Overexpression of the Bacteriophage T4 motB Gene Alters H-NS Dependent Repression of Specific Host DNA
Abstract
1. Introduction
2. Materials and Methods
2.1. Strains
2.2. DNA
2.3. Proteins
2.4. In Silico Analyses of motB
2.5. DNase I Footprinting
2.6. Purification of Total RNA and RNA-seq Analyses
3. Results
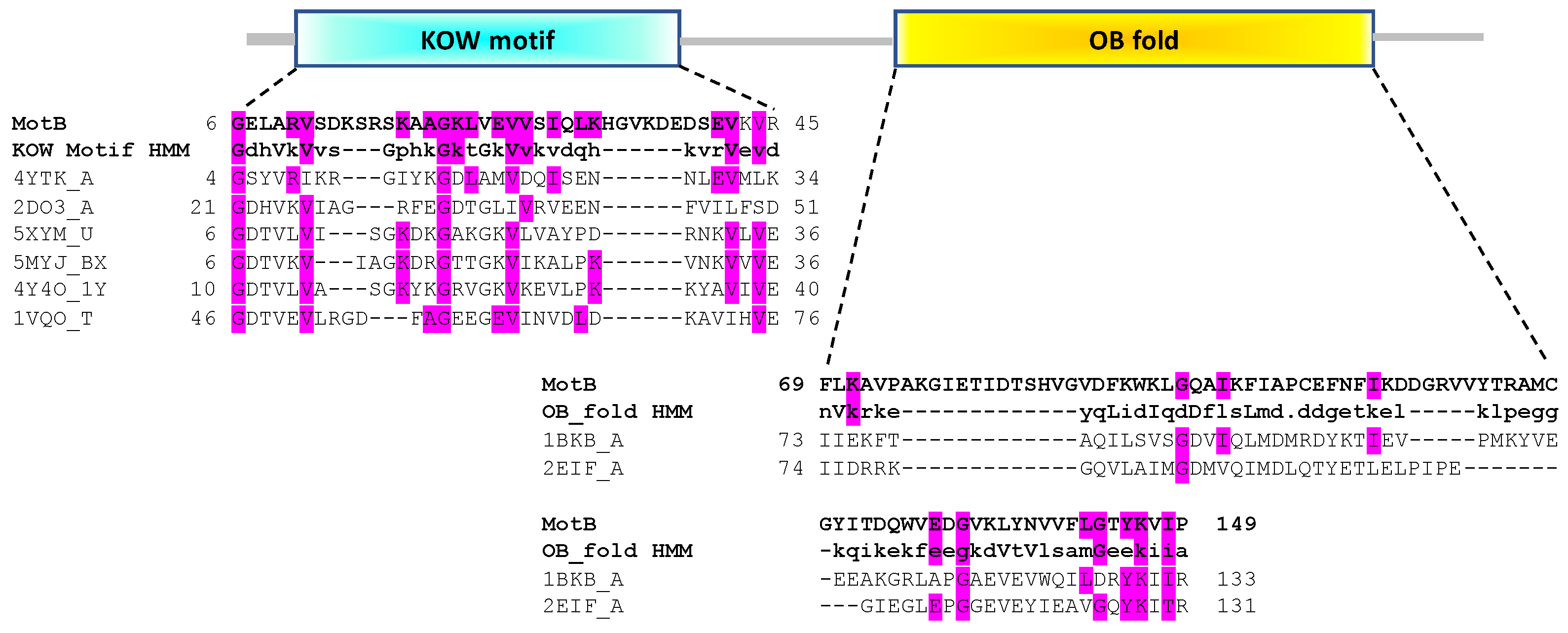
3.1. In Silico Analyses Predict That MotB Contains Both a KOW and an OB-Fold Domain
3.2. MotB Is Highly Conserved among T-Even Phage
3.3. MotB Affects the Interaction of H-NS with DNA
3.4. Expression of motB Results in Up-Regulation of 75 Host Genes, a Subset of Which Are Repressed by the Histone-Like Protein, H-NS
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hatfull, G.F.; Hendrix, R.W. Bacteriophages and their genomes. Curr. Opin. Virol. 2011, 1, 298–303. [Google Scholar] [CrossRef]
- Patterson-West, J.; Arroyo-Mendoza, M.; Hsieh, M.-L.; Harrison, D.; Walker, M.M.; Knipling, L.; Hinton, D.M. The Bacteriophage T4 MotB Protein, a DNA-Binding Protein, Improves Phage Fitness. Viruses 2018, 10, 343. [Google Scholar] [CrossRef] [PubMed]
- Hinton, D.M. Transcriptional control in the prereplicative phase of T4 development. Virol. J. 2010, 7, 289. [Google Scholar] [CrossRef] [PubMed]
- Pulitzer, J.F.; Colombo, M.; Ciaramella, M. New control elements of bacteriophage T4 pre-replicative transcription. J. Mol. Biol. 1985, 182, 249–263. [Google Scholar] [CrossRef]
- Wang, W.; Li, G.W.; Chen, C.; Xie, X.S.; Zhuang, X. Chromosome organization by a nucleoid-associated protein in live bacteria. Science 2011, 333, 1445–1449. [Google Scholar] [CrossRef] [PubMed]
- Dillon, S.C.; Dorman, C.J. Bacterial nucleoid-associated proteins, nucleoid structure and gene expression. Nat. Rev. Genet. 2010, 8, 185–195. [Google Scholar] [CrossRef] [PubMed]
- Ohniwa, R.L.; Muchaku, H.; Saito, S.; Wada, C.; Morikawa, K. Atomic Force Microscopy Analysis of the Role of Major DNA-Binding Proteins in Organization of the Nucleoid in Escherichia coli. PLoS ONE 2013, 8, e72954. [Google Scholar] [CrossRef] [PubMed]
- Song, D.; Loparo, J.J. Building bridges within the bacterial chromosome. Trends Genet. 2015, 31, 164–173. [Google Scholar] [CrossRef]
- Grainger, D.C.; Busby, S.J. Global Regulators of Transcription in Escherichia coli: Mechanisms of Action and Methods for Study. Adv. Clin. Chem. 2008, 65, 93–113. [Google Scholar] [CrossRef]
- Shen, B.; Landick, R. Transcription of Bacterial Chromatin. J. Mol. Biol. 2019, 431, 4040–4066. [Google Scholar] [CrossRef]
- Grainger, D.C. Structure and function of bacterial H-NS protein. Biochem. Soc. Trans. 2016, 44, 1561–1569. [Google Scholar] [CrossRef] [PubMed]
