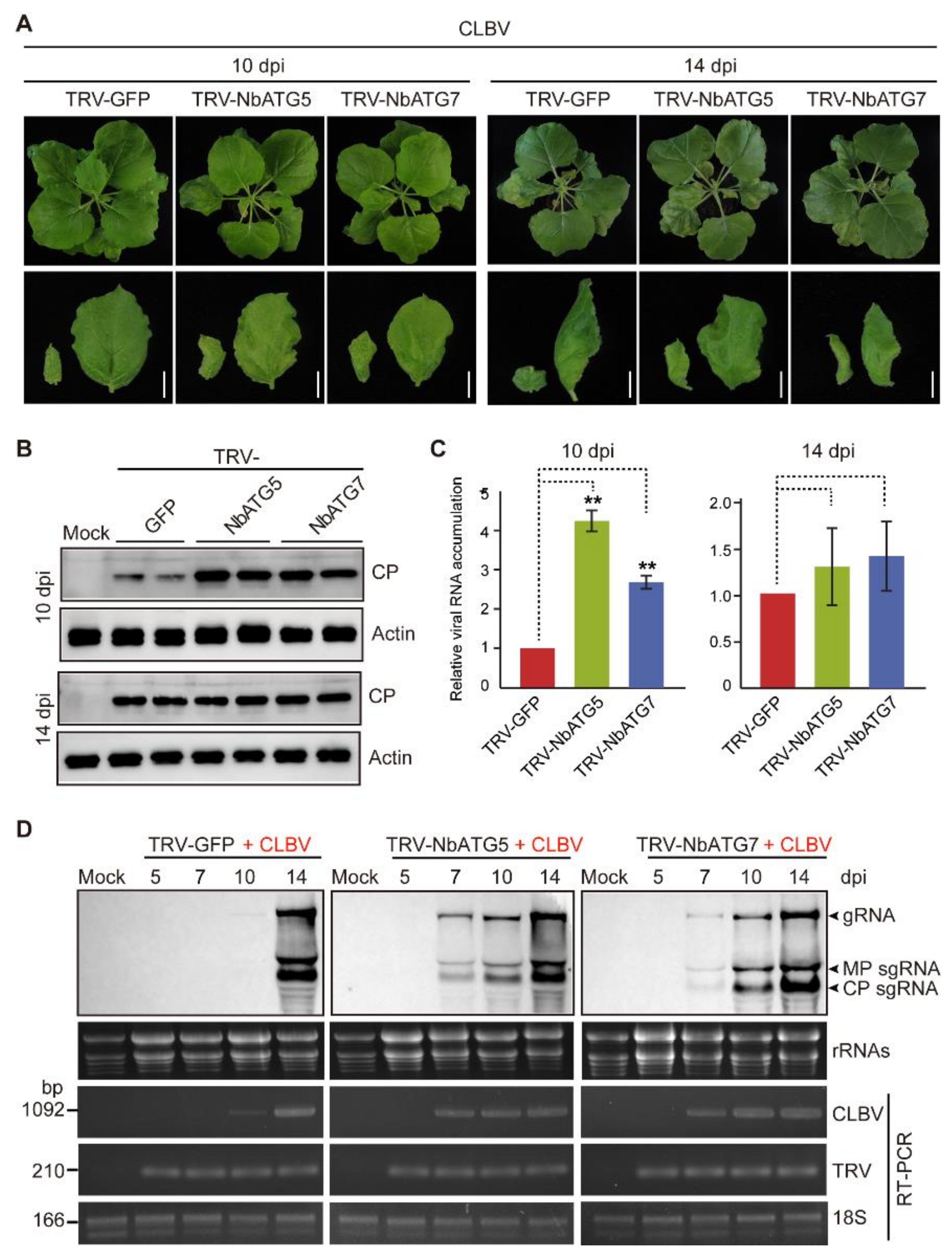
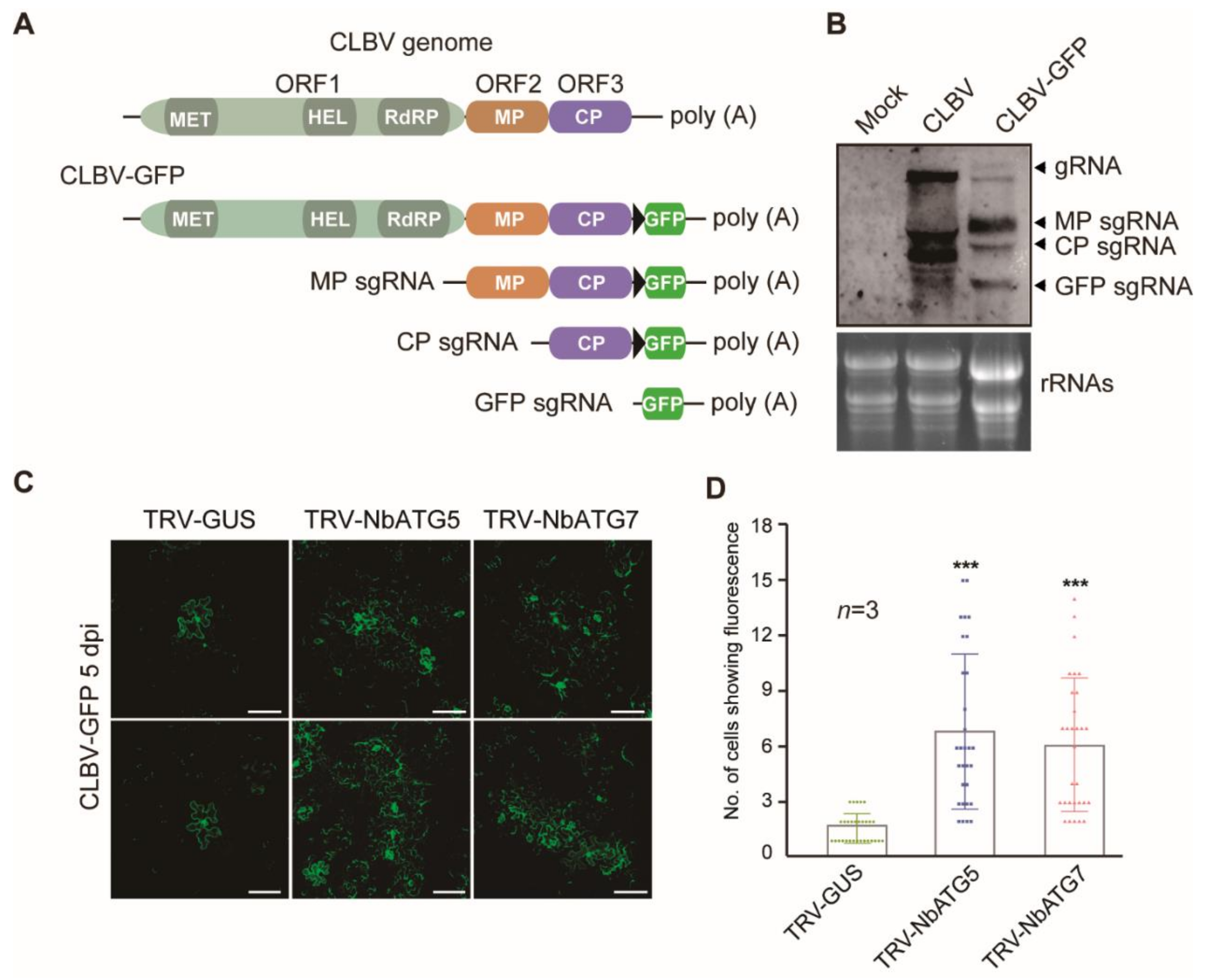
3.2. Autophagy Inhibits Systemic Infection of CLBV
3.3. Autophagy Inhibits Cell-to-Cell Movement of CLBV
3.4. CLBV MP Is Targeted for Autophagic Degradation
3.5. CLBV MP Interacts with NbATG8C1 and NbATG8i
3.6. N-terminal Region Containing AIM Is Important for Recruitment of CLBV MP to Autophagosomes
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Marshall, R.S.; Vierstra, R.D. Autophagy: The Master of Bulk and Selective Recycling. Annu. Rev. Plant Biol. 2018, 69, 173–208. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.; Chen, Y.; Tooze, S.A. Autophagy pathway: Cellular and molecular mechanisms. Autophagy 2018, 14, 207–215. [Google Scholar] [CrossRef] [PubMed]
- Boya, P.; Reggiori, F.; Codogno, P. Emerging regulation and functions of autophagy. Nat. Cell Biol. 2013, 15, 713–720. [Google Scholar] [CrossRef] [PubMed]
- Klionsky, D.J.; Codogno, P. The Mechanism and Physiological Function of Macroautophagy. J. Innate Immun. 2013, 5, 427–433. [Google Scholar] [CrossRef]
- Klionsky, D.J. The molecular machinery of autophagy: Unanswered questions. J. Cell Sci. 2005, 118, 7–18. [Google Scholar] [CrossRef]
- Massey, A.; Kiffin, R.; Cuervo, A.M. Pathophysiology of chaperone-mediated autophagy. Int. J. Biochem. Cell Biol. 2004, 36, 2420–2434. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Vierstra, R.D. Autophagy: A multifaceted intracellular system for bulk and selective recycling. Trends Plant Sci. 2012, 17, 526–537. [Google Scholar] [CrossRef]
- Xie, Z.; Klionsky, D.J. Autophagosome formation: Core machinery and adaptations. Nat. Cell Biol. 2007, 9, 1102–1109. [Google Scholar] [CrossRef] [PubMed]
- Lamb, C.; Yoshimori, T.; Tooze, S. The autophagosome: Origins unknown, biogenesis complex. Nat. Rev. Mol. Cell Biol. 2013, 14, 759–774. [Google Scholar] [CrossRef]
- Johansen, T.; Lamark, T. Selective autophagy mediated by autophagic adapter proteins. Autophagy 2011, 7, 279–296. [Google Scholar] [CrossRef] [PubMed]
- Kellner, R.; De la Concepcion, J.C.; Maqbool, A.; Kamoun, S.; Dagdas, Y.F. ATG8 Expansion: A Driver of Selective Autophagy Diversification? Trends Plant Sci. 2017, 22, 204–214. [Google Scholar] [CrossRef]
- Nakatogawa, H.; Ichimura, Y.; Ohsumi, Y. Atg8, a Ubiquitin-like Protein Required for Autophagosome Formation, Mediates Membrane Tethering and Hemifusion. Cell 2007, 130, 165–178. [Google Scholar] [CrossRef]
- Kaufmann, A.; Wollert, T. Scaffolding the expansion of autophagosomes. Autophagy 2014, 10, 1343–1345. [Google Scholar] [CrossRef][Green Version]
- Bu, F.; Yang, M.; Guo, X.; Huang, W.; Chen, L. Multiple Functions of ATG8 Family Proteins in Plant Autophagy. Front. Cell Dev. Biol. 2020, 8, 466. [Google Scholar] [CrossRef]
