Transcriptomic Analysis of Respiratory Tissue and Cell Line Models to Examine Glycosylation Machinery during SARS-CoV-2 Infection
Abstract
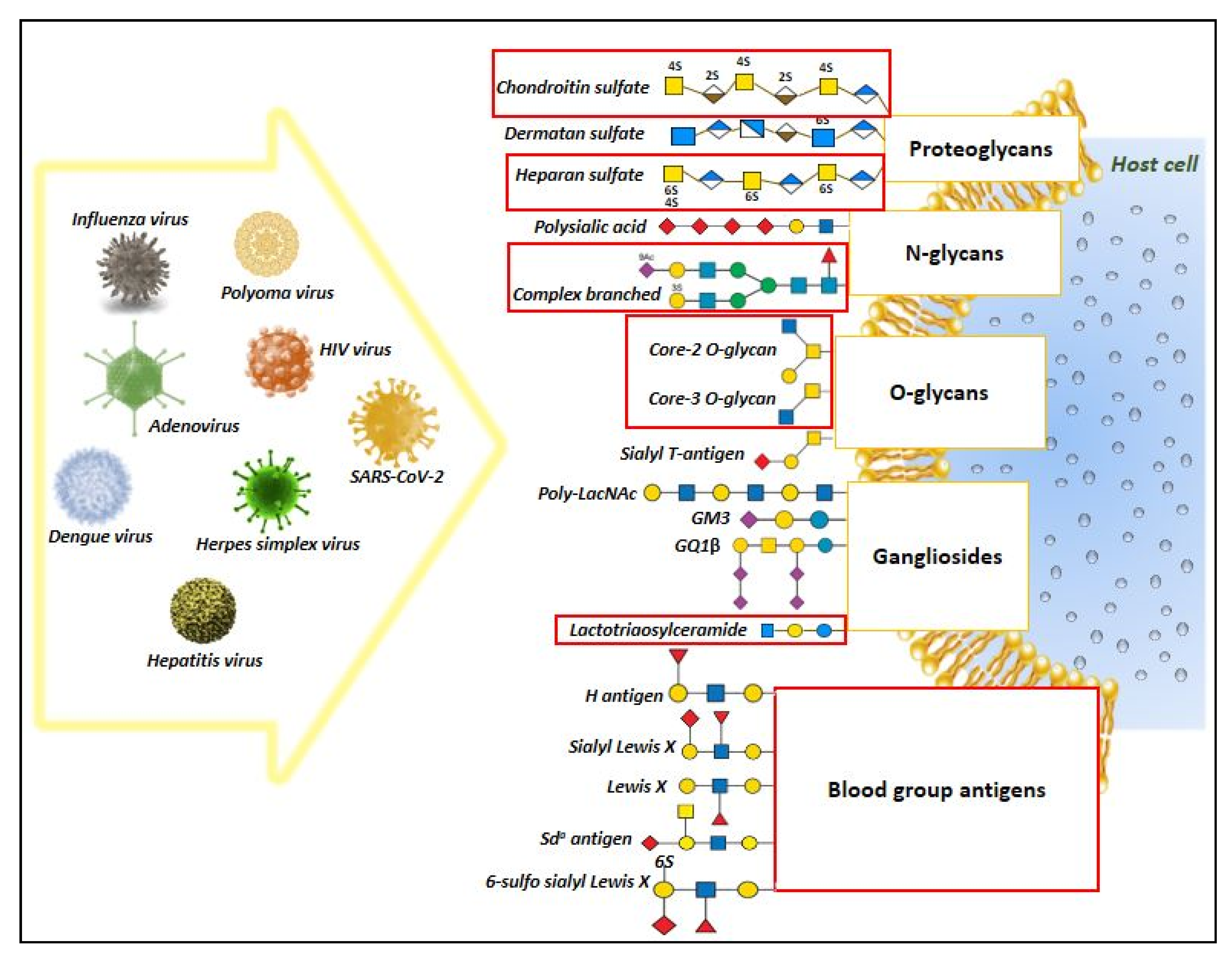
1. Introduction
2. Materials and Methods
2.1. Gene Expression Data Selection
2.2. Glycosylation Process Related Gene Set
2.3. Data Processing, Functional Enrichment Analysis and Network Visualization
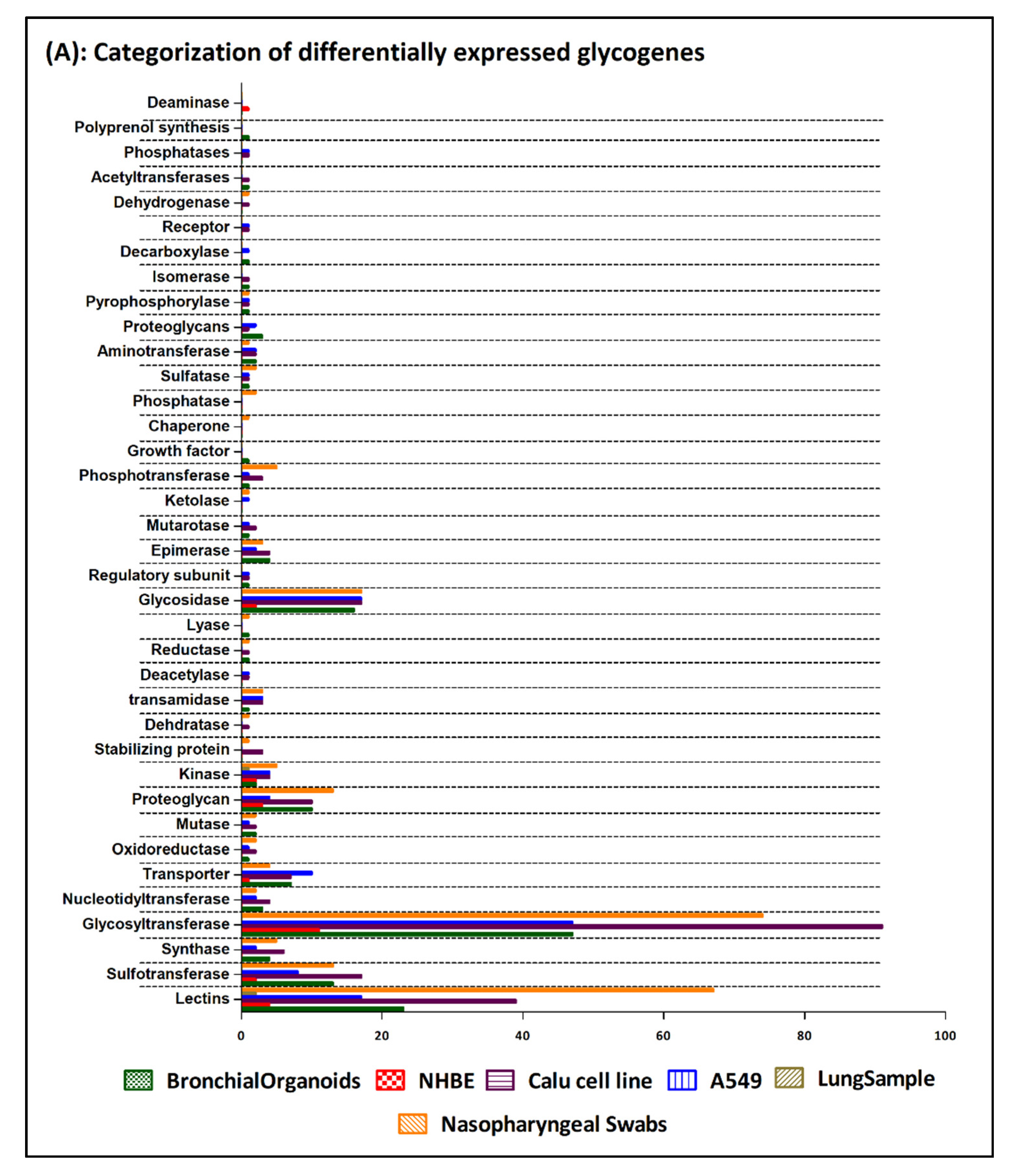
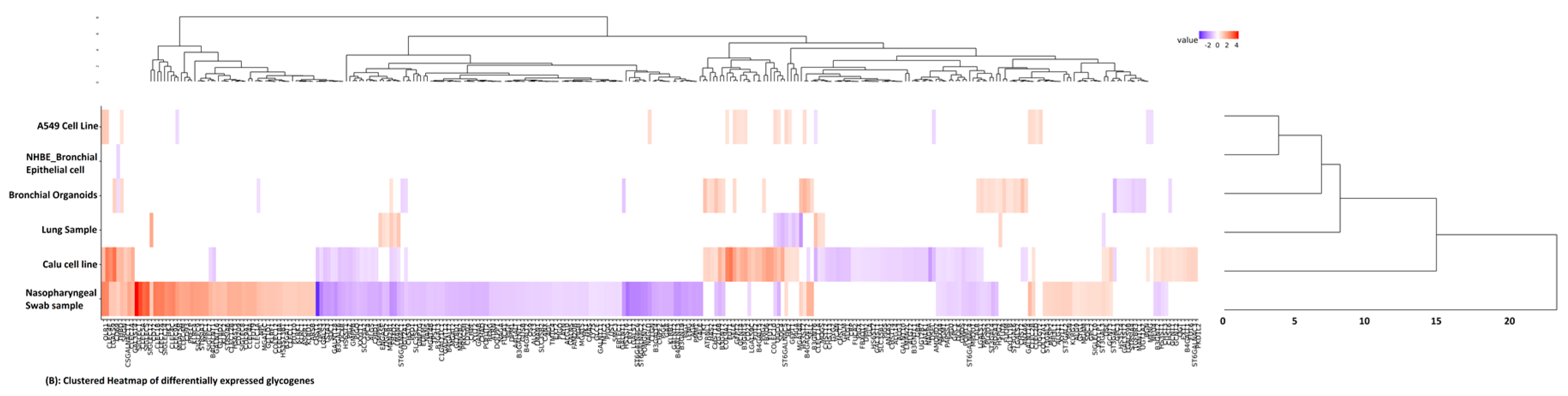
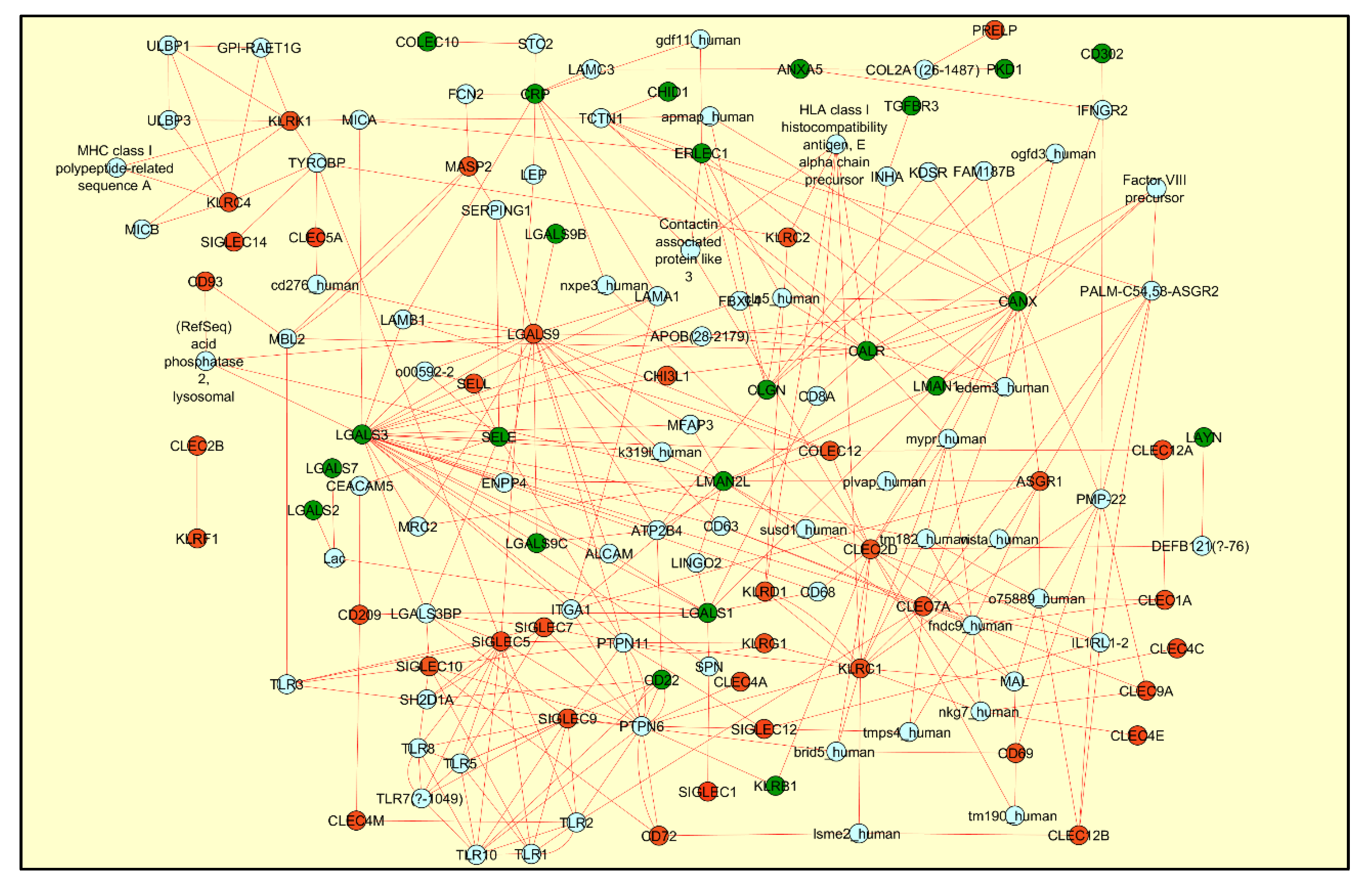
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Varki, A.; Freeze, H.H.; Manzi, A.E. Overview of Glycoconjugate Analysis. Curr. Protoc. Protein Sci. 2009, 57, 12.1.1–12.1.10. [Google Scholar] [CrossRef]
- Jennewein, M.F.; Alter, G. The Immunoregulatory Roles of Antibody Glycosylation. Trends Immunol. 2017, 38, 358–372. [Google Scholar] [CrossRef] [PubMed]
- Schnaar, R.L. Glycans and glycan-binding proteins in immune regulation: A concise introduction to glycobiology for the allergist. J. Allergy Clin. Immunol. 2015, 135, 609–615. [Google Scholar] [CrossRef]
- Shang, J.; Ye, G.; Shi, K.; Wan, Y.; Luo, C.; Aihara, H.; Geng, Q.; Auerbach, A.; Li, F. Structural basis of receptor recognition by SARS-CoV-2. Nat. Cell Biol. 2020, 581, 221–224. [Google Scholar] [CrossRef]
- Kai, H.; Kai, M. Interactions of coronaviruses with ACE2, angiotensin II, and RAS inhibitors—lessons from available evi-dence and insights into COVID-19. Hypertens. Res. 2020, 43, 648–654. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, Y.; Allen, J.D.; Wrapp, D.; McLellan, J.S.; Crispin, M. Site-specific glycan analysis of the SARS-CoV-2 spike. Sci-ence 2020, 369, 330–333. [Google Scholar] [CrossRef]
- Zhao, P.; Praissman, J.L.; Grant, O.C.; Cai, Y.; Xiao, T.; Rosenbalm, K.E.; Aoki, K.; Kellman, B.P.; Bridger, R.; Barouch, D.H.; et al. Virus-Receptor Interactions of Glycosylated SARS-CoV-2 Spike and Human ACE2 Receptor. Cell Host Microbe 2020, 28, 586–601.e6. [Google Scholar] [CrossRef] [PubMed]
- Shang, J.; Wan, Y.; Luo, C.; Ye, G.; Geng, Q.; Auerbach, A.; Li, F. Cell entry mechanisms of SARS-CoV-2. Proc. Natl. Acad. Sci. USA 2020, 117, 11727–11734. [Google Scholar] [CrossRef]
- Grant, O.C.; Montgomery, D.; Ito, K.; Woods, R.J. Analysis of the SARS-CoV-2 spike protein glycan shield reveals implica-tions for immune recognition. Sci. Rep. 2020, 10, 1–11. [Google Scholar] [CrossRef]
- Han, J.; Perez, J.T.; Chen, C.; Li, Y.; Benitez, A.; Kandasamy, M.; Lee, Y.; Andrade, J.; Tenoever, B.; Manicassamy, B. Ge-nome-wide CRISPR/Cas9 Screen Identifies Host Factors Essential for Influenza Virus Replication. Cell Rep. 2018, 23, 596–607. [Google Scholar] [CrossRef]