- Winardhi, R.S.; Yan, J.; Kenney, L.J. H-NS Regulates Gene Expression and Compacts the Nucleoid: Insights from Single-Molecule Experiments. Biophys. J. 2015, 109, 1321–1329. [Google Scholar] [CrossRef] [PubMed]
- Pfeifer, E.; Hünnefeld, M.; Popa, O.; Frunzke, J. Impact of Xenogeneic Silencing on Phage–Host Interactions. J. Mol. Biol. 2019, 431, 4670–4683. [Google Scholar] [CrossRef] [PubMed]
- Navarre, W.W. H-NS as a defence system. In Bacterial Chromatin; Dame, R.T., Dorman, C.J., Eds.; Springer: Dordrecht, The Netherlands, 2010; pp. 251–322. [Google Scholar]
- Liu, Q.; Richardson, C.C. Gene 5.5 protein of bacteriophage T7 inhibits the nucleoid protein H-NS of Escherichia coli. Proc. Natl. Acad. Sci. USA 1993, 90, 1761–1765. [Google Scholar] [CrossRef] [PubMed]
- Ali, S.S.; Beckett, E.; Bae, S.J.; Navarre, W.W. The 5.5 Protein of Phage T7 Inhibits H-NS through Interactions with the Central Oligomerization Domain. J. Bacteriol. 2011, 193, 4881–4892. [Google Scholar] [CrossRef]
- Ho, C.-H.; Wang, H.-C.; Ko, T.-P.; Chang, Y.-C.; Wang, A.H.-J. The T4 Phage DNA Mimic Protein Arn Inhibits the DNA Binding Activity of the Bacterial Histone-like Protein H-NS. J. Biol. Chem. 2014, 289, 27046–27054. [Google Scholar] [CrossRef]
- Jeong, H.; Barbe, V.; Lee, C.H.; Vallenet, D.; Yu, D.S.; Choi, S.-H.; Couloux, A.; Lee, S.-W.; Yoon, S.H.; Cattolico, L.; et al. Genome Sequences of Escherichia coli B strains REL606 and BL21(DE3). J. Mol. Biol. 2009, 394, 644–652. [Google Scholar] [CrossRef]
- Studier, F.W.; Rosenberg, A.H.; Dunn, J.J.; Dubendorff, J.W. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 1990, 185, 60–89. [Google Scholar] [CrossRef]
- Bonocora, R.P.; Caignan, G.; Woodrell, C.; Werner, M.H.; Hinton, D.M. A basic/hydrophobic cleft of the T4 activator MotA interacts with the C-terminus of E.coli sigma70 to activate middle gene transcription. Mol. Microbiol. 2008, 69, 331–343. [Google Scholar] [CrossRef]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef]
- Papadopoulos, J.S.; Agarwala, R. COBALT: Constraint-based alignment tool for multiple protein sequences. Bioinformatics 2007, 23, 1073–1079. [Google Scholar] [CrossRef] [PubMed]
- Sokal, R.R.; Michener, C.D. A statistical method for evaluating systematic relationships. Univ. Kans. Sci. Bull. 1958, 38, 1409–1438. [Google Scholar]
- Waterhouse, A.M.; Procter, J.B.; Martin, D.M.A.; Clamp, M.; Barton, G.J. Jalview Version 2--a multiple sequence alignment editor and analysis workbench. Bioinformatics 2009, 25, 1189–1191. [Google Scholar] [CrossRef] [PubMed]
- Hildebrand, A.; Remmert, M.; Biegert, A.; Söding, J. Fast and accurate automatic structure prediction with HHpred. Proteins Struct. Funct. Bioinform. 2009, 77, 128–132. [Google Scholar] [CrossRef]