- Rogov, V.; Dotsch, V.; Johansen, T.; Kirkin, V. Interactions between Autophagy Receptors and Ubiquitin-like Proteins Form the Molecular Basis for Selective Autophagy. Mol. Cell 2014, 53, 167–178. [Google Scholar] [CrossRef]
- Birgisdottir, A.; Lamark, T.; Johansen, T. The LIR motif-crucial for selective autophagy. J. Cell Sci. 2013, 126, 3237–3247. [Google Scholar] [CrossRef]
- Haxim, Y.; Ismayil, A.; Jia, Q.; Wang, Y.; Zheng, X.; Chen, T.; Qian, L.; Liu, N.; Wang, Y.; Han, S.; et al. Autophagy functions as an antiviral mechanism against geminiviruses in plants. eLife 2017, 6, e23897. [Google Scholar] [CrossRef]
- Li, F.; Zhang, M.; Zhang, C.; Zhou, X. Nuclear autophagy degrades a geminivirus nuclear protein to restrict viral infection in solanaceous plants. New Phytol. 2019, 225, 1746–1761. [Google Scholar] [CrossRef]
- Kushwaha, N.K.; Hafrén, A.; Hofius, D. Autophagy-virus interplay in plants: From antiviral recognition to proviral manipulation. Mol. Plant Pathol. 2019, 20, 1211–1216. [Google Scholar] [CrossRef]
- Huang, X.; Chen, S.; Yang, X.; Yang, X.; Zhang, T.; Zhou, G. Friend or Enemy: A Dual Role of Autophagy in Plant Virus Infection. Front. Microbiol. 2020, 11, 736. [Google Scholar] [CrossRef]
- Li, F.; Zhang, C.; Tang, Z.; Zhang, L.; Dai, Z.; Lyu, S.; Li, Y.; Hou, X.; Bernards, M.; Wang, A. A plant RNA virus activates selective autophagy in a UPR--dependent manner to promote virus infection. New Phytol. 2020, 228, 622–639. [Google Scholar] [CrossRef] [PubMed]
- Wong, H.H.; Sanyal, S. Manipulation of autophagy by (+) RNA viruses. Semin. Cell Dev. Biol. 2020, 101, 3–11. [Google Scholar] [CrossRef] [PubMed]
- Cheng, X.; Wang, A. The Potyvirus Silencing Suppressor Protein VPg Mediates Degradation of SGS3 via Ubiquitination and Autophagy Pathways. J. Virol. 2017, 91, 01478-16. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Zhao, N.; Li, Z.; Xu, X.; Wang, Y.; Yang, X.; Liu, S.-S.; Wang, A.; Zhou, X. A calmodulin-like protein suppresses RNA silencing and promotes geminivirus infection by degrading SGS3 via the autophagy pathway in Nicotiana benthamiana. PLoS Pathog. 2017, 13, e1006213. [Google Scholar] [CrossRef]
- Fu, S.; Xu, Y.; Li, C.; Li, Y.; Wu, J.; Zhou, X. Rice Stripe Virus Interferes with S-acylation of Remorin and Induces Its Autophagic Degradation to Facilitate Virus Infection. Mol. Plant. 2018, 11, 269–287. [Google Scholar] [CrossRef]
- Huang, Y.-P.; Hsiao, Y.-J.; Li, S.-C.; Hsu, Y.-H.; Tsai, C.-H. Autophagy is involved in assisting the replication of Bamboo mosaic virus in Nicotiana benthamiana. J. Exp. Bot. 2019, 70, 4657–4670. [Google Scholar] [CrossRef]
- Michaeli, S.; Clavel, M.; Lechner, E.; Viotti, C.; Wu, J.; Dubois, M.; Hacquard, T.; Derrien, B.; Izquierdo, E.; Lecorbeiller, M.; et al. The viral F-box protein P0 induces an ER-derived autophagy degradation pathway for the clearance of membrane-bound AGO1. Proc. Natl. Acad. Sci. USA 2019, 116, 22872–22883. [Google Scholar] [CrossRef]