- Watanabe, Y.; Bowden, T.A.; Wilson, I.A.; Crispin, M. Exploitation of glycosylation in enveloped virus pathobiology. Biochim. Biophys. Acta Gen. Subj. 2019, 1863, 1480–1497. [Google Scholar] [CrossRef] [PubMed]
- Vigerust, D.J.; Shepherd, V.L. Virus glycosylation: Role in virulence and immune interactions. Trends Microbiol. 2007, 15, 211–218. [Google Scholar] [CrossRef] [PubMed]
- Bagdonaite, I.; Wandall, H.H. Global aspects of viral glycosylation. Glycobiology 2018, 28, 443–467. [Google Scholar] [CrossRef] [PubMed]
- Sadat, M.A.; Moir, S.; Chun, T.-W.; Lusso, P.; Kaplan, G.; Wolfe, L.; Memoli, M.J.; He, M.; Vega, H.; Kim, L.J.; et al. Glycosyla-tion, Hypogammaglobulinemia, and Resistance to Viral Infections. N. Engl. J. Med. 2014, 370, 1615–1625. [Google Scholar] [CrossRef]
- Irvine, E.B.; Alter, G. Understanding the role of antibody glycosylation through the lens of severe viral and bacterial dis-eases. Glycobiology 2020, 30, 241–253. [Google Scholar] [CrossRef]
- Wang, Q.; Fang, P.; He, R.; Li, M.; Yu, H.; Zhou, L.; Yi, Y.; Wang, F.; Rong, Y.; Zhang, Y.; et al. O-GlcNAc transferase pro-motes influenza A virus–induced cytokine storm by targeting interferon regulatory factor–5. Sci. Adv. 2020, 6, eaaz7086. [Google Scholar] [CrossRef]
- Heindel, D.W.; Koppolu, S.; Zhang, Y.; Kasper, B.; Meche, L.; Vaiana, C.A.; Bissel, S.J.; Carter, C.E.; Kelvin, A.A.; Elaish, M.; et al. Glycomic analysis of host response reveals high mannose as a key mediator of influenza severity. Proc. Natl. Acad. Sci. USA 2020, 117, 26926–26935. [Google Scholar] [CrossRef]
- Mahan, A.E.; Jennewein, M.F.; Suscovich, T.; Dionne, K.; Tedesco, J.; Chung, A.W.; Streeck, H.; Pau, M.; Schuitemaker, H.; Francis, D.; et al. Antigen-Specific Antibody Glycosylation Is Regulated via Vaccination. PLoS Pathog. 2016, 12, e1005456. [Google Scholar] [CrossRef]
- Li, Q.; Wu, J.; Nie, J.; Zhang, L.; Hao, H.; Liu, S.; Zhao, C.; Zhang, Q.; Liu, H.; Nie, L.; et al. The Impact of Mutations in SARS-CoV-2 Spike on Viral Infectivity and Antigenicity. Cell 2020, 182, 1284–1294.e9. [Google Scholar] [CrossRef]
- Dewald, J.H.; Colomb, F.; Bobowski-Gerard, M.; Groux-Degroote, S.; Delannoy, P. Role of Cytokine-Induced Glycosylation Changes in Regulating Cell Interactions and Cell Signaling in Inflammatory Diseases and Cancer. Cells 2016, 5, 43. [Google Scholar] [CrossRef]
- Reily, C.; Stewart, T.J.; Renfrow, M.B.; Novak, J. Glycosylation in health and disease. Nat. Rev. Nephrol. 2019, 15, 346–366. [Google Scholar] [CrossRef] [PubMed]
- Rhim, A.D.; Stoykova, L.; Glick, M.C.; Scanlin, T.F. Terminal glycosylation in cystic fibrosis (CF): A review emphasizing the airway epithelial cell. Glycoconj. J. 2001, 18, 649–659. [Google Scholar] [CrossRef] [PubMed]
