PHIDA: A High Throughput Turbidimetric Data Analytic Tool to Compare Host Range Profiles of Bacteriophages Isolated Using Different Enrichment Methods
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Growth Conditions
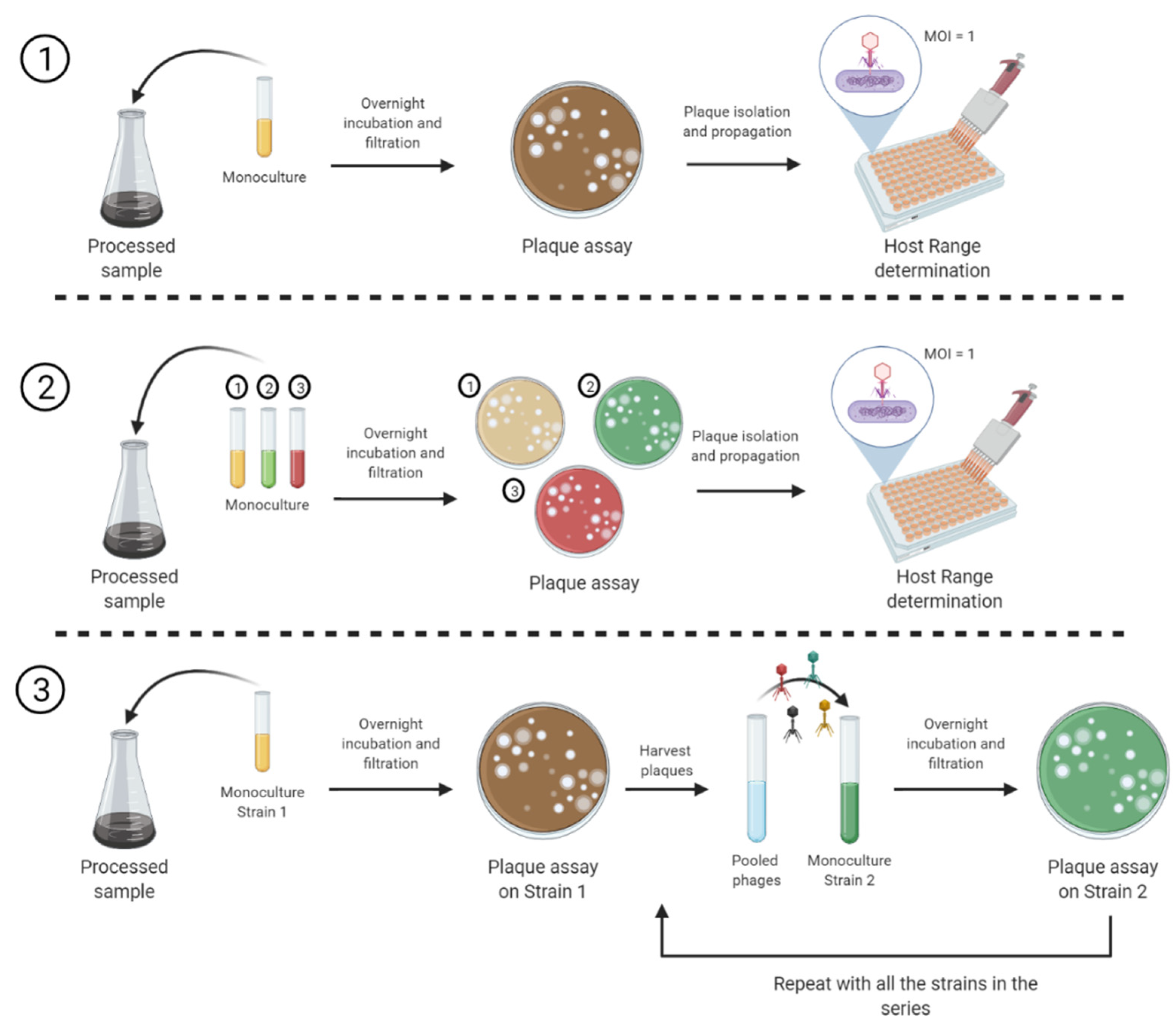
2.2. Sample Enrichment for Phage Isolation
2.3. Host Range Pattern Determination
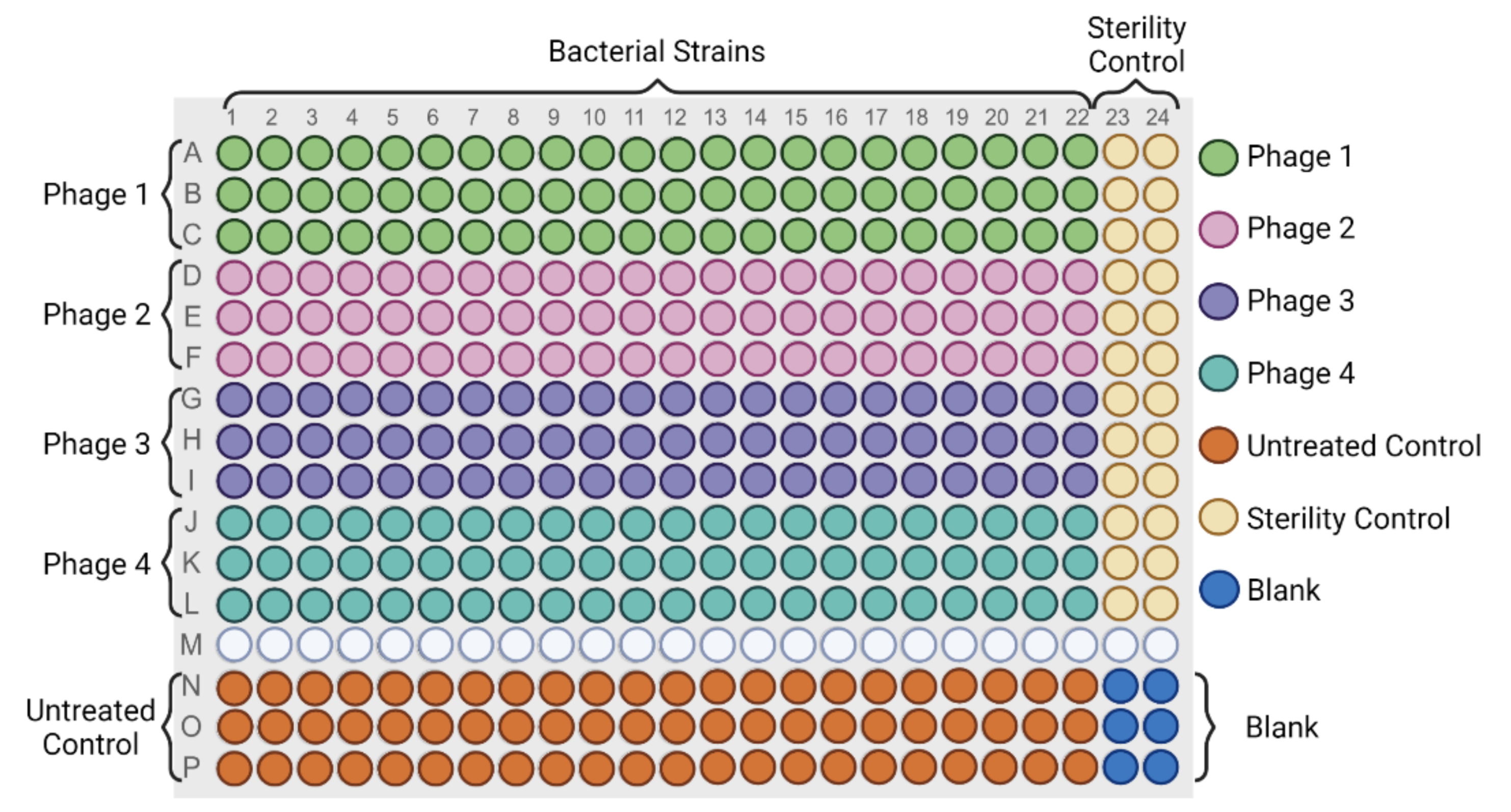
2.3.1. High Throughput Host Range Characterization in Liquid Culture
2.3.2. PHIDA: Data Analytics Tool for Efficient Host Range Determination
2.3.3. Implementation of the Data Analytics Tool
2.3.4. Definition of the Host Range Index
3. Results
3.1. Development of The High Throughput Host Range Assay and PHIDA
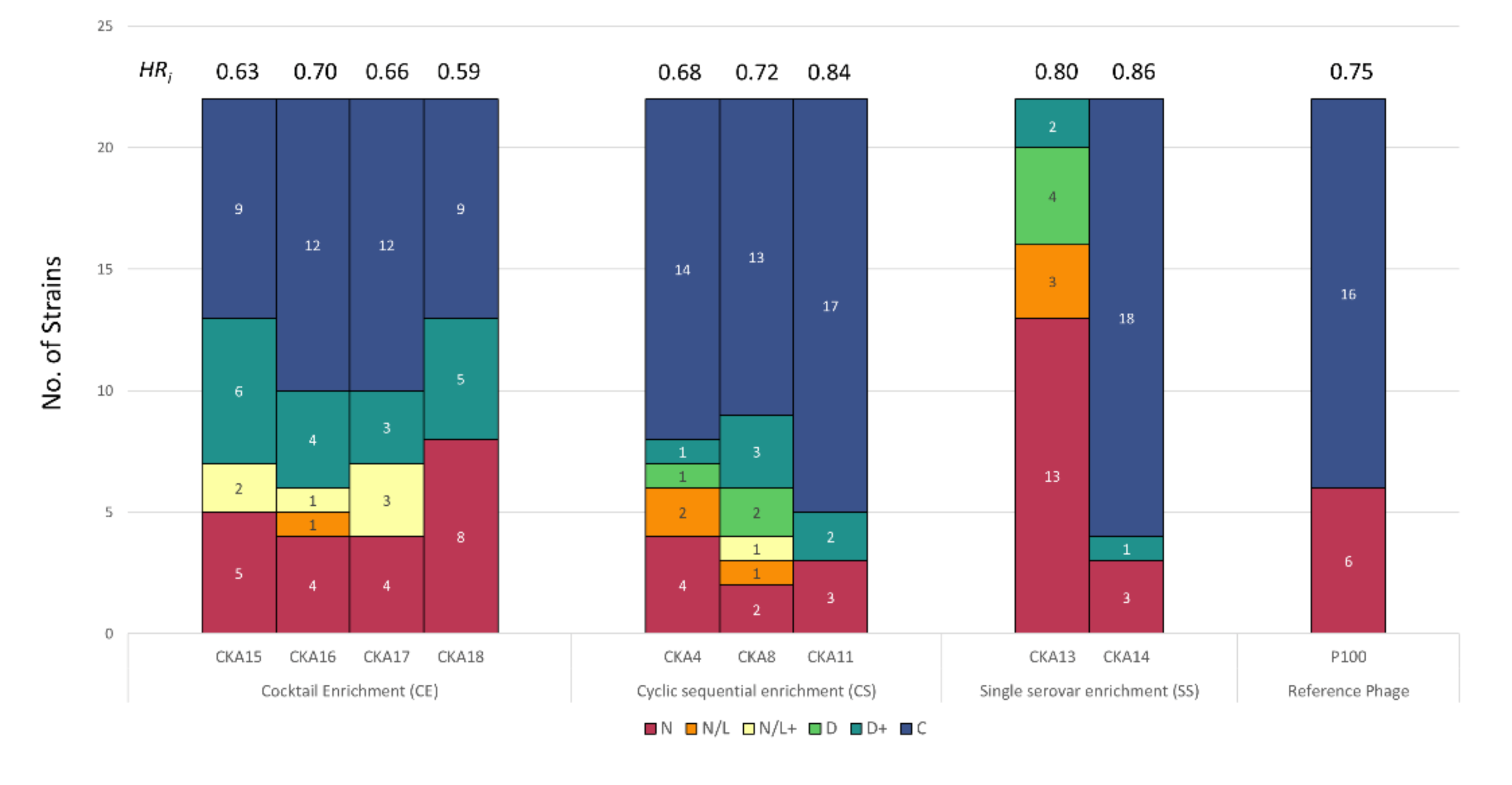
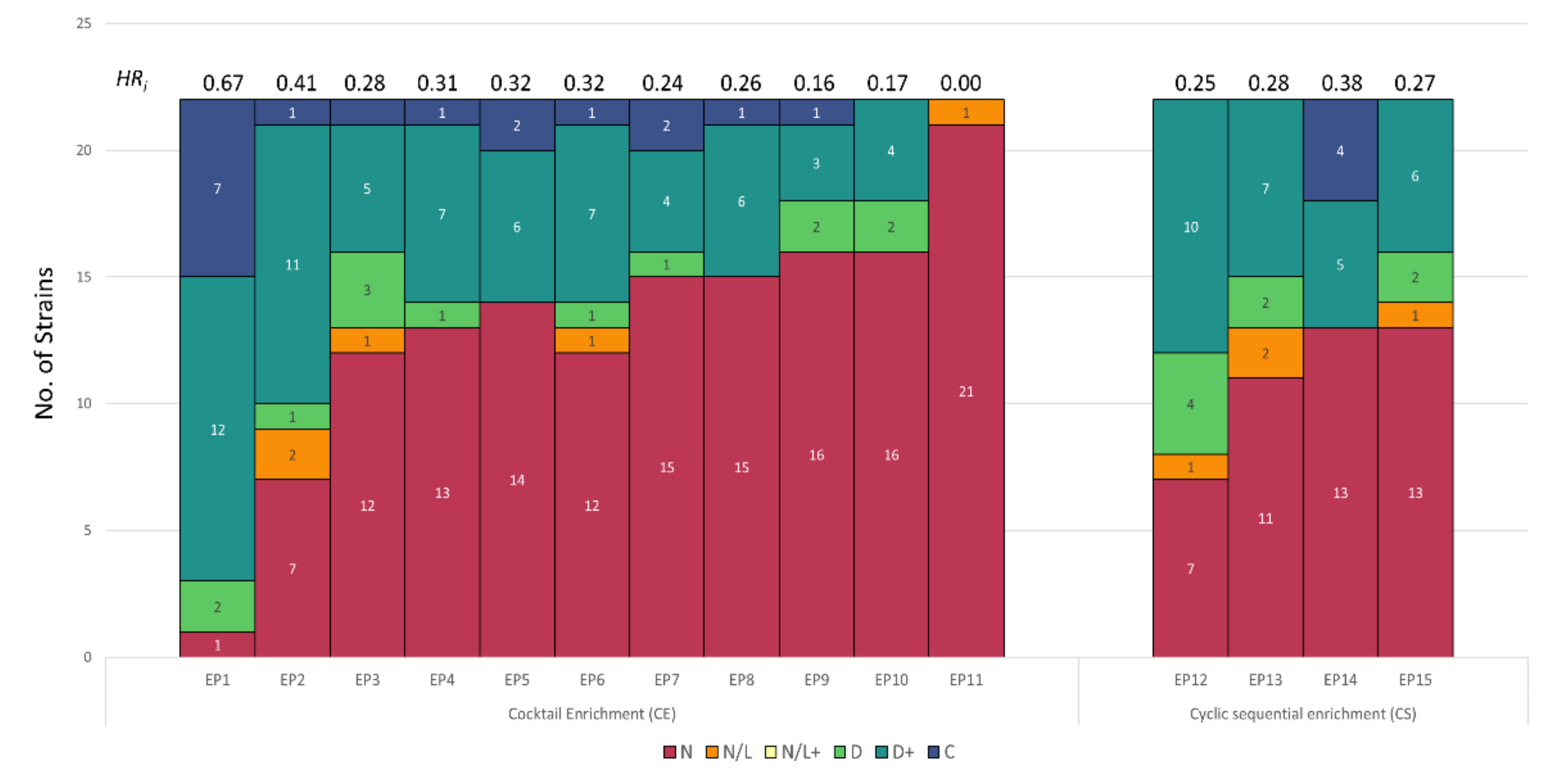
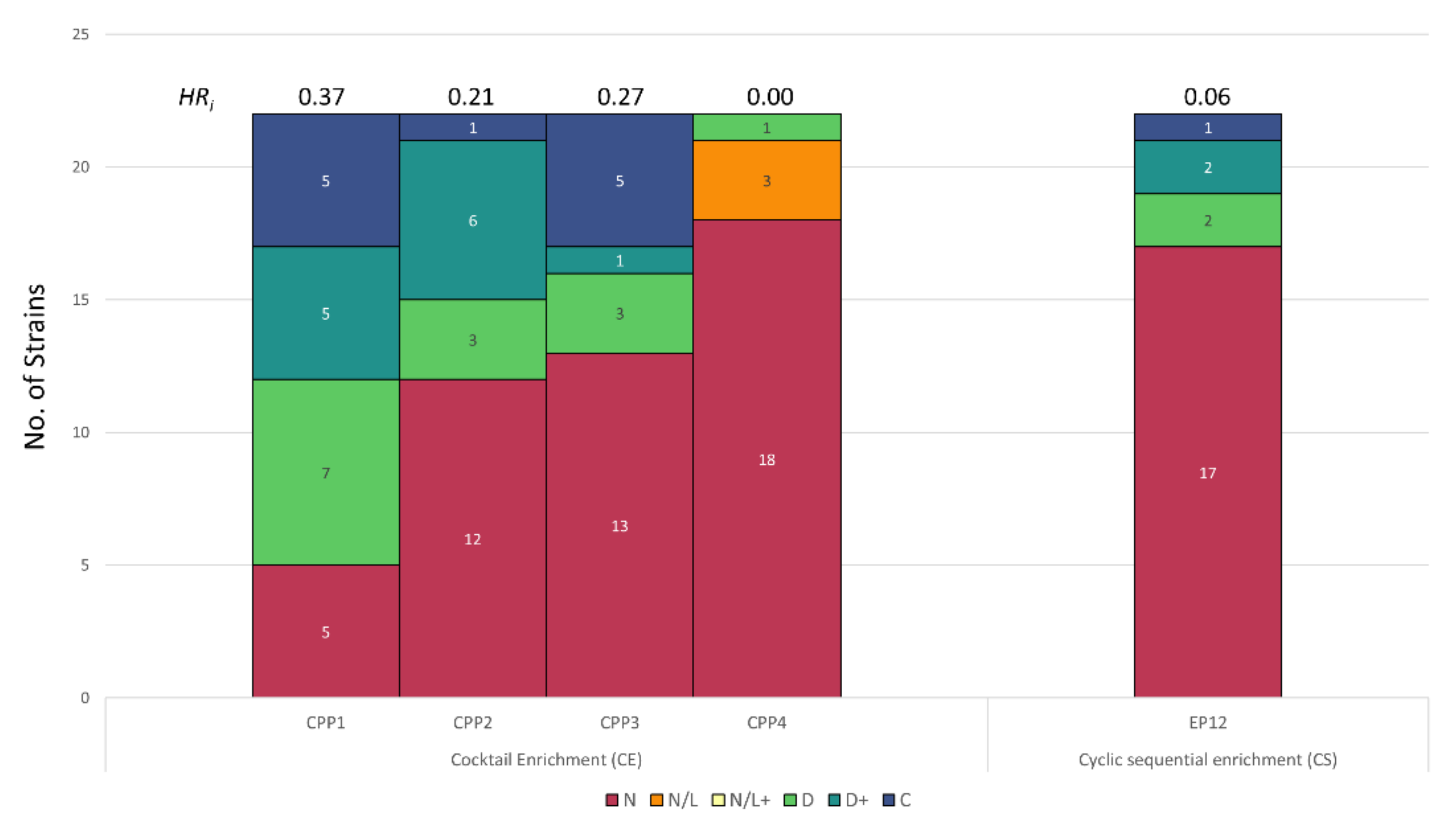
3.2. Host Range Pattern/Profile of the Isolated Listeria spp. Phages
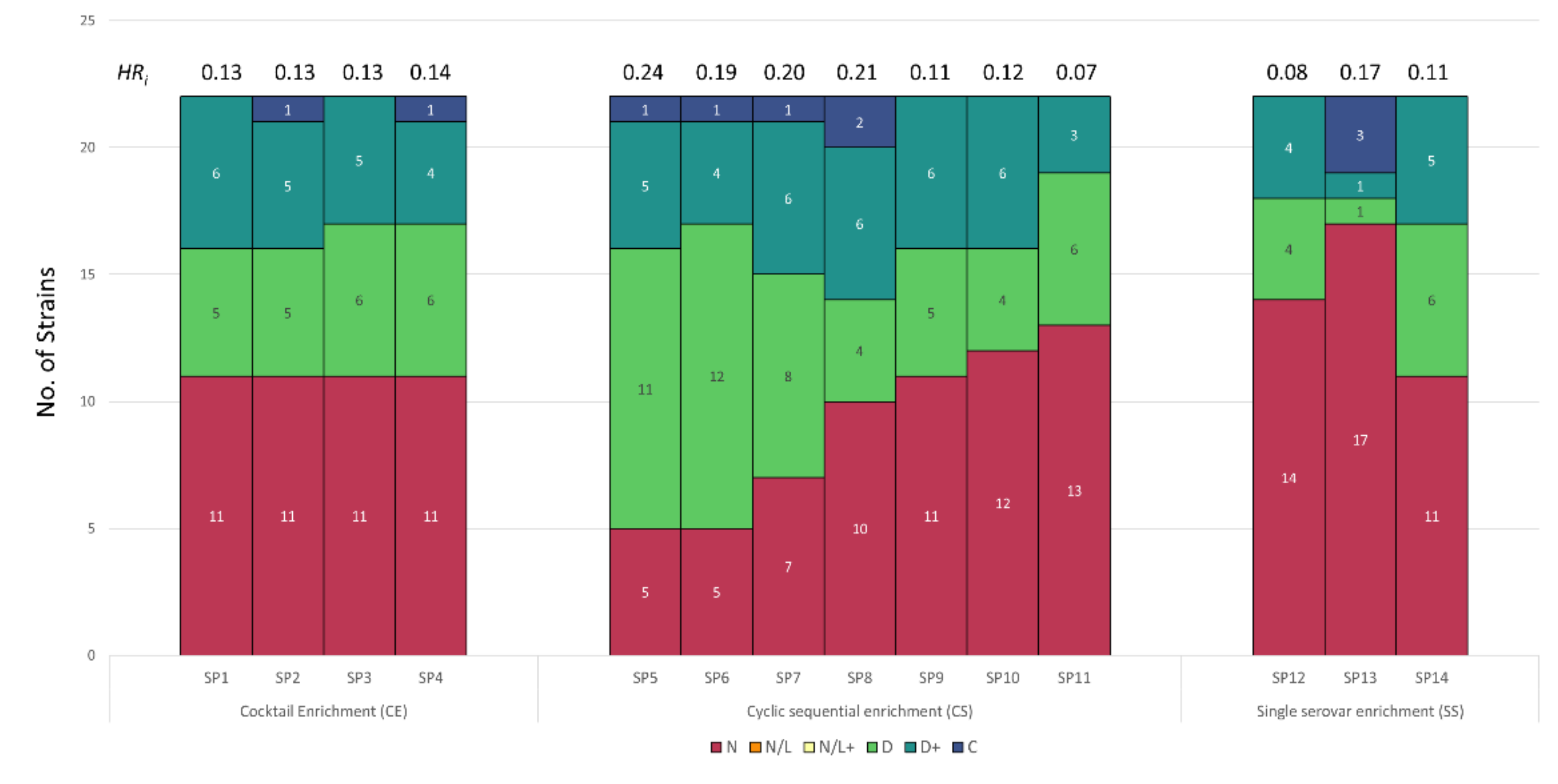
3.3. Host Range Pattern/Profile of the Isolated Salmonella spp. Phages
3.4. Host Range Pattern/Profile of the Isolated Pseudomonas spp. Phages
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Labrie, S.J.; Samson, J.E.; Moineau, S. Bacteriophage resistance mechanisms. Nat. Rev. Microbiol. 2010, 8, 317–327. [Google Scholar] [CrossRef]
- Mushegian, A.R. Are There 1031 Virus Particles on Earth, or More, or Fewer? J. Bacteriol. 2020, 202, e00052-20. [Google Scholar] [CrossRef] [PubMed]
- Gigante, A.; Atterbury, R.J. Veterinary use of bacteriophage therapy in intensively-reared livestock. Virol. J. 2019, 16, 155. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, D.M.; Koskella, B.; Lin, H.C. Phage therapy: An alternative to antibiotics in the age of multi-drug resistance. World J. Gastrointest. Pharmacol. Ther. 2017. [Google Scholar] [CrossRef]
- Moye, Z.D.; Woolston, J.; Sulakvelidze, A. Bacteriophage applications for food production and processing. Viruses 2018, 10, 205. [Google Scholar] [CrossRef] [Green Version]
- Wei, S.; Chelliah, R.; Rubab, M.; Oh, D.H.; Uddin, M.J.; Ahn, J. Bacteriophages as potential tools for detection and control of salmonella spp. Food Syst. 2019, 7, 570. [Google Scholar] [CrossRef] [Green Version]
- Summer, N.S.; Summer, E.J.; Gill, J.; Young, R. Phage remediation of microbe-induced corrosion. In Proceedings of the 17th International Corrosion Congress: Corrosion Control in the Service of Society, Las Vegas, NV, USA, 6–10 October 2008. [Google Scholar]
