Cross-Resistance of the Codling Moth against Different Isolates of Cydia pomonella Granulovirus Is Caused by Two Different but Genetically Linked Resistance Mechanisms
Abstract
1. Introduction
2. Materials and Methods
2.1. Viruses and Insects
2.2. Resistance Testing
2.3. Instar-Specific Assays
2.4. Budded Virus Preparation
2.5. Quantitative PCR
2.6. Intra-Hemocoelic BV Injections
2.7. Occlusion-Derived Virus Production and Labeling with R-18
2.8. ODV and Fluorescence Dequenching Assay
2.9. Statistical Analysis
3. Results
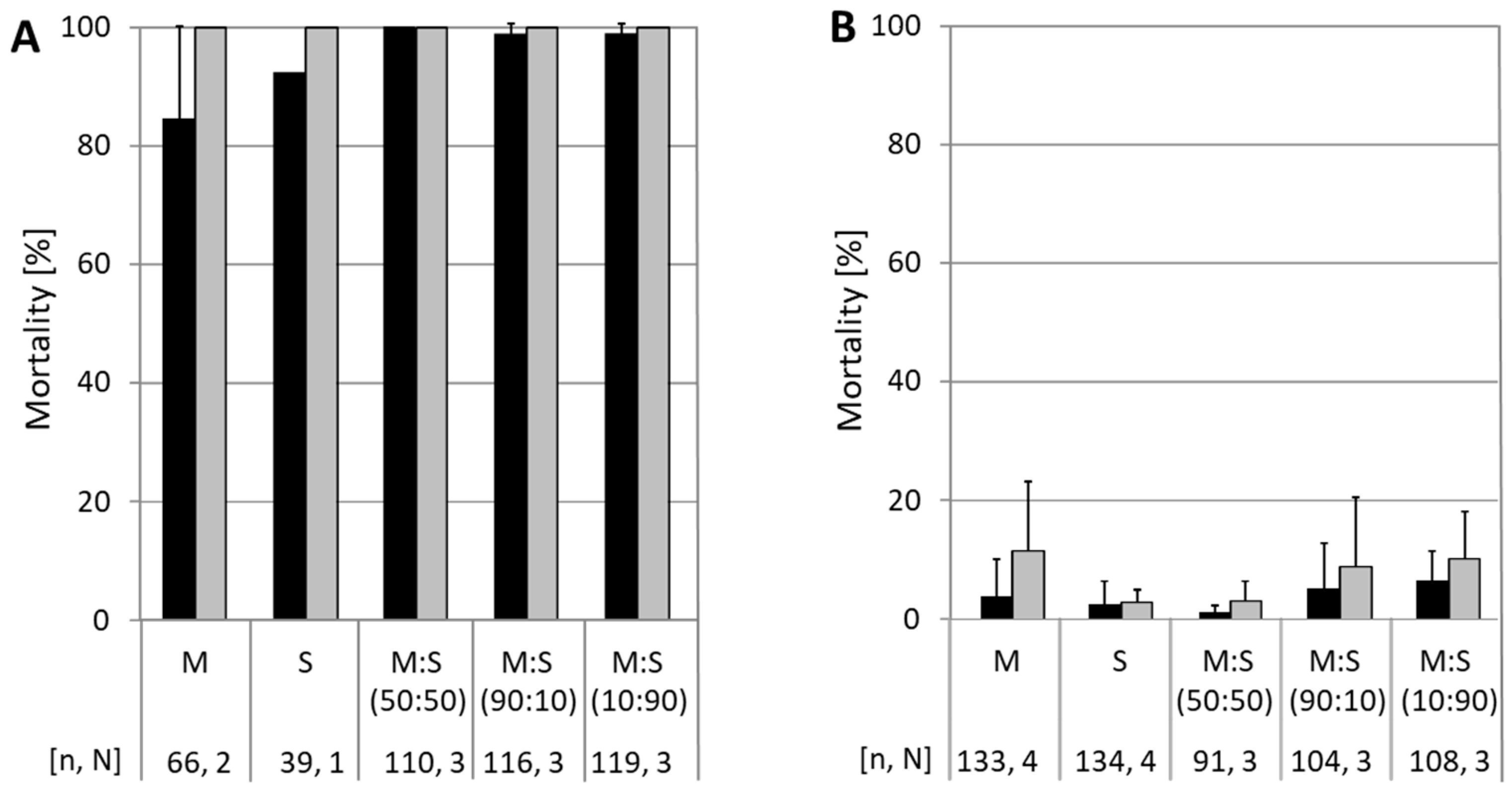
3.1. Mortality of CpS and CpR5M Larvae on Different CpGV Isolates
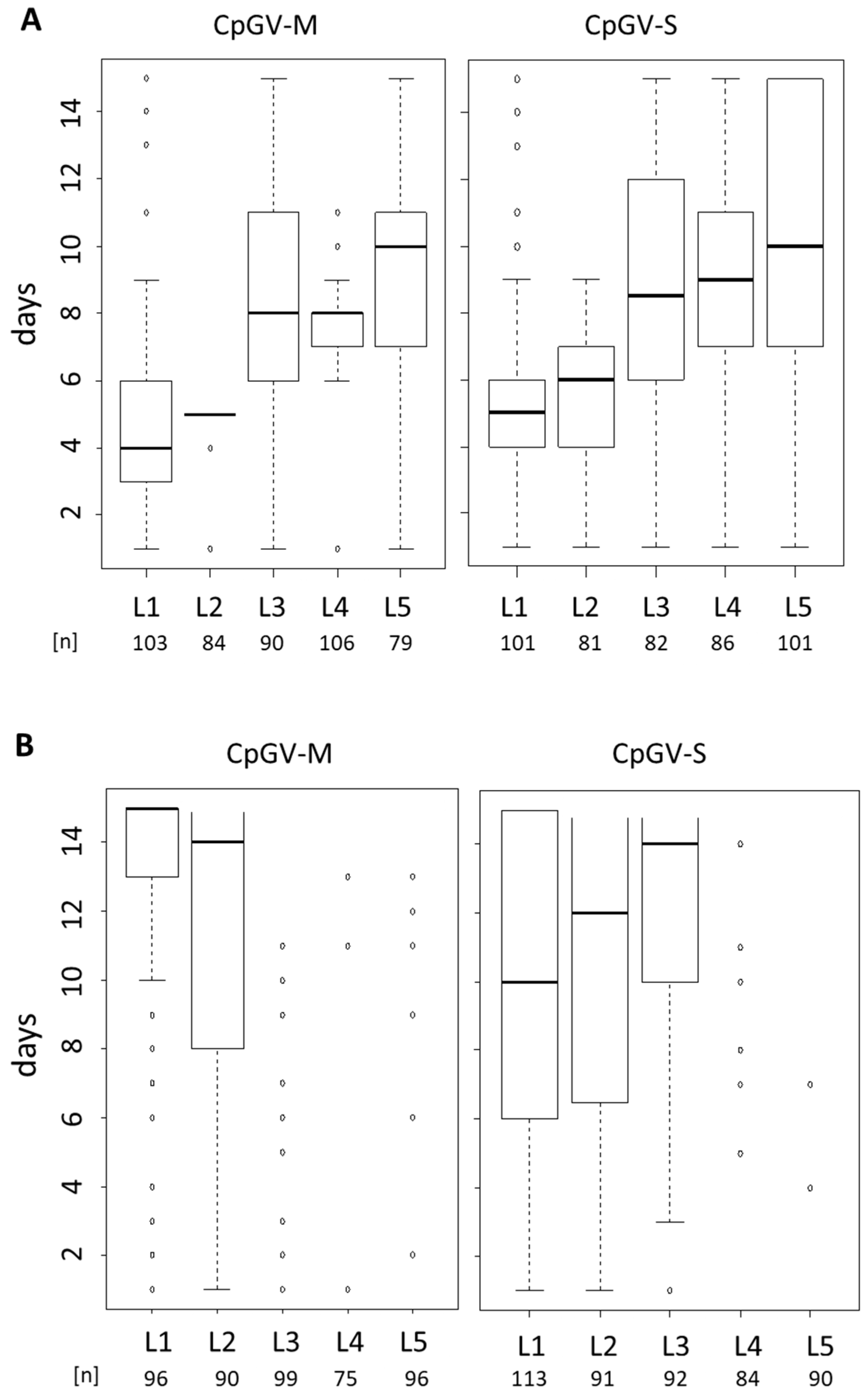
3.2. Instar-Specific Assay
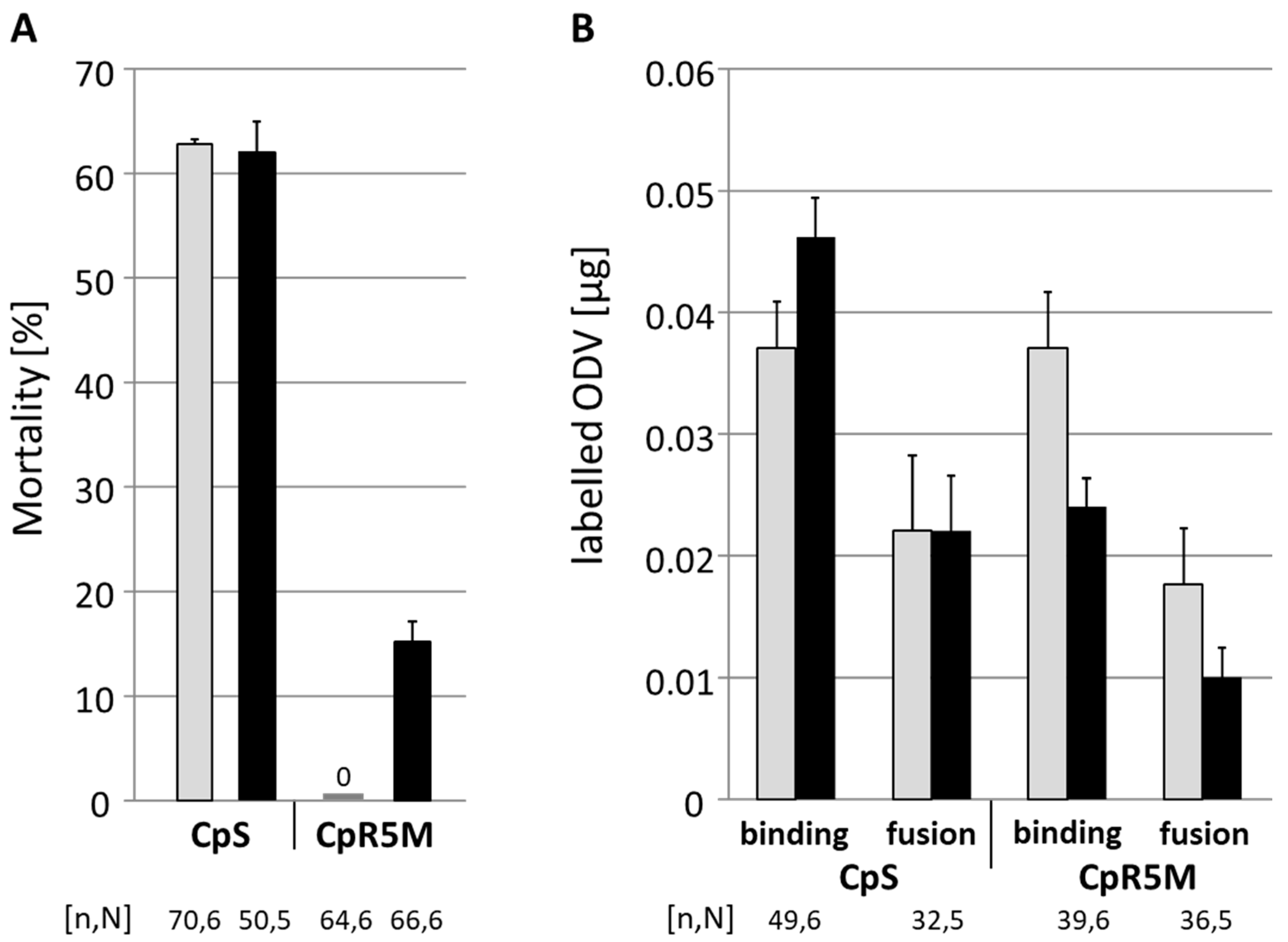
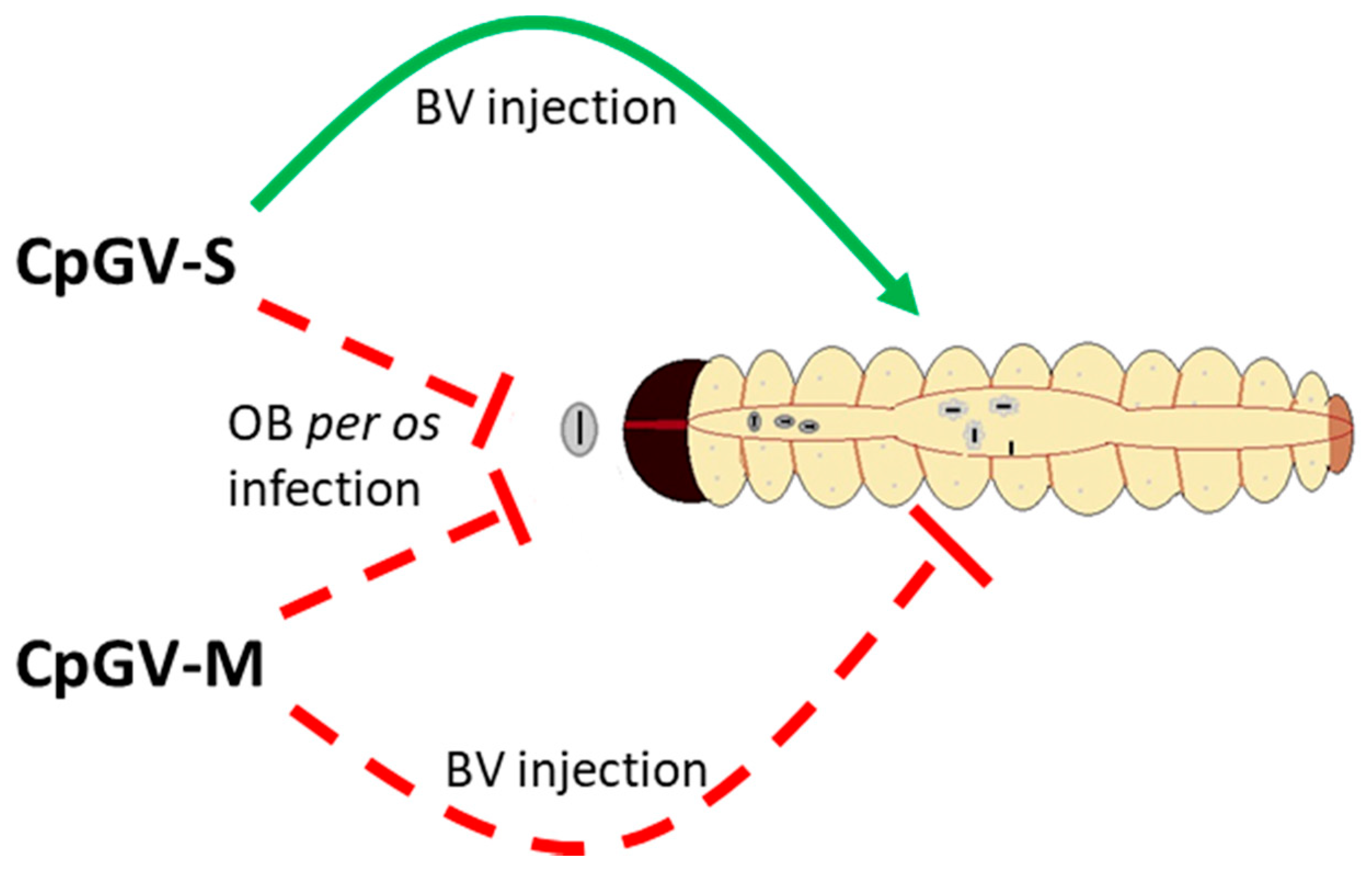
3.3. BV Injection Assay
3.4. ODV Infection Test and Fluorescence Dequenching Assay
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Herniou, E.A.; Olszewski, J.A.; Cory, J.S.; O’Reilly, D.R. The genome sequence and evolution of baculoviruses. Ann. Rev. Entomol. 2003, 48, 211–234. [Google Scholar] [CrossRef]
- Moscardi, F. Assessment of the application of baculoviruses for control of Lepidoptera. Annu. Rev. Entomol. 1999, 44, 257–289. [Google Scholar] [CrossRef]
- Lacey, L.A.; Grzywacz, D.; Shapiro-Ilan, D.I.; Frutos, R.; Brownbridge, M.; Goettel, M.S. Insect pathogens as biological control agents: Back to the future. J. Invertebr. Pathol. 2005, 132, 1–41. [Google Scholar] [CrossRef]
- Tanada, Y. A granulosis virus of the codling moth, Carpocapsa pomonella (Linnaeus) (Olethreutidae, Lepidoptera). J. Insect Pathol. 1964, 6, 378–380. [Google Scholar]
- Harvey, J.P.; Volkman, L.E. Biochemical and biological variation of Cydia pomonella (codling moth) granulosis virus. Virology 1983, 124, 21–34. [Google Scholar] [CrossRef]
- Crook, N.E.; Spencer, R.A.; Payne, C.C.; Leisy, D.J. Variation in Cydia pomonella granulosis virus isolates and physical maps of the DNA from three variants. J. Gen. Virol. 1985, 66, 2423–2430. [Google Scholar] [CrossRef]