- DeCoux, A.; Tian, Y.; DeLeon-Pennell, K.Y.; Nguyen, N.T.; Brás, L.E.D.C.; Flynn, E.R.; Cannon, P.L.; Griswold, M.E.; Jin, Y.-F.; Puskarich, M.A.; et al. Plasma Glycoproteomics Reveals Sepsis Outcomes Linked to Distinct Proteins in Common Pathways. Crit. Care Med. 2015, 43, 2049–2058. [Google Scholar] [CrossRef] [PubMed]
- Štambuk, T.; Dilber, D.; Kifer, D.; Selak, N.; Keser, T.; Ljubičić, Đ.; Dugac, A.V.; Lauc, G.; Rumora, L.; Gornik, O. N-glycosylation patterns of plasma proteins and immunoglobulin G in chronic obstructive pulmonary disease. J. Transl. Med. 2018, 16, 1–15. [Google Scholar] [CrossRef]
- Lyons, J.J.; Milner, J.D.; Rosenzweig, S.D. Glycans Instructing Immunity: The Emerging Role of Altered Glycosylation in Clinical Immunology. Front. Pediatr. 2015, 3, 54. [Google Scholar] [CrossRef]
- McCarthy, C.; Dunlea, D.M.; Saldova, R.; Henry, M.; Coleman, O.; McElvaney, O.J.; Marsh, B.; Rudd, P.M.; Reeves, E.P.; McElvaney, N.G. Glycosylation Repurposes Alpha-1 Antitrypsin for Resolution of Community-acquired Pneumonia. Am. J. Respir. Crit. Care Med. 2018, 197, 1346–1349. [Google Scholar] [CrossRef]
- Carter, C.L.; Parker, G.A.; Hankey, K.G.; Farese, A.M.; MacVittie, T.J.; Kane, M.A. MALDI-MSI spatially maps N-glycan al-terations to histologically distinct pulmonary pathologies following irradiation. Sci. Rep. 2020, 10, 1–14. [Google Scholar] [CrossRef]
- Mohanty, S.K.; Satapathy, A.; Naidu, M.M.; Mukhopadhyay, S.; Sharma, S.; Barton, L.M.; Stroberg, E.; Duval, E.J.; Pradhan, D.; Tzankov, A.; et al. Severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) and coronavirus disease 19 (COVID-19)—Anatomic pathology perspective on current knowledge. Diagn. Pathol. 2020, 15, 1–17. [Google Scholar] [CrossRef]
- Oommen, A.M.; Somaiya, N.; Vijayan, J.; Kumar, S.; Venkatachalam, S.; Joshi, L. GlycoGAIT: A web database to browse glycogenes and lectins under gastric inflammatory diseases. J. Theor. Biol. 2016, 406, 93–98. [Google Scholar] [CrossRef]
- Kuehn, H.; Liberzon, A.; Reich, M.; Mesirov, J.P. Using GenePattern for Gene Expression Analysis. Curr. Protoc. Bioinforma. 2008, 22, 7.12.1–7.12.39. [Google Scholar] [CrossRef]
- Blankenberg, D.; Hillman-Jackson, J. Analysis of Next-Generation Sequencing Data Using Galaxy. In Stem Cell Transcriptional Networks; Humana Press: New York, NY, USA, 2014; Volume 1150, pp. 21–43. [Google Scholar] [CrossRef]
- Raudvere, U.; Kolberg, L.; Kuzmin, I.; Arak, T.; Adler, P.; Peterson, H.; Vilo, J. g:Profiler: A web server for functional en-richment analysis and conversions of gene lists (2019 update). Nucleic Acids Res. 2019, 47, W191–W198. [Google Scholar] [CrossRef] [PubMed]