- Nakahara, K.S.; Masuta, C.; Yamada, S.; Shimura, H.; Kashihara, Y.; Wada, T.S.; Meguro, A.; Goto, K.; Tadamura, K.; Sueda, K.; et al. Tobacco calmodulin-like protein provides secondary defense by binding to and directing degradation of virus RNA silencing suppressors. Proc. Natl. Acad. Sci. USA 2012, 109, 10113–10118. [Google Scholar] [CrossRef]
- Li, F.; Zhang, C.; Li, Y.; Wu, G.-W.; Hou, X.; Zhou, X.; Wang, A. Beclin1 restricts RNA virus infection in plants through suppression and degradation of the viral polymerase. Nat. Commun. 2018, 9, 1–17. [Google Scholar] [CrossRef]
- Jiang, L.; Lu, Y.; Zheng, X.; Yang, X.; Chen, Y.; Zhang, T.; Zhao, X.; Wang, S.; Zhao, X.; Song, X.; et al. The plant protein NbP3IP directs degradation of Rice stripe virus p3 silencing suppressor protein to limit virus infection through interaction with the autophagy--related protein NbATG8. New Phytol. 2021, 229, 1036–1051. [Google Scholar] [CrossRef]
- Hafrén, A.; Üstün, S.; Hochmuth, A.; Svenning, S.; Johansen, T.; Hofius, D. Turnip Mosaic Virus Counteracts Selective Autophagy of the Viral Silencing Suppressor HCpro. Plant. Physiol. 2018, 176, 649–662. [Google Scholar] [CrossRef] [PubMed]
- Hafrén, A.; Macia, J.-L.; Love, A.J.; Milner, J.J.; Drucker, M.; Hofius, D. Selective autophagy limits cauliflower mosaic virus infection by NBR1-mediated targeting of viral capsid protein and particles. Proc. Natl. Acad. Sci. USA 2017, 114, E2026–E2035. [Google Scholar] [CrossRef] [PubMed]
- Ismayil, A.; Yang, M.; Haxim, Y.; Wang, Y.; Li, J.; Han, L.; Wang, Y.; Zheng, X.; Wei, X.; Nagalakshmi, U.; et al. Cotton leaf curl Multan virus βC1 Protein Induces Autophagy by Disrupting the Interaction of Autophagy-Related Protein 3 with Glyceraldehyde-3-Phosphate Dehydrogenases[OPEN]. Plant. Cell 2020, 32, 1124–1135. [Google Scholar] [CrossRef] [PubMed]
- Lefkowitz, E.J.; Dempsey, D.M.; Hendrickson, R.C.; Orton, R.J.; Siddell, S.G.; Smith, D.B. Virus taxonomy: The database of the International Committee on Taxonomy of Viruses (ICTV). Nucleic Acids Res. 2018, 46, D708–D717. [Google Scholar] [CrossRef]
- Guardo, M.; Sorrentino, G.; Marletta, T.; Caruso, A. First Report of Citrus leaf blotch virus on Kumquat in Italy. Plant. Dis. 2007, 91, 1054. [Google Scholar] [CrossRef]
- Wang, J.; Zhu, D.; Tan, Y.; Zong, X.; Wei, M.H.; Liu, Q. First Report of Citrus leaf blotch virus in Sweet Cherry. Plant. Dis. 2016, 100, 1027. [Google Scholar] [CrossRef]
- Cao, M.; Yu, Y.-Q.; Tian, X.; Yang, F.Y.; Li, R.H.; Zhou, C.Y. First Report of Citrus leaf blotch virus in Lemon in China. Plant. Dis. 2017, 101, 1561. [Google Scholar] [CrossRef]