- Rezapanah, M.; Shojai-Estabragh, S.; Huber, J.; Jehle, J.A. Molecular characterization of new isolates of Cydia pomonella granulovirus from Iran. J. Pest. Sci. 2008, 81, 187–191. [Google Scholar] [CrossRef]
- Arneodo, J.D.; De Anna, J.; Salvador, R.; Farinon, M.; Quintana, G.; Sciocco-Cap, A. Prospection and molecular analysis of CpGV isolates infecting Cydia pomonella at different geographical locations in Argentina. Ann. Appl. Biol. 2015, 166, 67–74. [Google Scholar] [CrossRef]
- Motsoeneng, B.; Jukes, M.D.; Knox, C.M.; Hill, M.P.; Moore, S.D. Genome analysis of a novel south African Cydia pomonella granulovirus (CpGV-SA) with resistance-breaking potential. Viruses 2019, 11, 658. [Google Scholar] [CrossRef]
- Fan, J.; Wennmann, J.T.; Wang, D.; Jehle, J.A. Novel diversity and virulence patterns found in new isolates of Cydia pomonella granulovirus from China. Appl. Environ. Microbiol. 2019, 86, e02000-19. [Google Scholar] [CrossRef]
- Fan, J.; Wennmann, J.T.; Wang, D.; Jehle, J.A. Single nucleotide polymorphism (SNP) frequencies and distribution reveal complex genetic composition of seven novel natural isolates of Cydia pomonella granulovirus. Virology 2020, 541, 32–40. [Google Scholar] [CrossRef]
- Fan, J.; Jehle, J.A.; Wennmann, J.T. Population structure of Cydia pomonella granulovirus isolates revealed by quantitative analysis of genetic variation. Virus Evol. 2020, 6, veaa073. [Google Scholar] [CrossRef]
- Luque, T.; Finch, R.; Crook, N.; O’Reilly, D.R.; Winstanley, D. The complete sequence of the Cydia pomonella granulovirus genome. J. Gen. Virol. 2001, 82, 2531–2547. [Google Scholar] [CrossRef]
- Gebhardt, M.M.; Eberle, K.E.; Radtke, P.; Jehle, J.A. Baculovirus resistance in codling moth is virus isolate-dependent and the consequence of a mutation in viral gene pe38. Proc. Natl. Acad. Sci. USA 2014, 111, 15711–15716. [Google Scholar] [CrossRef]