- Ross, A.; Ward, S.; Hyman, P. More is better: Selecting for broad host range bacteriophages. Front. Microbiol. 2016. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brovko, L.Y.; Anany, H.; Griffiths, M.W. Bacteriophages for Detection and Control of Bacterial Pathogens in Food and Food-Processing Environment. Adv. Food Nutr. Res. 2012, 67, 241–288. [Google Scholar]
- Adams, M.H. Bacteriophages; Interscience Publishers: New York, NY, USA, 1959. [Google Scholar]
- Hyman, P.; Abedon, S.T. Bacteriophage host range and bacterial resistance. Adv. Appl. Microbiol. 2010, 70, 217–248. [Google Scholar] [CrossRef]
- Díaz-Muñoz, S.L.; Koskella, B. Bacteria-phage interactions in natural environments. Adv. Appl. Microbiol. 2014, 89, 135–183. [Google Scholar] [CrossRef]
- Mapes, A.C.; Trautner, B.W.; Liao, K.S.; Ramig, R.F. Development of expanded host range phage active on biofilms of multi-drug resistant Pseudomonas aeruginosa. Bacteriophage 2016. [Google Scholar] [CrossRef] [Green Version]
- Nilsson, A.S. Phage therapy-constraints and possibilities. Upsala J. Med. Sci. 2014, 119, 192–198. [Google Scholar] [CrossRef]
- Young, R.; Gill, J.J. Phage therapy redux—What is to be done? Science 2015, 350, 1163–1164. [Google Scholar] [CrossRef] [PubMed]
- Islam, M.S.; Zhou, Y.; Liang, L.; Nime, I.; Liu, K.; Yan, T.; Wang, X.; Li, J. Application of a Phage Cocktail for Control of Salmonella in Foods and Reducing Biofilms. Viruses 2019, 11, 841. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schofield, D.; Sharp, N.J.; Westwater, C. Phage-based platforms for the clinical detection of human bacterial pathogens. Bacteriophage 2012. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bai, J.; Kim, Y.T.; Ryu, S.; Lee, J.H. Biocontrol and rapid detection of food-borne pathogens using bacteriophages and endolysins. Front. Microbiol. 2016. [Google Scholar] [CrossRef] [PubMed]
- Meile, S.; Sarbach, A.; Du, J.; Schuppler, M.; Saez, C.; Loessner, M.J.; Kilcher, S. Engineered Reporter Phages for Rapid Bioluminescence-Based Detection and Differentiation of Viable Listeria Cells. Appl. Environ. Microbiol. 2020, 86. [Google Scholar] [CrossRef] [Green Version]
- Kortright, K.E.; Chan, B.K.; Koff, J.T.L.; Turner, P.E. Phage Therapy: A Renewed Approach to Combat Antibiotic-Resistant Bacteria. Cell Host Microbe 2019. [Google Scholar] [CrossRef] [Green Version]
- Melo, L.D.R.; Sillankorva, S.; Ackermann, H.W.; Kropinski, A.M.; Azeredo, J.; Cerca, N. Isolation and characterization of a new Staphylococcus epidermidis broad-spectrum bacteriophage. J. Gen. Virol. 2014. [Google Scholar] [CrossRef]