- Zimmermann, L.; Stephens, A.; Nam, S.-Z.; Rau, D.; Kübler, J.; Lozajic, M.; Gabler, F.; Söding, J.; Lupas, A.N.; Alva, V. A Completely Reimplemented MPI Bioinformatics Toolkit with a New HHpred Server at its Core. J. Mol. Biol. 2018, 430, 2237–2243. [Google Scholar] [CrossRef]
- Kuchibhatla, D.B.; Sherman, W.A.; Chung, B.Y.W.; Cook, S.; Schneider, G.; Eisenhaber, B.; Karlin, D.G. Powerful Sequence Similarity Search Methods and In-Depth Manual Analyses Can Identify Remote Homologs in Many Apparently "Orphan" Viral Proteins. J. Virol. 2014, 88, 10–20. [Google Scholar] [CrossRef]
- El-Gebali, S.; Mistry, J.; Bateman, A.; Eddy, S.R.; Luciani, A.; Potter, S.C.; Qureshi, M.; Richardson, L.J.; Salazar, G.A.; Smart, A.; et al. The Pfam protein families database in 2019. Nucleic Acids Res. 2018, 47, D427–D432. [Google Scholar] [CrossRef]
- Murzin, A.G.; Brenner, S.E.; Hubbard, T.; Chothia, C. SCOP: A structural classification of proteins database for the investigation of sequences and structures. J. Mol. Biol. 1995, 247, 536–540. [Google Scholar] [CrossRef]
- Chandonia, J.-M.; Fox, N.K.; Brenner, S.E. SCOPe: Classification of large macromolecular structures in the structural classification of proteins—extended database. Nucleic Acids Res. 2019, 47, D475–D481. [Google Scholar] [CrossRef]
- Fox, N.K.; Brenner, S.E.; Chandonia, J.-M. SCOPe: Structural Classification of Proteins—extended, integrating SCOP and ASTRAL data and classification of new structures. Nucleic Acids Res. 2014, 42, D304–D309. [Google Scholar] [CrossRef]
- Hinton, D.M. Transcript analyses of the uvsX-40-41 region of bacteriophage T4. Changes in the RNA as infection proceeds. J. Biol. Chem. 1989, 264, 14432–14439. [Google Scholar] [CrossRef]
- Shishkin, A.A.; Giannoukos, G.; Kucukural, A.; Ciulla, D.; Busby, M.; Surka, C.; Chen, J.; Bhattacharyya, R.P.; Rudy, R.F.; Patel, M.M.; et al. Simultaneous generation of many RNA-seq libraries in a single reaction. Nat. Methods 2015, 12, 323–325. [Google Scholar] [CrossRef] [PubMed]
- Patterson-West, J.; James, T.D.; Fernández-Coll, L.; Iben, J.R.; Moon, K.; Knipling, L.; Cashel, M.; Hinton, D.M. The E. coli Global Regulator DksA Reduces Transcription during T4 Infection. Viruses 2018, 10, 308. [Google Scholar] [CrossRef]
- Sanchez-Vazquez, P.; Dewey, C.N.; Kitten, N.; Ross, W.; Gourse, R.L. Genome-wide effects on Escherichia coli transcription from ppGpp binding to its two sites on RNA polymerase. Proc. Natl. Acad. Sci. USA 2019, 116, 8310–8319. [Google Scholar] [CrossRef] [PubMed]
- Kyrpides, N.C.; Woese, C.R.; Ouzounis, C.A. KOW: A novel motif linking a bacterial transcription factor with ribosomal proteins. Trends Biochem. Sci. 1996, 21, 425–426. [Google Scholar] [CrossRef]
- Murzin, A.G. OB (oligonucleotide/oligosaccharide binding)-fold: Common structural and functional solution for non-homologous sequences. EMBO J. 1993, 12, 861–867. [Google Scholar] [CrossRef] [PubMed]