- Liu, H.; Song, S.; Wu, W.; Mi, W.; Shen, C.; Bai, B.; Wu, Y. Distribution and molecular characterization of Citrus leaf blotch virus from Actinidia in Shaanxi province, China. Eur. J. Plant. Pathol. 2019, 154, 855–862. [Google Scholar] [CrossRef]
- Galipienso, L.; Navarro, L.; Ballester-Olmos, J.; Pina, J.; Moreno, P.; Guerri, J. Host range and symptomatology of a graft-transmissible pathogen causing bud union crease of citrus on trifoliate rootstocks. Plant. Pathol. 2000, 49, 308–314. [Google Scholar] [CrossRef]
- Vives, M.C.; Martín, S.; Ambrós, S.; Renovell, A.; Navarro, L.; Pina, J.A.; Moreno, P.; Guerri, J. Development of a full-genome cDNA clone of Citrus leaf blotch virus and infection of citrus plants. Mol. Plant Pathol. 2008, 9, 787–797. [Google Scholar] [CrossRef]
- Guardo, M.; Oriana, P.; Castellano, M.A.; Savino, V.; Caruso, A. A new herbaceous host of citrus leaf blotch virus. J. Plant Pathol. 2009, 91, 485–488. [Google Scholar]
- Agüero, J.; Vives, M.C.; Velázquez, K.; Ruiz-Ruiz, S.; Juárez, J.; Navarro, L.; Moreno, P.; Guerri, J. Citrus leaf blotch virus invades meristematic regions in Nicotiana benthamiana and citrus. Mol. Plant. Pathol. 2013, 14, 610–616. [Google Scholar] [CrossRef]
- Vives, M.; Galipienso, L.; Navarro, L.; Moreno, P.; Guerri, J. The Nucleotide Sequence and Genomic Organization of Citrus Leaf Blotch Virus: Candidate Type Species for a New Virus Genus. Virology 2001, 287, 225–233. [Google Scholar] [CrossRef] [PubMed]
- Vives, M.C.; Galipienso, L.; Navarro, L.; Moreno, P.; Guerri, J. Characterization of Two Kinds of Subgenomic RNAs Produced by Citrus Leaf Blotch Virus. Virology 2002, 295, 328–336. [Google Scholar] [CrossRef] [PubMed]
- Renovell, Á.; Gago, S.; Ruiz-Ruiz, S.; Velázquez, K.; Navarro, L.; Moreno, P.; Vives, M.C.; Guerri, J. Mapping the subgenomic RNA promoter of the Citrus leaf blotch virus coat protein gene by Agrobacterium-mediated inoculation. Virology 2010, 406, 360–369. [Google Scholar] [CrossRef]
- Renovell, Á.; Vives, M.C.; Ruiz-Ruiz, S.; Navarro, L.; Moreno, P.; Guerri, J. The Citrus leaf blotch virus movement protein acts as silencing suppressor. Virus Genes 2011, 44, 131–140. [Google Scholar] [CrossRef]
- Zhang, X.-Y.; Dong, S.-W.; Xiang, H.-Y.; Chen, X.-R.; Li, D.-W.; Yu, J.-L.; Han, C.-G. Development of three full-length infectious cDNA clones of distinct brassica yellows virus genotypes for agrobacterium-mediated inoculation. Virus Res. 2015, 197, 13–16. [Google Scholar] [CrossRef]
- Agüero, J.; Ruiz-Ruiz, S.; Vives, M.D.C.; Velázquez, K.; Navarro, L.; Peña, L.; Moreno, P.; Guerri, J. Development of Viral Vectors Based on Citrus leaf blotch virus to Express Foreign Proteins or Analyze Gene Function in Citrus Plants. Mol. Plant.-Microbe Interact. 2012, 25, 1326–1337. [Google Scholar] [CrossRef]
