Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) in a Dog in Connecticut in February 2021
Abstract
:1. Introduction
2. Materials and Methods
2.1. RNA Extraction and Reverse Transcription Real-Time PCR (RT-qPCR)
2.2. Histopathology and RNAscope® in Situ Hybridization
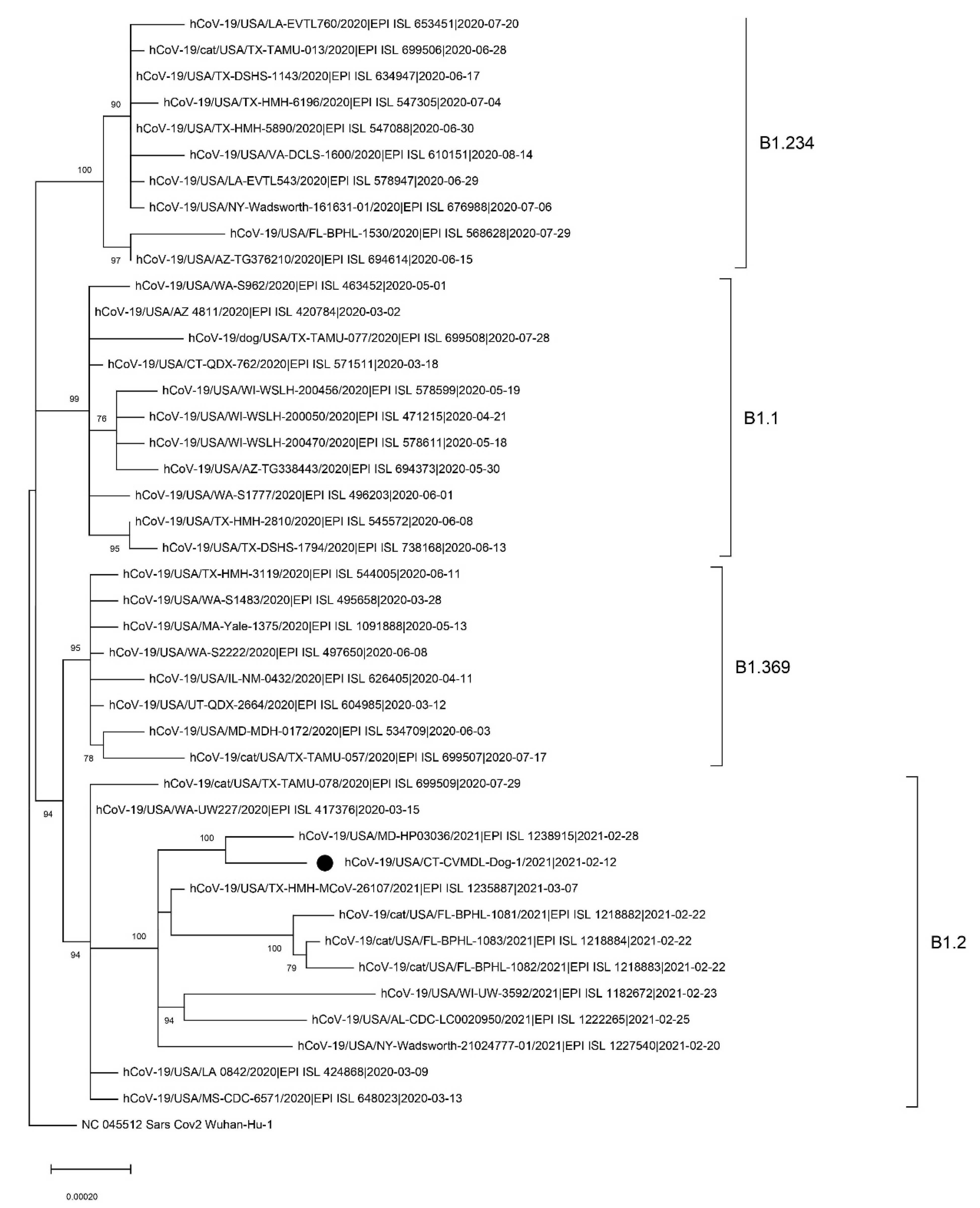
2.3. Complete Genome Sequencing and Phylogenetic Analysis
3. Results and Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhu, N.; Zhang, D.; Wang, W.; Li, X.; Yang, B.; Song, J.; Zhao, X.; Huang, B.; Shi, W.; Lu, R.; et al. A Novel Coronavirus from Patients with Pneumonia in China, 2019. N. Engl. J. Med. 2020, 382, 727–733. [Google Scholar] [CrossRef] [PubMed]
- Mahdy, M.A.A.; Younis, W.; Ewaida, Z. An Overview of SARS-CoV-2 and Animal Infection. Front. Vet. Sci 2020, 7, 596391. [Google Scholar] [CrossRef] [PubMed]
- McAloose, D.; Laverack, M.; Wang, L.; Killian, M.L.; Caserta, L.C.; Yuan, F.; Mitchell, P.K.; Queen, K.; Mauldin, M.R.; Cronk, B.D.; et al. From People to Panthera: Natural SARS-CoV-2 Infection in Tigers and Lions at the Bronx Zoo. Mbio 2020, 11, e02220-20. [Google Scholar] [CrossRef] [PubMed]
- Shriner, S.A.; Ellis, J.W.; Root, J.J.; Roug, A.; Stopak, S.R.; Wiscomb, G.W.; Zierenberg, J.R.; Ip, H.S.; Torchetti, M.K.; DeLiberto, T.J. SARS-CoV-2 Exposure in Escaped Mink, Utah, USA. Emerg. Infect. Dis. 2021, 27, 988–990. [Google Scholar] [CrossRef] [PubMed]
- USDA-APHIS. Cases of SARS-CoV-2 in Animals in the United States. 2021. Available online: https://www.aphis.usda.gov/aphis/ourfocus/animalhealth/sa_one_health/sars-cov-2-animals-us (accessed on 9 March 2021).
- CDC. CDC 2019-Novel Coronavirus (2019-nCoV) Real-Time RT-PCR Diagnostic Panel. Available online: https://www.cdc.gov/coronavirus/2019-ncov/lab/virus-requests.html (accessed on 1 February 2021).