- Meyer, P.A.; Li, S.; Zhang, M.; Yamada, K.; Takagi, Y.; Hartzog, G.A.; Fu, J. Structures and Functions of the Multiple KOW Domains of Transcription Elongation Factor Spt5. Mol. Cell. Biol. 2015, 35, 3354–3369. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kang, J.Y.; Mooney, R.A.; Nedialkov, Y.; Saba, J.; Mishanina, T.V.; Artsimovitch, I.; Landick, R.; Darst, S.A. Structural Basis for Transcript Elongation Control by NusG Family Universal Regulators. Cell 2018, 173, 1650–1662.e14. [Google Scholar] [CrossRef]
- Mooney, R.A.; Schweimer, K.; Roesch, P.; Gottesman, M.; Landick, R. Two Structurally Independent Domains of E. coli NusG Create Regulatory Plasticity via Distinct Interactions with RNA Polymerase and Regulators. J. Mol. Biol. 2009, 391, 341–358. [Google Scholar] [CrossRef]
- Zuber, P.K.; Hahn, L.; Reinl, A.; Schweimer, K.; Knauer, S.H.; Gottesman, M.E.; Rösch, P.; Wöhrl, B.M. Structure and nucleic acid binding properties of KOW domains 4 and 6–7 of human transcription elongation factor DSIF. Sci. Rep. 2018, 8, 1–12. [Google Scholar] [CrossRef]
- Flynn, R.L.; Zou, L. Oligonucleotide/oligosaccharide-binding fold proteins: A growing family of genome guardians. Crit. Rev. Biochem. Mol. Biol. 2010, 45, 266–275. [Google Scholar] [CrossRef] [PubMed]
- Lavigne, R.; Darius, P.; Summer, E.J.; Seto, D.; Mahadevan, P.; Nilsson, A.S.; Ackermann, H.-W.; Kropinski, A.M. Classification of Myoviridae bacteriophages using protein sequence similarity. BMC Microbiol. 2009, 9, 224. [Google Scholar] [CrossRef] [PubMed]
- Lucht, J.M.; Dersch, P.; Kempf, B.; Bremer, E. Interactions of the nucleoid-associated DNA-binding protein H-NS with the regulatory region of the osmotically controlled proU operon of Escherichia coli. J. Biol. Chem. 1994, 269, 6578. [Google Scholar] [CrossRef]
- Bouffartigues, E.; Buckle, M.; Badaut, C.; Travers, A.; Rimsky, S. H-NS cooperative binding to high-affinity sites in a regulatory element results in transcriptional silencing. Nat. Struct. Mol. Biol. 2007, 14, 441–448. [Google Scholar] [CrossRef]
- Galli, E.; Gerdes, K. FtsZ-ZapA-ZapB interactome of Escherichia coli. J. Bacteriol. 2012, 194, 292–302. [Google Scholar] [CrossRef]
- Eguchi, Y.; Utsumi, R. Introduction to Bacterial Signal Transduction Networks. Neurotransm. Interact. Cogn. Funct. 2008, 631, 1–6. [Google Scholar] [CrossRef]
- Tucker, D.L.; Tucker, N.; Conway, T. Gene Expression Profiling of the pH Response in Escherichia coli. J. Bacteriol. 2002, 184, 6551–6558. [Google Scholar] [CrossRef]
- Dorman, C.J.; Bhriain, N.N. CRISPR-Cas, DNA Supercoiling, and Nucleoid-Associated Proteins. Trends Microbiol. 2020, 28, 19–27. [Google Scholar] [CrossRef]