- Imoto, A.; Aida, M. A ClearSee-Based Clearing Protocol for 3D Visualization of Arabidopsis thaliana Embryos. Plants 2021, 10, 190. [Google Scholar] [CrossRef]
- Voinnet, O.; Lederer, C.; Baulcombe, D.C. A Viral Movement Protein Prevents Spread of the Gene Silencing Signal in Nicotiana benthamiana. Cell 2016, 166, 780. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, X.; Niu, S.; Han, C.; Yu, J.; Li, D. Nuclear localization of Beet black scorch virus capsid protein and its interaction with importin α. Virus Res. 2011, 155, 307–315. [Google Scholar] [CrossRef]
- Zhang, X.; Yang, Y.; Zhao, M.; Yang, L.; Jiang, J.; Walcott, R.; Yang, S.; Zhao, T. Acidovorax citrulli Type III Effector AopP Suppresses Plant Immunity by Targeting the Watermelon Transcription Factor WRKY6. Front. Plant. Sci. 2020, 11, 579218. [Google Scholar] [CrossRef]
- Sun, L.; Andika, I.; Kondo, H.; Chen, J. Identification of the amino acid residues and domains in the cysteine-rich protein of Chinese wheat mosaic virus that are important for RNA silencing suppression and subcellular localization. Mol. Plant Pathol. 2013, 14, 265–278. [Google Scholar] [CrossRef]
- Walker, I.H.; Hsieh, P.-C.; Riggs, P.D. Mutations in maltose-binding protein that alter affinity and solubility properties. Appl. Microbiol. Biotechnol. 2010, 88, 187–197. [Google Scholar] [CrossRef]
- Liu, Y.; Schiff, M.; Dinesh-Kumar, S.P. Virus-induced gene silencing in tomato. Plant. J. 2002, 31, 777–786. [Google Scholar] [CrossRef]
- Voinnet, O.; Pinto, Y.M.; Baulcombe, D.C. Suppression of gene silencing: A general strategy used by diverse DNA and RNA viruses of plants. Proc. Natl. Acad. Sci. USA 1999, 96, 14147–14152. [Google Scholar] [CrossRef]
- Sun, L.; Nuss, D.L.; Suzuki, N. Synergism between a mycoreovirus and a hypovirus mediated by the papain-like protease p29 of the prototypic hypovirus CHV1-EP713. J. Gen. Virol. 2006, 87, 3703–3714. [Google Scholar] [CrossRef]
- Hofius, D.; Schultz-Larsen, T.; Joensen, J.; Tsitsigiannis, D.I.; Petersen, N.H.; Mattsson, O.; Jørgensen, L.B.; Jones, J.; Mundy, J.; Petersen, M. Autophagic Components Contribute to Hypersensitive Cell Death in Arabidopsis. Cell 2009, 137, 773–783. [Google Scholar] [CrossRef]
- Kabeya, Y.; Mizushima, N.; Ueno, T.; Yamamoto, A.; Kirisako, T.; Noda, T.; Kominami, E.; Ohsumi, Y.; Yoshimori, T. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000, 19, 5720–5728. [Google Scholar] [CrossRef]
- Álvarez, C.; García, I.; Moreno, I.; Pérez-Pérez, M.E.; Crespo, J.L.; Romero, L.C.; Gotor, C. Cysteine-Generated Sulfide in the Cytosol Negatively Regulates Autophagy and Modulates the Transcriptional Profile in Arabidopsis. Plant. Cell 2012, 24, 4621–4634. [Google Scholar] [CrossRef]