- UNIPORT. Retrieve/ID Mapping. Available online: https://www.uniprot.org/uploadlists/ (accessed on 7 August 2020).
- The UniProt Consortium UniProt: A worldwide hub of protein knowledge. Nucleic Acids Res. 2019, 47, D506–D515. [CrossRef] [PubMed]
- Jassal, B.; Matthews, L.; Viteri, G.; Gong, C.; Lorente, P.; Fabregat, A.; Sidiropoulos, K.; Cook, J.; Gillespie, M.; Haw, R.; et al. The reactome pathway knowledgebase. Nucleic Acids Res. 2020, 48, D498–D503. [Google Scholar] [CrossRef] [PubMed]
- Kamburov, A.; Wierling, C.; Lehrach, H.; Herwig, R. ConsensusPathDB—a database for integrating human functional in-teraction networks. Nucleic Acids Res. 2008, 37, D623–D628. [Google Scholar] [CrossRef]
- Su, G.; Morris, J.H.; Demchak, B.; Bader, G.D. Biological Network Exploration with Cytoscape 3. Curr. Protoc. Bioinform. 2014, 47, 8.13.1–8.13.24. [Google Scholar] [CrossRef]
- Rabinovich, G.A.; Croci, D.O. Regulatory Circuits Mediated by Lectin-Glycan Interactions in Autoimmunity and Cancer. Immunity 2012, 36, 322–335. [Google Scholar] [CrossRef]
- Rönnberg E, Pejler G. Serglycin: The master of the mast cell. Methods Mol Biol. 2012, 836, 201–217. [Google Scholar] [CrossRef]
- Theoharides, T.C. Potential Association of Mast Cells with COVID-19. Ann. Allergy Asthma Immunol. 2020, 5, S1081–S1206. [Google Scholar] [CrossRef]
- Clausen, T.M.; Sandoval, D.R.; Spliid, C.B.; Pihl, J.; Perrett, H.R.; Painter, C.D.; Narayanan, A.; Majowicz, S.A.; Kwong, E.M.; McVicar, R.N.; et al. SARS-CoV-2 Infection Depends on Cellular Heparan Sulfate and ACE2. Cell 2020, 183, 1043–1057.e15. [Google Scholar] [CrossRef]
- Mahmudpour, M.; Roozbeh, J.; Keshavarz, M.; Farrokhi, S.; Nabipour, I. COVID-19 cytokine storm: The anger of inflamma-tion. Cytokine 2020, 133, 155151. [Google Scholar] [CrossRef]
- Tandon, R.; Chew, G.M.; Byron, M.M.; Borrow, P.; Niki, T.; Hirashima, M.; Barbour, J.D.; Norris, P.J.; Lanteri, M.C.; Martin, J.N.; et al. Galectin-9 Is Rapidly Released During Acute HIV-1 Infection and Remains Sustained at High Levels Despite Viral Suppression Even in Elite Controllers. AIDS Res. Hum. Retrovir. 2014, 30, 654–664. [Google Scholar] [CrossRef] [PubMed]
- Amanat, F.; Krammer, F. SARS-CoV-2 Vaccines: Status Report. Immunity 2020, 52, 583–589. [Google Scholar] [CrossRef] [PubMed]
- Funk, C.D.; Laferrière, C.; Ardakani, A. A Snapshot of the Global Race for Vaccines Targeting SARS-CoV-2 and the COVID-19 Pandemic. Front. Pharmacol. 2020, 11, 937. [Google Scholar] [CrossRef] [PubMed]
- Kuri-Cervantes, L.; Pampena, M.B.; Meng, W.; Rosenfeld, A.M.; Ittner, C.A.; Weisman, A.R.; Agyekum, R.S.; Mathew, D.; Baxter, A.E.; Vella, L.A.; et al. Comprehensive mapping of immune perturbations associated with severe COVID-19. Sci. Immunol. 2020, 5, eabd7114. [Google Scholar] [CrossRef]