- Eberle, K.E.; Sayed, S.; Rezapanah, M.; Shojai-Estabragh, S.; Jehle, J.A. Diversity and evolution of the Cydia pomonella granulovirus. J. Gen. Virol. 2009, 90, 662–671. [Google Scholar] [CrossRef]
- Federici, B.A. Baculovirus pathogenesis. In The Baculoviruses; Miller, L.K., Ed.; New York Plenum Press: New York, NY, USA, 1997. [Google Scholar]
- Engelhard, E.K.; Kammorgan, L.N.; Washburn, J.O.; Volkman, L.E. The insect tracheal system—A conduit for the systemic spread of Autographa californica M nuclear polyhedrosis virus. Proc. Natl. Acad. Sci. USA 1994, 91, 3224–3227. [Google Scholar] [CrossRef]
- Flipsen, J.T.; Martens, J.W.; Van Oers, M.M.; Vlak, J.M.; Van Lent, J.W. Passage of Autographa californica nuclear polyhedrosis virus through the midgut epithelium of Spodoptera exigua larvae. Virology 1995, 208, 328–335. [Google Scholar] [CrossRef]
- Huber, J. Use of Baculoviruses in Pest Management Programs. In The Biology of Baculoviruses; Granados, R.R., Federici, B.A., Eds.; CRC Press: Boca Raton, FL, USA, 1986; Volume 2, pp. 182–202. [Google Scholar]
- Lacey, L.A.; Thompson, D.; Vincent, C.; Arthurs, S.P. Codling moth granulovirus: A comprehensive review. Biocontrol Sci. Technol. 2008, 18, 639–663. [Google Scholar] [CrossRef]
- Fritsch, E.; Undorf-Spahn, K.; Kienzle, J.; Zebitz, C.P.; Huber, J. Codling moth granulovirus: Variations in the susceptibility of local codling moth populations. Nachr. Dtsch. Pflanzenschutzd. 2005, 57, 29–34. [Google Scholar]
- Sauphanor, B.; Berling, M.; Toubon, J.F.; Reyes, M.; Delnatte, J.; Allemoz, P. Carpocapse des pommes: Cas de résistance au virus de la granulose en vergers biologiques. Phytoma Def. Veg. 2006, 590, 24–27. [Google Scholar]
- Berling, M.; Blachère-López, C.; Soubabère, O.; Léry, X.; Bonhomme, A.; Sauphanor, B.; López-Ferber, M. Cydia pomonella granulovirus genotypes overcome virus resistance in the codling moth and improve virus efficiency by selection against resistant hosts. Appl. Environ. Microbiol. 2009, 75, 925–930. [Google Scholar] [CrossRef]