- Contento, A.L.; Xiong, Y.; Bassham, D.C. Visualization of autophagy in Arabidopsis using the fluorescent dye monodansylcadaverine and a GFP-AtATG8e fusion protein. Plant J. 2005, 42, 598–608. [Google Scholar] [CrossRef]
- Li, F.; Chung, T.; Pennington, J.G.; Federico, M.L.; Kaeppler, H.F.; Kaeppler, S.; Otegui, M.S.; Vierstra, R.D. Autophagic Recycling Plays a Central Role in Maize Nitrogen Remobilization. Plant. Cell 2015, 27, 1389–1408. [Google Scholar] [CrossRef] [PubMed]
- Zhu, T.; Zou, L.; Li, Y.; Yao, X.; Xu, F.; Deng, X.; Zhang, D.; Lin, H. Mitochondrial alternative oxidase-dependent autophagy involved in ethylene-mediated drought tolerance in Solanum lycopersicum. Plant. Biotechnol. J. 2018, 16, 2063–2076. [Google Scholar] [CrossRef]
- Kirisako, T.; Ichimura, Y.; Okada, H.; Kabeya, Y.; Mizushima, N.; Yoshimori, T.; Ohsumi, M.; Takao, T.; Noda, T.; Ohsumi, Y. The Reversible Modification Regulates the Membrane-Binding State of Apg8/Aut7 Essential for Autophagy and the Cytoplasm to Vacuole Targeting Pathway. J. Cell Biol. 2000, 151, 263–276. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Yu, B.; Zhao, J.; Guo, J.; Li, Y.; Han, S.; Huang, L.; Du, Y.; Hong, Y.; Tang, D.; et al. Autophagy Contributes to Leaf Starch Degradation. Plant. Cell 2013, 25, 1383–1399. [Google Scholar] [CrossRef]
- Benitez-Alfonso, Y.; Faulkner, C.; Ritzenthaler, C.; Maule, A.J. Plasmodesmata: Gateways to Local and Systemic Virus Infection. Mol. Plant. Microbe Interact. 2010, 23, 1403–1412. [Google Scholar] [CrossRef]
- Hipper, C.; Brault, V.; Ziegler-Graff, V.; Revers, F. Viral and Cellular Factors Involved in Phloem Transport of Plant Viruses. Front. Plant. Sci. 2013, 4, 154. [Google Scholar] [CrossRef]
- Hu, C.-D.; Kerppola, T.K. Simultaneous visualization of multiple protein interactions in living cells using multicolor fluorescence complementation analysis. Nat. Biotechnol. 2003, 21, 539–545. [Google Scholar] [CrossRef]
- Kalvari, I.; Tsompanis, S.; Mulakkal, N.C.; Osgood, R.; Johansen, T.; Nezis, I.; Promponas, V. iLIR. Autophagy 2014, 10, 913–925. [Google Scholar] [CrossRef]
- Calil, I.P.; Fontes, E.P.B. Plant immunity against viruses: Antiviral immune receptors in focus. Ann. Bot. 2016, 119, mcw200–723. [Google Scholar] [CrossRef]
- Li, F.; Wang, A. RNA-Targeted Antiviral Immunity: More Than Just RNA Silencing. Trends Microbiol. 2019, 27, 792–805. [Google Scholar] [CrossRef]
- Ismayil, A.; Yang, M.; Liu, Y. Role of autophagy during plant-virus interactions. Semin. Cell Dev. Biol. 2020, 101, 36–40. [Google Scholar] [CrossRef]
- Leary, A.; Savage, Z.; Tumtas, Y.; Bozkurt, T.O. Contrasting and emerging roles of autophagy in plant immunity. Curr. Opin. Plant. Biol. 2019, 52, 46–53. [Google Scholar] [CrossRef]
- Yang, M.; Ismayil, A.; Liu, Y. Autophagy in Plant-Virus Interactions. Annu. Rev. Virol. 2020, 7, 403–419. [Google Scholar] [CrossRef]