- Zhang, F.; Gan, R.; Zhen, Z.; Hu, X.; Li, X.; Zhou, F.; Liu, Y.; Chen, C.; Xie, S.; Zhang, B.; et al. Adaptive immune responses to SARS-CoV-2 infection in severe versus mild individuals. Signal Transduct. Target. Ther. 2020, 5, 1–11. [Google Scholar] [CrossRef]
- Giamarellos-Bourboulis, E.J.; Netea, M.G.; Rovina, N.; Akinosoglou, K.; Antoniadou, A.; Antonakos, N.; Damoraki, G.; Gkavogianni, T.; Adami, M.-E.; Katsaounou, P.; et al. Complex Immune Dysregulation in COVID-19 Patients with Severe Respiratory Failure. Cell Host Microbe 2020, 27, 992–1000.e3. [Google Scholar] [CrossRef] [PubMed]
- Long, Q.-X.; Liu, B.-Z.; Deng, H.-J.; Wu, G.-C.; Deng, K.; Chen, Y.-K.; Liao, P.; Qiu, J.-F.; Lin, Y.; Cai, X.-F.; et al. Antibody responses to SARS-CoV-2 in patients with COVID-19. Nat. Med. 2020, 26, 845–848. [Google Scholar] [CrossRef]
- Liu, J.; Li, S.; Liu, J.; Liang, B.; Wang, X.; Wang, H.; Li, W.; Tong, Q.; Yi, J.; Zhao, L.; et al. Longitudinal characteristics of lymphocyte responses and cytokine profiles in the peripheral blood of SARS-CoV-2 infected patients. EBioMedicine 2020, 55, 102763. [Google Scholar] [CrossRef]
- Merad, M.; Martin, J.C. Author Correction: Pathological inflammation in patients with COVID-19: A key role for monocytes and macrophages. Nat. Rev. Immunol. 2020, 20, 448. [Google Scholar] [CrossRef]
- Xu, Z.; Shi, L.; Wang, Y.; Zhang, J.; Huang, L.; Zhang, C.; Liu, S.; Zhao, P.; Liu, H.; Zhu, L.; et al. Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Respir. Med. 2020, 8, 420–422. [Google Scholar] [CrossRef]
- Chen, Z.; Wherry, E.J. T cell responses in patients with COVID-19. Nat. Rev. Immunol. 2020, 20, 529–536. [Google Scholar] [CrossRef] [PubMed]
- Imai, Y.; Kuba, K.; Rao, S.; Huan, Y.; Guo, F.; Guan, B.; Yang, P.; Sarao, R.; Wada, T.; Leong-Poi, H.; et al. Angioten-sin-converting enzyme 2 protects from severe acute lung failure. Nat. Cell Biol. 2005, 436, 112–116. [Google Scholar] [CrossRef]
- Zhang, X.; Yang, J.; Yu, X.; Cheng, S.; Gan, H.; Xia, Y. Angiotensin II-Induced Early and Late Inflammatory Responses through NOXs and MAPK Pathways. Inflammation 2017, 40, 154–165. [Google Scholar] [CrossRef] [PubMed]
- Tian, S.; Hu, W.; Niu, L.; Liu, H.; Xu, H.; Xiao, S. Pulmonary Pathology of Early-Phase 2019 Novel Coronavirus (COVID-19) Pneumonia in Two Patients with Lung Cancer. J. Thorac. Oncol. 2020, 15, 700–704. [Google Scholar] [CrossRef] [PubMed]
- Costela-Ruiz, V.J.; Illescas-Montes, R.; Puerta-Puerta, J.M.; Ruiz, C.; Melguizo-Rodríguez, L. SARS-CoV-2 infection: The role of cytokines in COVID-19 disease. Cytokine Growth Factor Rev. 2020, 54, 62–75. [Google Scholar] [CrossRef]