- El-Dougdoug, N.K.; Cucic, S.; Abdelhamid, A.G.; Brovko, L.; Kropinski, A.M.; Griffiths, M.W.; Anany, H. Control of Salmonella Newport on cherry tomato using a cocktail of lytic bacteriophages. Int. J. Food Microbiol. 2019, 293, 60–71. [Google Scholar] [CrossRef]
- Yu, P.; Mathieu, J.; Li, M.; Dai, Z.; Alvarez, P.J.J. Isolation of polyvalent bacteriophages by sequential multiple-host approaches. Appl. Environ. Microbiol. 2016, 82, 808–815. [Google Scholar] [CrossRef] [Green Version]
- Montso, P.K.; Mlambo, V.; Ateba, C.N. Characterization of Lytic Bacteriophages Infecting Multidrug-Resistant Shiga Toxigenic Atypical Escherichia coli O177 Strains Isolated From Cattle Feces. Front. Public Health 2019, 7, 355. [Google Scholar] [CrossRef]
- Mirzaei, M.K.; Nilsson, A.S. Isolation of phages for phage therapy: A comparison of spot tests and efficiency of plating analyses for determination of host range and efficacy. PLoS ONE 2015, 10, e0118557. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xie, Y.; Wahab, L.; Gill, J.J. Development and Validation of a Microtiter Plate-Based Assay for Determination of Bacteriophage Host Range and Virulence. Viruses 2018, 10, 189. [Google Scholar] [CrossRef] [Green Version]
- Viazis, S.; Akhtar, M.; Feirtag, J.; Brabban, A.; Diez-Gonzalez, F. Isolation and characterization of lytic bacteriophages against enterohaemorrhagic Escherichia coli. J. Appl. Microbiol. 2011, 110, 1323–1331. [Google Scholar] [CrossRef] [PubMed]
- Kutter, E. Phage host range and efficiency of plating. In Bacteriophages; Springer: Berlin/Heidelberg, Germany, 2009; pp. 141–149. [Google Scholar]
- Kropinski, A.M. Practical advice on the one-step growth curve. In Bacteriophages; Springer: Berlin/Heidelberg, Germany, 2018; pp. 41–47. [Google Scholar]
- Storms, Z.J.; Teel, M.R.; Mercurio, K.; Sauvageau, D. The virulence index: A metric for quantitative analysis of phage virulence. Phage 2020, 1, 27–36. [Google Scholar] [CrossRef] [Green Version]
- Trudelle, D.M.; Bryan, D.W.; Hudson, L.K.; Denes, T.G. Cross-resistance to phage infection in Listeria monocytogenes serotype 1/2a mutants. Food Microbiol. 2019, 84, 103239. [Google Scholar] [CrossRef]
- Danovaro, R.; Middelboe, M. Separation of free virus particles from sediments in aquatic systems. Man. Aquat. Viral Ecol. ASLO 2010, 74–81. [Google Scholar]
- Sambrook, J.; Fritsch, E.F.; Maniatis, T. Molecular Cloning: A Laboratory Manual; Cold Spring Harbor Laboratory Press: Long Island, NY, USA, 1989. [Google Scholar]