- Yang, M.; Zhang, Y.; Xie, X.; Yue, N.; Li, J.; Wang, X.-B.; Han, C.; Yu, J.; Liu, Y.; Li, D. Barley stripe mosaic virus γb Protein Subverts Autophagy to Promote Viral Infection by Disrupting the ATG7-ATG8 Interaction. Plant. Cell 2018, 30, 1582–1595. [Google Scholar] [CrossRef]
- Lucas, W.J. Plant viral movement proteins: Agents for cell-to-cell trafficking of viral genomes. Virology 2006, 344, 169–184. [Google Scholar] [CrossRef]
- Reichel, C.; Beachy, R.N. Degradation of Tobacco Mosaic Virus Movement Protein by the 26S Proteasome. J. Virol. 2000, 74, 3330–3337. [Google Scholar] [CrossRef]
- Drugeon, G.; Jupin, I. Stability in vitro of the 69K movement protein of Turnip yellow mosaic virus is regulated by the ubiquitin-mediated proteasome pathway. J. Gen. Virol. 2002, 83, 3187–3197. [Google Scholar] [CrossRef]
- Becker, F.; Buschfeld, E.; Schell, J.; Bachmair, A. Altered response to viral infection by tobacco plants perturbed in ubiquitin system. Plant J. 1993, 3, 875–881. [Google Scholar] [CrossRef]
- Citovsky, V.; Zaltsman, A.; Kozlovsky, S.V.; Gafni, Y.; Krichevsky, A. Proteasomal degradation in plant–pathogen interactions. Semin. Cell Dev. Biol. 2009, 20, 1048–1054. [Google Scholar] [CrossRef] [PubMed]
- Ye, C.; Dickman, M.B.; Whitham, S.A.; Payton, M.; Verchot, J. The Unfolded Protein Response Is Triggered by a Plant Viral Movement Protein. Plant. Physiol. 2011, 156, 741–755. [Google Scholar] [CrossRef] [PubMed]
- Chiu, M.-H.; Chen, I.-H.; Baulcombe, D.; Tsai, C.-H. The silencing suppressor P25 of Potato virus X interacts with Argonaute1 and mediates its degradation through the proteasome pathway. Mol. Plant. Pathol. 2010, 11, 641–649. [Google Scholar] [CrossRef]
- Yaegashi, H.; Takahashi, T.; Isogai, M.; Kobori, T.; Ohki, S.; Yoshikawa, N. Apple chlorotic leaf spot virus 50 kDa movement protein acts as a suppressor of systemic silencing without interfering with local silencing in Nicotiana benthamiana. J. Gen. Virol. 2007, 88, 316–324. [Google Scholar] [CrossRef]
- Deng, X.; Kelloniemi, J.; Haikonen, T.; Vuorinen, A.L.; Elomaa, P.; Teeri, T.H.; Valkonen, J.P.T. Modification of Tobacco rattle virus RNA1 to Serve as a VIGS Vector Reveals That the 29K Movement Protein Is an RNA Silencing Suppressor of the Virus. Mol. Plant. Microbe Interact. 2013, 26, 503–514. [Google Scholar] [CrossRef] [PubMed]
- Fusaro, A.F.; Barton, D.A.; Nakasugi, K.; Jackson, C.; Kalischuk, M.L.; Kawchuk, L.M.; Vaslin, M.F.S.; Correa, R.L.; Waterhouse, P.M. The Luteovirus P4 Movement Protein Is a Suppressor of Systemic RNA Silencing. Viruses 2017, 9, 294. [Google Scholar] [CrossRef] [PubMed]
- Siré, C.; Bangratz-Reyser, M.; Fargette, D.; Brugidou, C. Genetic diversity and silencing suppression effects of Rice yellow mottle virus and the P1 protein. Virol. J. 2008, 5, 55. [Google Scholar] [CrossRef]