- Jose, R.J.; Manuel, A. COVID-19 cytokine storm: The interplay between inflammation and coagulation. Lancet Respir. Med. 2020, 8, e46–e47. [Google Scholar] [CrossRef]
- Goshua, G.; Pine, A.B.; Meizlish, M.L.; Chang, C.-H.; Zhang, H.; Bahel, P.; Baluha, A.; Bar, N.; Bona, R.D.; Burns, A.J.; et al. Endotheliopathy in COVID-19-associated coagulopathy: Evidence from a single-centre, cross-sectional study. Lancet Haematol. 2020, 7, e575–e582. [Google Scholar] [CrossRef]
- Groux-Degroote, S.; Cavdarli, S.; Uchimura, K.; Allain, F.; Delannoy, P. Glycosylation changes in inflammatory diseases. In Advances in Protein Chemistry and Structural Biology; Academic Press: Cambridge, MA, USA, 2020; Volume 119, pp. 111–156. [Google Scholar] [CrossRef]
- Awasthi, M.; Gulati, S.; Sarkar, D.P.; Tiwari, S.; Kateriya, S.; Ranjan, P.; Verma, S.K. The Sialoside-Binding Pocket of SARS-CoV-2 Spike Glycoprotein Structurally Resembles MERS-CoV. Viruses 2020, 12, 909. [Google Scholar] [CrossRef]
- Liu, L.; Chopra, P.; Li, X.; Wolfert, M.A.; Tompkins, S.M.; Boons, G.J. SARS-CoV-2 spike protein binds heparan sulfate in a length- and sequence-dependent manner. bioRxiv 2020. [Google Scholar] [CrossRef]
- Robson, B. Bioinformatics studies on a function of the SARS-CoV-2 spike glycoprotein as the binding of host sialic acid glycans. Comput. Biol. Med. 2020, 122, 103849. [Google Scholar] [CrossRef]
- Zhang, Q.; Chen, C.Z.; Swaroop, M.; Xu, M.; Wang, L.; Lee, J.; Wang, A.Q.; Pradhan, M.; Hagen, N.; Chen, L.; et al. Heparan sulfate assists SARS-CoV-2 in cell entry and can be targeted by approved drugs in vitro. Cell Discov. 2020, 6, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Breiman, A.; Ruvën-Clouet, N.; Le Pendu, J. Harnessing the natural anti-glycan immune response to limit the transmission of enveloped viruses such as SARS-CoV-2. PLOS Pathog. 2020, 16, e1008556. [Google Scholar] [CrossRef] [PubMed]
- Silva-Filho, J.C.; De Melo, C.G.F.; De Oliveira, J.L. The influence of ABO blood groups on COVID-19 susceptibility and se-verity: A molecular hypothesis based on carbohydrate-carbohydrate interactions. Med. Hypotheses 2020, 144, 110155. [Google Scholar] [CrossRef] [PubMed]
- Saku, A.; Hirose, K.; Ito, T.; Iwata, A.; Sato, T.; Kaji, H.; Tamachi, T.; Suto, A.; Goto, Y.; Domino, S.E.; et al. Fucosyltransferase 2 induces lung epithelial fucosylation and exacerbates house dust mite–induced airway inflammation. J. Allergy Clin. Immunol. 2019, 144, 698–709.e9. [Google Scholar] [CrossRef] [PubMed]
- Schottelius, A.J.; Hamann, A.; Asadullah, K. Role of fucosyltransferases in leukocyte trafficking: Major impact for cutaneous immunity. Trends Immunol. 2003, 24, 101–104. [Google Scholar] [CrossRef]