- Sambrook, J.; Russell, D.W. Purification of Bacteriophage λ Particles by Centrifugation through a Glycerol Step Gradient. CSH Protoc. 2006, 2006. [Google Scholar] [CrossRef] [PubMed]
- Kropinski, A.M.; Mazzocco, A.; Waddell, T.E.; Lingohr, E.; Johnson, R.P. Enumeration of bacteriophages by double agar overlay plaque assay. Methods Mol. Biol. 2009. [Google Scholar] [CrossRef]
- Jeukens, J.; Boyle, B.; Kukavica-Ibrulj, I.; Ouellet, M.M.; Aaron, S.D.; Charette, S.J.; Fothergill, J.T.L.; Tucker, N.P.; Winstanley, C.; Levesque, R.C. Comparative Genomics of Isolates of a Pseudomonas aeruginosa Epidemic Strain Associated with Chronic Lung Infections of Cystic Fibrosis Patients. PLoS ONE 2014, 9, e87611. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hyman, P. Phages for Phage Therapy: Isolation, Characterization, and Host Range Breadth. Pharmaceuticals 2019, 12, 35. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Islam, M.S.; Zhou, Y.; Liang, L.; Nime, I.; Yan, T.; Willias, S.P.; Mia, M.Z.; Bei, W.; Connerton, I.F.; Fischetti, V.A.; et al. Application of a Broad Range Lytic Phage LPST94 for Biological Control of Salmonella in Foods. Microorganisms 2020, 8, 247. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, J.-W.; Siletzky, R.M.; Kathariou, S. Host Ranges of Listeria Specific Bacteriophages from the Turkey Processing Plant Environment in the United States. Appl. Environ. Microbiol. 2008, 74, 6623–6630. [Google Scholar] [CrossRef] [Green Version]
- Oda, M.; Morita, M.; Unno, H.; Tanji, Y. Rapid detection of Escherichia coli O157:H7 by using green fluorescent protein-labeled PP01 bacteriophage. Appl. Environ. Microbiol. 2004, 70, 527–534. [Google Scholar] [CrossRef] [Green Version]
- Huang, C.; Virk, S.M.; Shi, J.; Zhou, Y.; Willias, S.P.; Morsy, M.K.; Abdelnabby, H.E.; Liu, J.; Wang, X.; Li, J. Isolation, Characterization, and Application of Bacteriophage LPSE1 Against Salmonella enterica in Ready to Eat (RTE) Foods. Front. Microbiol. 2018, 9, 1046. [Google Scholar] [CrossRef]
- Ramesh, N.; Archana, L.; Madurantakam Royam, M.; Manohar, P.; Eniyan, K. Effect of various bacteriological media on the plaque morphology of Staphylococcus and Vibrio phages. Access Microbiol. 2019, 1, e000036. [Google Scholar] [CrossRef]
- Abedon, S.T.; Yin, J. Bacteriophage plaques: Theory and analysis. Methods Mol. Biol. 2009, 501, 161–174. [Google Scholar] [CrossRef]
- Gallet, R.; Kannoly, S.; Wang, I.-N. Effects of bacteriophage traits on plaque formation. BMC Microbiol. 2011, 11, 181. [Google Scholar] [CrossRef] [Green Version]