- Zichová, T.; Stará, J.; Kundu, J.K.; Eberle, K.E.; Jehle, J.A. Resistance to Cydia pomonella granulovirus follows a geographically widely distributed inheritance type within Europe. BioControl 2013, 58, 525–534. [Google Scholar] [CrossRef]
- Schmitt, A.; Bisutti, I.L.; Ladurner, E.; Benuzzi, M.; Sauphanor, B.; Kienzle, J.; Zingg, D.; Undorf-Spahn, K.; Fritsch, E.; Huber, J.; et al. The occurrence and distribution of resistance of codling moth to Cydia pomonella granulovirus in Europe. J. Appl. Entomol. 2013, 137, 641–649. [Google Scholar] [CrossRef]
- Jehle, J.A.; Schulze-Bopp, S.; Undorf-Spahn, K.; Fritsch, E. Evidence for a second type of resistance against Cydia pomonella granulovirus in field populations of codling moths. Appl. Environ. Microbiol. 2017, 83, e02330-16. [Google Scholar] [CrossRef] [PubMed]
- Asser-Kaiser, S.; Fritsch, E.; Undorf-Spahn, K.; Kienzle, J.; Eberle, K.E.; Gund, N.A.; Reineke, A.; Zebitz, C.P.; Heckel, D.G.; Huber, J.; et al. Rapid emergence of baculovirus resistance in codling moth due to dominant, sex-linked inheritance. Science 2007, 317, 1916–1918. [Google Scholar] [CrossRef] [PubMed]
- Eberle, K.E.; Asser-Kaiser, S.; Sayed, S.M.; Nguyen, H.T.; Jehle, J.A. Overcoming the resistance of codling moth against conventional Cydia pomonella granulovirus (CpGV-M) by a new isolate CpGV-I12. J. Invertebr. Pathol. 2008, 98, 293–298. [Google Scholar] [CrossRef] [PubMed]
- Graillot, B.; Berling, M.; Blachere-Lopez, C.; Siegwart, M.; Besse, S.; Lopez-Ferber, M. Progressive adaptation of a CpGV isolate to codling moth populations resistant to CpGV-M. Viruses 2014, 6, 5135–5144. [Google Scholar] [CrossRef] [PubMed]