- Tinoco, R.; Carrette, F.; Henriquez, M.L.; Fujita, Y.; Bradley, L.M. Fucosyltransferase Induction during Influenza Virus Infec-tion Is Required for the Generation of Functional Memory CD4+ T Cells. J. Immunol. 2018, 200, 2690–2702. [Google Scholar] [CrossRef]
- Marth, J.D.; Grewal, P.K. Mammalian glycosylation in immunity. Nat. Rev. Immunol. 2008, 8, 874–887. [Google Scholar] [CrossRef]
- Zhuo, Y.; Bellis, S.L. Emerging Role of α2,6-Sialic Acid as a Negative Regulator of Galectin Binding and Function. J. Biol. Chem. 2010, 286, 5935–5941. [Google Scholar] [CrossRef]
- Walther, T.; Karamanska, R.; Chan, R.W.Y.; Chan, M.C.W.; Jia, N.; Air, G.; Hopton, C.; Wong, M.P.; Dell, A.; Peiris, J.S.M.; et al. Glycomic Analysis of Human Respiratory Tract Tissues and Correlation with Influenza Virus Infection. PLoS Pathog. 2013, 9, e1003223. [Google Scholar] [CrossRef]
- Jia, N.; Byrd-Leotis, L.; Matsumoto, Y.; Gao, C.; Wein, A.N.; Lobby, J.L.; Kohlmeier, J.E.; Steinhauer, D.A.; Cummings, R.D. The Human Lung Glycome Reveals Novel Glycan Ligands for Influenza A Virus. Sci. Rep. 2020, 10, 1–14. [Google Scholar] [CrossRef]
- Kirkeby, S.; Martel, C.; Aasted, B. Infection with human H1N1 influenza virus affects the expression of sialic acids of meta-plastic mucous cells in the ferret airways. Virus Res. 2009, 144, 225–232. [Google Scholar] [CrossRef] [PubMed]
- Oslund, K.L.; Baumgarth, N. Influenza-induced innate immunity: Regulators of viral replication, respiratory tract patholo-gy & adaptive immunity. Futur. Virol. 2011, 6, 951–962. [Google Scholar] [CrossRef]
- Davril, M.; DeGroote, S.; Humbert, P.; Galabert, C.; Dumur, V.; Lafitte, J.-J.; Lamblin, G.; Roussel, P. The sialylation of bronchial mucins secreted by patients suffering from cystic fibrosis or from chronic bronchitis is related to the severity of airway infection. Glycobiology 1999, 9, 311–321. [Google Scholar] [CrossRef] [PubMed]
- Dall’Olio, F.; Malagolini, N.; Chiricolo, M.; Trinchera, M.; Harduin-Lepers, A. The expanding roles of the Sda/Cad carbohy-drate antigen and its cognate glycosyltransferase B4GALNT2. Biochim. Biophys. Acta Gen. Subj. 2014, 1840, 443–453. [Google Scholar] [CrossRef] [PubMed]
- Middleton, E.A.; He, X.-Y.; Denorme, F.; Campbell, R.A.; Ng, D.; Salvatore, S.P.; Mostyka, M.; Baxter-Stoltzfus, A.; Borczuk, A.C.; Loda, M.; et al. Neutrophil extracellular traps contribute to immunothrombosis in COVID-19 acute respiratory dis-tress syndrome. Blood 2020, 136, 1169–1179. [Google Scholar] [CrossRef] [PubMed]
- Masselli, E.; Vaccarezza, M.; Carubbi, C.; Pozzi, G.; Presta, V.; Mirandola, P.; Vitale, M. NK cells: A double edge sword against SARS-CoV-2. Adv. Biol. Regul. 2020, 77, 100737. [Google Scholar] [CrossRef] [PubMed]