- Tokman, J.I.; Kent, D.J.; Wiedmann, M.; Denes, T. Temperature Significantly Affects the Plaquing and Adsorption Efficiencies of Listeria Phages. Front. Microbiol. 2016, 7, 631. [Google Scholar] [CrossRef]
- Moller, A.G.; Winston, K.; Ji, S.; Wang, J.; Davis, M.N.H.; Solís-Lemus, C.R.; Read, T.D.; Dunman, P. Genes Influencing Phage Host Range in Staphylococcus aureus on a Species-Wide Scale. Msphere 2021, 6, e01263-20. [Google Scholar] [CrossRef]
- Klumpp, J.; Loessner, M.J. Listeria phages: Genomes, evolution, and application. Bacteriophage 2013, 3, e26861. [Google Scholar] [CrossRef] [Green Version]
- Gray, M.J.; Zadoks, R.N.; Fortes, E.D.; Dogan, B.; Cai, S.; Chen, Y.; Scott, V.N.; Gombas, D.E.; Boor, K.J.; Wiedmann, M. Listeria monocytogenes isolates from foods and humans form distinct but overlapping populations. Appl. Environ. Microbiol. 2004, 70, 5833–5841. [Google Scholar] [CrossRef] [Green Version]
- Anany, H.E.-S.M. Biocontrol of Foodborne Bacterial Pathogens Using Immobilized Bacteriophages. Ph.D. Dissertation, University of Guelph, Guelph, ON, Canada, 2010. [Google Scholar]
- Wendlinger, G.; Loessner, M.J.; Scherer, S. Bacteriophage receptors on Listeria monocytogenes cells are the N-acetylglucosamine and rhamnose substituents of teichoic acids or the peptidoglycan itself. Microbiology 1996, 142 Pt 4, 985–992. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Habann, M.; Leiman, P.G.; Vandersteegen, K.; Van den Bossche, A.; Lavigne, R.; Shneider, M.M.; Bielmann, R.; Eugster, M.R.; Loessner, M.J.; Klumpp, J. Listeria phage A511, a model for the contractile tail machineries of SPO1-related bacteriophages. Mol. Microbiol. 2014, 92, 84–99. [Google Scholar] [CrossRef] [Green Version]
- Sumrall, E.T.; Shen, Y.; Keller, A.P.; Rismondo, J.; Pavlou, M.; Eugster, M.R.; Boulos, S.; Disson, O.; Thouvenot, P.; Kilcher, S. Phage resistance at the cost of virulence: Listeria monocytogenes serovar 4b requires galactosylated teichoic acids for InlB-mediated invasion. PLoS Pathog. 2019, 15, e1008032. [Google Scholar] [CrossRef] [Green Version]
- Di, H.; Ye, L.; Yan, H.; Meng, H.; Yamasak, S.; Shi, L. Comparative analysis of CRISPR loci in different Listeria monocytogenes lineages. Biochem. Biophys. Res. Commun. 2014, 454, 399–403. [Google Scholar] [CrossRef] [PubMed]
- Susskind, M.M.; Wright, A.; Botstein, D. Superinfection exclusion by P22 prophage in lysogens of Salmonella typhimurium: IV. Genetics and physiology of sieB exclusion. Virology 1974, 62, 367–384. [Google Scholar] [CrossRef]