- Wang, F.; Flanagan, J.; Su, N.; Wang, L.C.; Bui, S.; Nielson, A.; Wu, X.; Vo, H.T.; Ma, X.J.; Luo, Y. RNAscope: A novel in situ RNA analysis platform for formalin-fixed, paraffin-embedded tissues. J. Mol. Diagn. 2012, 14, 22–29. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, H. Minimap2: Pairwise alignment for nucleotide sequences. Bioinformatics 2018, 34, 3094–3100. [Google Scholar] [CrossRef] [PubMed]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef] [PubMed]
- Kannan, S.R.; Spratt, A.N.; Quinn, T.P.; Heng, X.; Lorson, C.L.; Sonnerborg, A.; Byrareddy, S.N.; Singh, K. Infectivity of SARS-CoV-2: There Is Something More than D614G? J. Neuroimmune Pharm. 2020, 15, 574–577. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Chen, J.; Gao, K.; Hozumi, Y.; Yin, C.; Wei, G.W. Analysis of SARS-CoV-2 mutations in the United States suggests presence of four substrains and novel variants. Commun. Biol. 2021, 4, 228. [Google Scholar] [CrossRef] [PubMed]
- Moura, I.B.; Buckley, A.M.; Wilcox, M.H. Can SARS-CoV-2 be transmitted via faeces? Curr. Opin. Gastroenterol. 2021. [Google Scholar] [CrossRef] [PubMed]

| Sample/Tissue | SARS-CoV-2 RT-qPCR (Ct Value) | ISH RNAscope | ||
|---|---|---|---|---|
| N1 Probe | N2 Probe | Sense Probe Signal | Antisense Probe Signal | |
| Nasal swab | 21.15 | 23.15 | - | - |
| Oral swab | 37.5 | 43.08 | - | - |
| Rectal swab | 39.1 | 38 | - | - |
| Lung tissue | 38.7 | 39 | Detected | Detected |
| Heart tissue | 36.57 | 38.36 | Detected | Detected |
| Kidney tissue | 38.76 | 39.16 | Detected | Detected |
| Spleen tissue | not detected | not detected | - | - |
| Liver tissue | not detected | not detected | - | - |
| Intestine tissue | - | - | Detected | Detected |
| GISAID Accession | Virus | Location | Collection Date (YYYY-MM-DD) | Sequence Identity |
|---|---|---|---|---|
| EPI_ISL_1137193 | hCoV-19/USA/PA-MGEL-01496/2021 | USA/Pennsylvania | 2021-01-25 | 99.99% |
| EPI_ISL_853340 | hCoV-19/USA/PA-MGEL-01148/2020 | USA/Pennsylvania | 2020-12-02 | 99.99% |
| EPI_ISL_1137240 | hCoV-19/USA/PA-MGEL-00902/2020 | USA/Pennsylvania | 2020-10-30 | 99.98% |