- Graillot, B.; Bayle, S.; Blachere-Lopez, C.; Besse, S.; Siegwart, M.; Lopez-Ferber, M. Biological characteristics of experimental genotype mixtures of Cydia pomonella granulovirus (CpGV): Ability to control susceptible and resistant pest populations. Viruses 2016, 8, 147. [Google Scholar] [CrossRef] [PubMed]
- Asser-Kaiser, S.; Radtke, P.; El-Salamouny, S.; Winstanley, D.; Jehle, J.A. Baculovirus resistance in codling moth (Cydia pomonella L.) caused by early block of virus replication. Virology 2011, 410, 360–367. [Google Scholar] [CrossRef]
- Sauer, A.J.; Fritsch, E.; Undorf-Spahn, K.; Nguyen, H.T.; Frantisek, M.; Heckel, D.G.; Jehle, J.A. Novel resistance to Cydia pomonella granulovirus (CpGV) in codling moth shows autosomal and dominant inheritance and confers cross-resistance to different CpGV genome groups. PLoS ONE 2017, 12, e0179157. [Google Scholar] [CrossRef] [PubMed]
- Sauer, A.J.; Schulze-Bopp, S.; Fritsch, E.; Undorf-Spahn, K.; Jehle, J.A. A third type of resistance of codling moth against Cydia pomonella granulovirus (CpGV) shows a mixture of a Z-linked and autosomal inheritance pattern. Appl. Environ. Microbiol. 2017, 17, e01036-17. [Google Scholar] [CrossRef]
- Siegwart, M.; Maugin, S.; Besse, S.; Lopez-Ferber, M.; Hinsberger, A.; Gauffre, B. Le carpocapse des pommes résiste au virus de la granulose. Phytoma Def. Veg. 2020, 738, 45–50. [Google Scholar]
- Ivaldi-Sender, C. A simple technique for the permanent rearing of the oriental fruit moth, Grapholita molesta (Lepidoptera. Tortricidae) on an artificial medium. Techniques simples pour un elevage permanent de la tordeuse orientale, Grapholita molesta (Lepidoptera: Tortricidae) sur milieu artificiel. Ann. Zool. Ecol. Anim. 1974, 6, 337–343. [Google Scholar]
- Undorf-Spahn, K.; Fritsch, E.; Huber, J.; Kienzle, J.; Zebitz, C.P.; Jehle, J.A. High stability and no fitness costs of the resistance of codling moth to Cydia pomonella granulovirus (CpGV-M). J. Invertebr. Pathol. 2012, 111, 136–142. [Google Scholar] [CrossRef]