- Liu, B.; Knirel, Y.A.; Feng, L.; Perepelov, A.V.; Senchenkova, S.N.; Reeves, P.R.; Wang, L. Structural diversity in Salmonella O antigens and its genetic basis. FEMS Microbiol. Rev. 2014, 38, 56–89. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weill, P.A.D.G.F.-X. Antigenic Formulae of the Salmonella Serovars; WHO Collaborating Centre for Reference and Research on Salmonella: Paris, France, 2007; p. 167. [Google Scholar]
- Shin, H.; Lee, J.-H.; Kim, H.; Choi, Y.; Heu, S.; Ryu, S. Receptor diversity and host interaction of bacteriophages infecting Salmonella enterica serovar Typhimurium. PLoS ONE 2012, 7, e43392. [Google Scholar] [CrossRef] [PubMed]
- Hudson, H.P.; Lindberg, A.A.; Stocker, B.A.D. Lipopolysaccharide Core Defects in Salmonella typhimurium Mutants Which Are Resistant to Felix O Phage but Retain Smooth Character. Microbiology 1978, 109, 97–112. [Google Scholar] [CrossRef] [Green Version]
- Santos, S.B.; Kropinski, A.M.; Ceyssens, P.J.; Ackermann, H.W.; Villegas, A.; Lavigne, R.; Krylov, V.N.; Carvalho, C.M.; Ferreira, E.C.; Azeredo, J. Genomic and proteomic characterization of the broad-host-range Salmonella phage PVP-SE1: Creation of a new phage genus. J. Virol. 2011, 85, 11265–11273. [Google Scholar] [CrossRef] [Green Version]
- Casjens, S.R.; Gilcrease, E.B.; Winn-Stapley, D.A.; Schicklmaier, P.; Schmieger, H.; Pedulla, M.L.; Ford, M.E.; Houtz, J.M.; Hatfull, G.F.; Hendrix, R.W. The Generalized Transducing Salmonella Bacteriophage ES18: Complete Genome Sequence and DNA Packaging Strategy. J. Bacteriol. 2005, 187, 1091–1104. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marti, R.; Zurfluh, K.; Hagens, S.; Pianezzi, J.; Klumpp, J.; Loessner, M.J. Long tail fibres of the novel broad-host-range T-even bacteriophage S16 specifically recognize Salmonella OmpC. Mol. Microbiol. 2013, 87, 818–834. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Nie, T.; Lin, F.; Connerton, I.F.; Lu, Z.; Zhou, S.; Hang, H. Resistance mechanisms adopted by a Salmonella Typhimurium mutant against bacteriophage. Virus Res. 2019, 273, 197759. [Google Scholar] [CrossRef] [PubMed]
- Fox, K.L.; Srikhanta, Y.N.; Jennings, M.P. Phase variable type III restriction-modification systems of host-adapted bacterial pathogens. Mol. Microbiol. 2007, 65, 1375–1379. [Google Scholar] [CrossRef]
- Santander, J.; Robeson, J. Phage-resistance of Salmonella enterica serovar Enteritidis and pathogenesis in Caenorhabditis elegans is mediated by the lipopolysaccharide. Electron. J. Biotechnol. 2007, 10. [Google Scholar] [CrossRef] [Green Version]