| EPI_ISL_1202236 | hCoV-19/USA/TX-HMH-MCoV-16526/2020 | USA/Texas | 2020-11-03 | 99.98% |
| EPI_ISL_1094242 | hCoV-19/USA/FL-CDC-2-3847117/2021 | USA/Florida | 2021-01-20 | 99.98% |
| EPI_ISL_1080077 | hCoV-19/USA/TX-HMH-MCoV-25439/2021 | USA/Texas | 2021-01-21 | 99.98% |
| EPI_ISL_1075269 | hCoV-19/USA/TX-HMH-MCoV-20826/2021 | USA/Texas | 2021-01-02 | 99.98% |
| EPI_ISL_1049241 | hCoV-19/USA/UT-UPHL-2102150865/2021 | USA/Utah | 2021-02-02 | 99.97% |
| EPI_ISL_783726 | hCoV-19/USA/TX-HMH-MCoV-19267/2020 | USA/Texas | 2020-11-24 | 99.97% |
| EPI_ISL_1095270 | hCoV-19/USA/AZ-CDC-2-3846423/2021 | USA/Arizona | 2021-01-14 | 99.97% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, D.-H.; Helal, Z.H.; Kim, J.; Hunt, A.; Barbieri, A.; Tocco, N.; Frasca, S., Jr.; Kerr, K.; Hyeon, J.-Y.; Chung, D.H.; et al. Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) in a Dog in Connecticut in February 2021. Viruses 2021, 13, 2141. https://doi.org/10.3390/v13112141
Lee D-H, Helal ZH, Kim J, Hunt A, Barbieri A, Tocco N, Frasca S Jr., Kerr K, Hyeon J-Y, Chung DH, et al. Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) in a Dog in Connecticut in February 2021. Viruses. 2021; 13(11):2141. https://doi.org/10.3390/v13112141
Chicago/Turabian StyleLee, Dong-Hun, Zeinab H. Helal, Junwon Kim, Amelia Hunt, Alyza Barbieri, Natalie Tocco, Salvatore Frasca, Jr., Kirklyn Kerr, Ji-Yeon Hyeon, David H. Chung, and et al. 2021. "Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) in a Dog in Connecticut in February 2021" Viruses 13, no. 11: 2141. https://doi.org/10.3390/v13112141
APA StyleLee, D.-H., Helal, Z. H., Kim, J., Hunt, A., Barbieri, A., Tocco, N., Frasca, S., Jr., Kerr, K., Hyeon, J.-Y., Chung, D. H., & Risatti, G. (2021). Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) in a Dog in Connecticut in February 2021. Viruses, 13(11), 2141. https://doi.org/10.3390/v13112141