- Abbott, W.S. A method of computing the effectiveness of an insecticide. J. Econ. Entomol. 1925, 18, 265–267. [Google Scholar] [CrossRef]
- Haas-Stapleton, E.J.; Washburn, J.O.; Volkman, L.E. P74 mediates specific binding of Autographa californica M nucleopolyhedrovirus occlusion-derived virus to primary cellular targets in the midgut epithelia of Heliothis virescens larvae. J. Virol. 2004, 78, 6786–6791. [Google Scholar] [CrossRef]
- Iwata, K.; Haas-Stapleton, E.J.; Kunimi, Y.; Inoue, M.N.; Nakai, M. Midgut based resistance to oral infection by a nucleopolyhedrovirus in the laboratory-selected strain of the small tea tortrix, Adoxophyses honmai (Lepidoptera: Tortricidae). J. Gen. Virol. 2017, 98, 296–304. [Google Scholar] [CrossRef]
- Haas-Stapleton, E.J.; Washburn, J.O.; Volkman, L.E. Spodoptera frugiperda resistance to oral infection by Autographa californica multiple nucleopolyhedrovirus linked to aberrant occlusion-derived virus binding in the midgut. J. Virol. 2005, 86, 1349–1355. [Google Scholar] [CrossRef]
- Wennmann, J.T.; Kohler, T.; Gueli Alletti, G.; Jehle, J.A. Mortality of cutworm larvae is not enhanced by Agrotis segetum granulovirus and Agrotis segetum Nucleopolyhedrovirus B coinfection relative to single infection by either virus. Appl. Environm. Microbiol. 2015, 81, 2893–2899. [Google Scholar] [CrossRef][Green Version]
- Graillot, B.; Blachere-López, C.; Besse, S.; Siegwart, M.; López-Ferber, M. Importance of the host phenotype on the preservation of the genetic diversity in codling moth granulovirus. Viruses 2019, 11, 621. [Google Scholar] [CrossRef] [PubMed]
- Grove, M.J.; Hoover, K. Intrastadial developmental resistance of third instar gypsy moths (Lymantria dispar L.) to L. dispar nucleopolyhedrovirus. Biol. Contr. 2007, 40, 355–361. [Google Scholar] [CrossRef]
- Jakubowska, A.K.; Lynn, D.E.; Herrero, S.; Vlak, J.M.; van Oers, M.M. Host-range expansion of Spodoptera exigua multiple nucleopolyhedrovirus to Agrotis segetum larvae when the midgut is bypassed. J. Gen. Virol. 2010, 91, 898–906. [Google Scholar] [CrossRef] [PubMed]