- Shariat, N.; Timme, R.E.; Pettengill, J.B.; Barrangou, R.; Dudley, E.G. Characterization and evolution of Salmonella CRISPR-Cas systems. Microbiology 2015, 161, 374–386. [Google Scholar] [CrossRef]
- Wahl, A.; Battesti, A.; Ansaldi, M. Prophages in Salmonella enterica: A driving force in reshaping the genome and physiology of their bacterial host? Mol. Microbiol. 2019, 111, 303–316. [Google Scholar] [CrossRef] [Green Version]
- Garrido-Sanz, D.; Meier-Kolthoff, J.P.; Göker, M.; Martín, M.; Rivilla, R.; Redondo-Nieto, M. Genomic and Genetic Diversity within the Pseudomonas fluorescens Complex. PLoS ONE 2016, 11, e0150183. [Google Scholar] [CrossRef] [Green Version]
- Anzai, Y.; Kim, H.; Park, J.Y.; Wakabayashi, H.; Oyaizu, H. Phylogenetic affiliation of the pseudomonads based on 16S rRNA sequence. Int. J. Syst. Evol. Microbiol. 2000, 50 Pt 4, 1563–1589. [Google Scholar] [CrossRef] [Green Version]
- Webb, J.S.; Lau, M.; Kjelleberg, S. Bacteriophage and phenotypic variation in Pseudomonas aeruginosa biofilm development. J. Bacteriol. 2004, 186, 8066–8073. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Liu, X.; Tang, K.; Wang, P.; Zeng, Z.; Guo, Y.; Wang, X. Excisionase in Pf filamentous prophage controls lysis-lysogeny decision-making in Pseudomonas aeruginosa. Mol. Microbiol. 2019, 111, 495–513. [Google Scholar] [CrossRef] [Green Version]
- Secor, P.R.; Burgener, E.B.; Kinnersley, M.; Jennings, L.K.; Roman-Cruz, V.; Popescu, M.; Van Belleghem, J.D.; Haddock, N.; Copeland, C.; Michaels, L.A.; et al. Pf Bacteriophage and Their Impact on Pseudomonas Virulence, Mammalian Immunity, and Chronic Infections. Front. Immunol. 2020, 11, 244. [Google Scholar] [CrossRef] [Green Version]
- James, C.E.; Fothergill, J.T.L.; Kalwij, H.; Hall, A.J.; Cottell, J.; Brockhurst, M.A.; Winstanley, C. Differential infection properties of three inducible prophages from an epidemic strain of Pseudomonas aeruginosa. BMC Microbiol. 2012, 12, 216. [Google Scholar] [CrossRef] [Green Version]
- Samson, J.E.; Magadán, A.H.; Sabri, M.; Moineau, S. Revenge of the phages: Defeating bacterial defences. Nat. Rev. Microbiol. 2013, 11, 675–687. [Google Scholar] [CrossRef] [PubMed]
- Trend, S.; Chang, B.J.; O’Dea, M.; Stick, S.M.; Kicic, A.; WAERP; AusREC; AREST CF. Use of a Primary Epithelial Cell Screening Tool to Investigate Phage Therapy in Cystic Fibrosis. Front. Pharmacol. 2018, 9. [Google Scholar] [CrossRef] [Green Version]