- Yosef, I.; Goren, M.G.; Qimron, U. Proteins and DNA elements essential for the CRISPR adaptation process in Escherichia coli. Nucleic Acids Res. 2012, 40, 5569–5576. [Google Scholar] [CrossRef]
- Ueda, T.; Takahashi, H.; Uyar, E.; Ishikawa, S.; Ogasawara, N.; Oshima, T. Functions of the Hha and YdgT Proteins in Transcriptional Silencing by the Nucleoid Proteins, H-NS and StpA, in Escherichia coli. DNA Res. 2013, 20, 263–271. [Google Scholar] [CrossRef]
- Ueguchi, C.; Suzuki, T.; Yoshida, T.; Tanaka, K.-I.; Mizuno, T. Systematic Mutational Analysis Revealing the Functional Domain Organization of Escherichia coli Nucleoid Protein H-NS. J. Mol. Biol. 1996, 263, 149–162. [Google Scholar] [CrossRef] [PubMed]
- Krell, K.; Gottesman, M.E.; Parks, J.S.; Eisenberg, M.A. Escape synthesis of the biotin operon in induced lambda b-2 lysogens. J. Mol. Biol. 1972, 68, 69–82. [Google Scholar] [CrossRef]
- Azam, T.A.; Ishihama, A. Twelve Species of the Nucleoid-associated Protein from Escherichia coli. J. Biol. Chem. 1999, 274, 33105–33113. [Google Scholar] [CrossRef] [PubMed]
- Navarre, W.W.; McClelland, M.; Libby, S.J.; Fang, F.C. Silencing of xenogeneic DNA by H-NS--facilitation of lateral gene transfer in bacteria by a defense system that recognizes foreign DNA. Genes Dev. 2007, 21, 1456–1471. [Google Scholar] [CrossRef]
- Stoebel, D.M.; Free, A.; Dorman, C.J. Anti-silencing: Overcoming H-NS-mediated repression of transcription in Gram-negative enteric bacteria. Microbiology 2008, 154, 2533–2545. [Google Scholar] [CrossRef]
- Yu, R.R.; DiRita, V.J. Regulation of gene expression in Vibrio cholerae by ToxT involves both antirepression and RNA polymerase stimulation. Mol. Microbiol. 2002, 43, 119–134. [Google Scholar] [CrossRef]
- Bustamante, V.H.; Santana, F.J.; Calva, E.; Puente, J.L. Transcriptional regulation of type III secretion genes in enteropathogenic Escherichia coli: Ler antagonizes H-NS-dependent repression. Mol. Microbiol. 2001, 39, 664–678. [Google Scholar] [CrossRef]
- Pao, C.C.; Speyer, J.F. Mutants of T7 bacteriophage inhibited by lambda prophage. Proc. Natl. Acad. Sci. USA 1975, 72, 3642–3646. [Google Scholar] [CrossRef]
- Lin, L. Study of Bacteriophage T7 Gene 5.9 and Gene 5.5. Ph.D. Thesis, State University of New York at Stony Brook, Stony Brook, NY, USA, 1992. [Google Scholar]
- Dole, S.; Nagarajavel, V.; Schnetz, K. The histone-like nucleoid structuring protein H-NS represses the Escherichia coli bgl operon downstream of the promoter. Mol. Microbiol. 2004, 52, 589–600. [Google Scholar] [CrossRef]
- Westra, E.R.; Pul, Ü.; Heidrich, N.; Jore, M.M.; Lundgren, M.; Stratmann, T.; Wurm, R.; Raine, A.; Mescher, M.; Van Heereveld, L.; et al. H-NS-mediated repression of CRISPR-based immunity in Escherichia coli K12 can be relieved by the transcription activator LeuO. Mol. Microbiol. 2010, 77, 1380–1393. [Google Scholar] [CrossRef]