- Chikhalya, A.; Stephens, K.D.; Archie, J.W.; Haas-Stapleton, E.J. Virulence and pathogenesis of the baculovirus Autographa californica M nucleopolyhedrovirus (AcMNPV) in Pseudoplusia includens larvae. Biol. Control 2013, 65, 101–108. [Google Scholar] [CrossRef]
- Wang, P.; Granados, R.R. Observations on the presence of the peritrophic membrane in larval Trichoplusia ni and its role in limiting baculovirus infection. J. Invertebr. Pathol. 1998, 72, 57–62. [Google Scholar] [CrossRef]
- Kuzio, J.; Jaques, R.; Faulkner, P. Identification of p74, a gene essential for virulence of baculovirus occlusion bodies. Virology 1989, 173, 759–763. [Google Scholar] [CrossRef]
- Ohkawa, T.; Washburn, J.O.; Sitapara, R.; Sid, E.; Volkman, L.E. Specific binding of Autographa californica M nucleopolyhedrovirus occlusion-derived virus to midgut cells of Heliothis virescens larvae is mediated by products of pif genes Ac119 and Ac022 but not by Ac115. J. Virol. 2005, 79, 15258–15264. [Google Scholar] [CrossRef]
- Sparks, W.O.; Harrison, R.L.; Bonning, B.C. Autographa californica multiple nucleopolyhedrovirus ODV-E56 is a per os infectivity factor, but is not essential for binding and fusion of occlusion-derived virus to the host midgut. Virology 2011, 409, 69–76. [Google Scholar] [CrossRef]
- Mu, J.F.; van Lent, J.W.; Smagghe, G.; Wang, Y.; Chen, X.W.; Vlak, J.M.; van Oers, M.M. Live imaging of baculovirus infection of midgut epithelium cells: A functional assay of per os infectivity factors. J. Gen. Virol. 2014, 95, 2531–2539. [Google Scholar] [CrossRef]
- Nakai, M.; Takahashi, K.; Iwata, K.; Koyanagi, J.; Ookuma, A.; Takatsuka, J.; Okuno, S.; Kunimi, Y. Acquired resistance to a nucleopolyhedrovirus in the smaller tea tortrix Adoxophyes honmai (Lepidoptera: Tortricidae) after selection by serial viral administration. J. Invertebr. Pathol. 2017, 145, 23–30. [Google Scholar] [CrossRef]