- Hiemstra, P.S.; Jr, P.B.M.; Bals, R. The innate immune function of airway epithelial cells in inflammatory lung disease. Eur. Respir. J. 2015, 45, 1150–1162. [Google Scholar] [CrossRef]
- Wortham, B.W.; Eppert, B.L.; Flury, J.L.; Garcia, S.M.; Donica, W.R.; Osterburg, A.; Joyce-Shaikh, B.; Cua, D.J.; Borchers, M.T. Cutting Edge: CLEC5A Mediates Macrophage Function and Chronic Obstructive Pulmonary Disease Pathologies. J. Immunol. 2016, 196, 3227–3231. [Google Scholar] [CrossRef]
- Chiffoleau, E. C-Type Lectin-Like Receptors as Emerging Orchestrators of Sterile Inflammation Represent Potential Thera-peutic Targets. Front. Immunol. 2018, 9, 227. [Google Scholar] [CrossRef]
- Pillai, S.; Netravali, I.A.; Cariappa, A.; Mattoo, H. Siglecs and Immune Regulation. Annu. Rev. Immunol. 2012, 30, 357–392. [Google Scholar] [CrossRef]
- Zheng, Q.; Hou, J.; Zhou, Y.; Yang, Y.; Xie, B.; Cao, X. Siglec1 suppresses antiviral innate immune response by inducing TBK1 degradation via the ubiquitin ligase TRIM27. Cell Res. 2015, 25, 1121–1136. [Google Scholar] [CrossRef] [PubMed]
- Vabret, N.; Britton, G.J.; Gruber, C.; Hegde, S.; Kim, J.; Kuksin, M.; Levantovsky, R.; Malle, L.; Moreira, A.; Park, M.D.; et al. Immunology of COVID-19: Current State of the Science. Immunity 2020, 52, 910–941. [Google Scholar] [CrossRef] [PubMed]
- Macauley, M.S.; Crocker, P.R.; Paulson, J.C. Siglec-mediated regulation of immune cell function in disease. Nat. Rev. Immu-nol. 2014, 14, 653–666. [Google Scholar] [CrossRef] [PubMed]
- Eyan, H.; Ekamiya, T.; Esuabjakyong, P.; Tsuji, N.M. Targeting C-Type Lectin Receptors for Cancer Immunity. Front. Immu-nol. 2015, 6, 408. [Google Scholar] [CrossRef] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Oommen, A.; Cunningham, S.; Joshi, L. Transcriptomic Analysis of Respiratory Tissue and Cell Line Models to Examine Glycosylation Machinery during SARS-CoV-2 Infection. Viruses 2021, 13, 82. https://doi.org/10.3390/v13010082
Oommen A, Cunningham S, Joshi L. Transcriptomic Analysis of Respiratory Tissue and Cell Line Models to Examine Glycosylation Machinery during SARS-CoV-2 Infection. Viruses. 2021; 13(1):82. https://doi.org/10.3390/v13010082
Chicago/Turabian StyleOommen, Anup, Stephen Cunningham, and Lokesh Joshi. 2021. "Transcriptomic Analysis of Respiratory Tissue and Cell Line Models to Examine Glycosylation Machinery during SARS-CoV-2 Infection" Viruses 13, no. 1: 82. https://doi.org/10.3390/v13010082
APA StyleOommen, A., Cunningham, S., & Joshi, L. (2021). Transcriptomic Analysis of Respiratory Tissue and Cell Line Models to Examine Glycosylation Machinery during SARS-CoV-2 Infection. Viruses, 13(1), 82. https://doi.org/10.3390/v13010082