- Cowan, J.; D’Acci, K.; Guttman, B.; Kutter, E. Gel analysis of T4 prereplicative proteins. In Molecular Biology of Bacteriophage T4; Karam, J., Ed.; American Society of Microbiology: Washington, DC, USA, 1994; pp. 520–527. [Google Scholar]
- Uzan, M.; Daubentoncarafa, Y.; Favre, R.; Defranciscis, V.; Brody, E. The T4 Mot Protein Functions as Part of a Pre-Replicative DNA-Protein Complex. J. Biol. Chem. 1985, 260, 633–639. [Google Scholar] [CrossRef]
- Warren, R.J.; Bose, S.K. Bacteriophage-induced inhibition of host functions. I. Degradation of Escherichia coli deoxyribonucleic acid after T4 infection. J. Virol. 1968, 2, 327–334. [Google Scholar] [CrossRef] [PubMed]
- Warner, H.R.; Snustad, P.; Jorgensen, S.E.; Koerner, J.F. Isolation of bacteriophage T4 mutants defective in the ability to degrade host deoxyribonucleic acid. J. Virol 1970, 5, 700–708. [Google Scholar] [CrossRef] [PubMed]
- van der Valk, R.A.; Vreede, J.; Qin, L.; Moolenaar, G.F.; Hofmann, A.; Goosen, N.; Dame, R.T. Mechanism of environmentally driven conformational changes that modulate H-NS DNA-bridging activity. eLife 2017, 6, e27369. [Google Scholar] [CrossRef]
- Amit, R.; Oppenheim, A.B.; Stavans, J. Increased Bending Rigidity of Single DNA Molecules by H-NS, a Temperature and Osmolarity Sensor. Biophys. J. 2003, 84, 2467–2473. [Google Scholar] [CrossRef]
- Sanson, B.; Hu, R.-M.; Troitskayadagger, E.; Mathy, N.; Uzan, M. Endoribonuclease RegB from bacteriophage T4 is necessary for the degradation of early but not middle or late mRNAs11Edited by M. Yaniv. J. Mol. Biol. 2000, 297, 1063–1074. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Patterson-West, J.; Tai, C.-H.; Son, B.; Hsieh, M.-L.; Iben, J.R.; Hinton, D.M. Overexpression of the Bacteriophage T4 motB Gene Alters H-NS Dependent Repression of Specific Host DNA. Viruses 2021, 13, 84. https://doi.org/10.3390/v13010084
Patterson-West J, Tai C-H, Son B, Hsieh M-L, Iben JR, Hinton DM. Overexpression of the Bacteriophage T4 motB Gene Alters H-NS Dependent Repression of Specific Host DNA. Viruses. 2021; 13(1):84. https://doi.org/10.3390/v13010084
Chicago/Turabian StylePatterson-West, Jennifer, Chin-Hsien Tai, Bokyung Son, Meng-Lun Hsieh, James R. Iben, and Deborah M. Hinton. 2021. "Overexpression of the Bacteriophage T4 motB Gene Alters H-NS Dependent Repression of Specific Host DNA" Viruses 13, no. 1: 84. https://doi.org/10.3390/v13010084
APA StylePatterson-West, J., Tai, C.-H., Son, B., Hsieh, M.-L., Iben, J. R., & Hinton, D. M. (2021). Overexpression of the Bacteriophage T4 motB Gene Alters H-NS Dependent Repression of Specific Host DNA. Viruses, 13(1), 84. https://doi.org/10.3390/v13010084