| Concentration (BV/Larvae) | Number of CpS Larvae | % Mortality |
|---|---|---|
| 50 | 14 | 19.6 |
| 500 | 15 | 58.3 |
| 5000 | 19 | 73.6 |
| 50,000 | 17 | 92.7 |
| 500,000 | 14 | 100.0 |
| Codling Moth Strain | BV Treatment | n, N | % Mortality * (±SD) 14 Days p.i. | Test for Significant Statistical Differences |
|---|---|---|---|---|
| CpR5M | CpGV-M | 61, 4 | 33.4 (±6.4) | A |
| CpGV-S | 42, 3 | 83.4 (±7.5) | B | |
| CpS | CpGV-M | 57, 4 | 73.2 (±10.9) | B |
| CpGV-S | 41, 3 | (±5.9) | B |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sauer, A.J.; Fritsch, E.; Undorf-Spahn, K.; Iwata, K.; Kleespies, R.G.; Nakai, M.; Jehle, J.A. Cross-Resistance of the Codling Moth against Different Isolates of Cydia pomonella Granulovirus Is Caused by Two Different but Genetically Linked Resistance Mechanisms. Viruses 2021, 13, 1952. https://doi.org/10.3390/v13101952
Sauer AJ, Fritsch E, Undorf-Spahn K, Iwata K, Kleespies RG, Nakai M, Jehle JA. Cross-Resistance of the Codling Moth against Different Isolates of Cydia pomonella Granulovirus Is Caused by Two Different but Genetically Linked Resistance Mechanisms. Viruses. 2021; 13(10):1952. https://doi.org/10.3390/v13101952
Chicago/Turabian StyleSauer, Annette J., Eva Fritsch, Karin Undorf-Spahn, Kento Iwata, Regina G. Kleespies, Madoka Nakai, and Johannes A. Jehle. 2021. "Cross-Resistance of the Codling Moth against Different Isolates of Cydia pomonella Granulovirus Is Caused by Two Different but Genetically Linked Resistance Mechanisms" Viruses 13, no. 10: 1952. https://doi.org/10.3390/v13101952
APA StyleSauer, A. J., Fritsch, E., Undorf-Spahn, K., Iwata, K., Kleespies, R. G., Nakai, M., & Jehle, J. A. (2021). Cross-Resistance of the Codling Moth against Different Isolates of Cydia pomonella Granulovirus Is Caused by Two Different but Genetically Linked Resistance Mechanisms. Viruses, 13(10), 1952. https://doi.org/10.3390/v13101952