- Powers, J.G.; Sit, T.L.; Heinsohn, C.; George, C.G.; Kim, K.-H.; Lommel, S.A. The Red clover necrotic mosaic virus RNA-2 encoded movement protein is a second suppressor of RNA silencing. Virology 2008, 381, 277–286. [Google Scholar] [CrossRef]
- Senshu, H.; Yamaji, Y.; Minato, N.; Shiraishi, T.; Maejima, K.; Hashimoto, M.; Miura, C.; Neriya, Y.; Namba, S. A Dual Strategy for the Suppression of Host Antiviral Silencing: Two Distinct Suppressors for Viral Replication and Viral Movement Encoded by Potato Virus M. J. Virol. 2011, 85, 10269–10278. [Google Scholar] [CrossRef] [PubMed]
- Kasschau, K.D.; Carrington, J.C. Long-Distance Movement and Replication Maintenance Functions Correlate with Silencing Suppression Activity of Potyviral HC-Pro. Virology 2001, 285, 71–81. [Google Scholar] [CrossRef] [PubMed]
- Bayne, E.; Rakitina, D.; Morozov, S.Y.; Baulcombe, D. Cell-to-cell movement of Potato Potexvirus X is dependent on suppression of RNA silencing. Plant. J. 2005, 44, 471–482. [Google Scholar] [CrossRef]
- Powers, J.G.; Sit, T.L.; Qu, F.; Morris, T.J.; Kim, K.-H.; Lommel, S.A. A Versatile Assay for the Identification of RNA Silencing Suppressors Based on Complementation of Viral Movement. Mol. Plant. Microbe Interact. 2008, 21, 879–890. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Chiba, S.; Hleibieh, K.; Delbianco, A.; Klein, E.; Ratti, C.; Ziegler-Graff, V.; Bouzoubaa, S.; Gilmer, D. The Benyvirus RNA Silencing Suppressor Is Essential for Long-Distance Movement, Requires Both Zinc-Finger and NoLS Basic Residues but Not a Nucleolar Localization for Its Silencing-Suppression Activity. Mol. Plant. Microbe Interact. 2013, 26, 168–181. [Google Scholar] [CrossRef] [PubMed]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Niu, E.; Liu, H.; Zhou, H.; Luo, L.; Wu, Y.; Andika, I.B.; Sun, L. Autophagy Inhibits Intercellular Transport of Citrus Leaf Blotch Virus by Targeting Viral Movement Protein. Viruses 2021, 13, 2189. https://doi.org/10.3390/v13112189
Niu E, Liu H, Zhou H, Luo L, Wu Y, Andika IB, Sun L. Autophagy Inhibits Intercellular Transport of Citrus Leaf Blotch Virus by Targeting Viral Movement Protein. Viruses. 2021; 13(11):2189. https://doi.org/10.3390/v13112189
Chicago/Turabian StyleNiu, Erbo, Huan Liu, Hongsheng Zhou, Lian Luo, Yunfeng Wu, Ida Bagus Andika, and Liying Sun. 2021. "Autophagy Inhibits Intercellular Transport of Citrus Leaf Blotch Virus by Targeting Viral Movement Protein" Viruses 13, no. 11: 2189. https://doi.org/10.3390/v13112189
APA StyleNiu, E., Liu, H., Zhou, H., Luo, L., Wu, Y., Andika, I. B., & Sun, L. (2021). Autophagy Inhibits Intercellular Transport of Citrus Leaf Blotch Virus by Targeting Viral Movement Protein. Viruses, 13(11), 2189. https://doi.org/10.3390/v13112189