- Meadow, P.; Wells, P.J.M. Receptor Sites for R-type Pyocins and Bacteriophage E79 in the Core Part of the Lipopolysaccharide of Pseudomonas aeruginosaPAC1. Microbiology 1978, 108, 339–343. [Google Scholar]
- Schwarzer, D.; Buettner, F.F.; Browning, C.; Nazarov, S.; Rabsch, W.; Bethe, A.; Oberbeck, A.; Bowman, V.D.; Stummeyer, K.; Mühlenhoff, M.; et al. A multivalent adsorption apparatus explains the broad host range of phage phi92: A comprehensive genomic and structural analysis. J. Virol. 2012, 86, 10384–10398. [Google Scholar] [CrossRef] [Green Version]
- Krüger, D.H.; Bickle, T.A. Bacteriophage survival: Multiple mechanisms for avoiding the deoxyribonucleic acid restriction systems of their hosts. Microbiol. Rev. 1983, 47, 345–360. [Google Scholar] [CrossRef] [PubMed]
- Touchon, M.; Rocha, E.P.C. The Small, Slow and Specialized CRISPR and Anti-CRISPR of Escherichia and Salmonella. PLoS ONE 2010, 5, e11126. [Google Scholar] [CrossRef] [PubMed] [Green Version]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the Her Majesty the Queen in Right of Canada as represented by the Minister of Agriculture and Agri-Food Canada. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Martinez-Soto, C.E.; Cucić, S.; Lin, J.T.; Kirst, S.; Mahmoud, E.S.; Khursigara, C.M.; Anany, H. PHIDA: A High Throughput Turbidimetric Data Analytic Tool to Compare Host Range Profiles of Bacteriophages Isolated Using Different Enrichment Methods. Viruses 2021, 13, 2120. https://doi.org/10.3390/v13112120
Martinez-Soto CE, Cucić S, Lin JT, Kirst S, Mahmoud ES, Khursigara CM, Anany H. PHIDA: A High Throughput Turbidimetric Data Analytic Tool to Compare Host Range Profiles of Bacteriophages Isolated Using Different Enrichment Methods. Viruses. 2021; 13(11):2120. https://doi.org/10.3390/v13112120
Chicago/Turabian StyleMartinez-Soto, Carlos E., Stevan Cucić, Janet T. Lin, Sarah Kirst, El Sayed Mahmoud, Cezar M. Khursigara, and Hany Anany. 2021. "PHIDA: A High Throughput Turbidimetric Data Analytic Tool to Compare Host Range Profiles of Bacteriophages Isolated Using Different Enrichment Methods" Viruses 13, no. 11: 2120. https://doi.org/10.3390/v13112120
APA StyleMartinez-Soto, C. E., Cucić, S., Lin, J. T., Kirst, S., Mahmoud, E. S., Khursigara, C. M., & Anany, H. (2021). PHIDA: A High Throughput Turbidimetric Data Analytic Tool to Compare Host Range Profiles of Bacteriophages Isolated Using Different Enrichment Methods. Viruses, 13(11), 2120. https://doi.org/10.3390/v13112120






