Oral Infection by Mucosal and Cutaneous Human Papillomaviruses in the Men Who Have Sex with Men from the OHMAR Study
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Population
2.2. Oral Sample Collection
2.3. Alpha, Beta, and Gamma HPV Detection and Genotyping
2.4. Next-Generation Sequencing for PV Characterization
2.5. Bioinformatic Analysis of NGS Data
2.6. Statistical Analysis
3. Results
3.1. Study Population
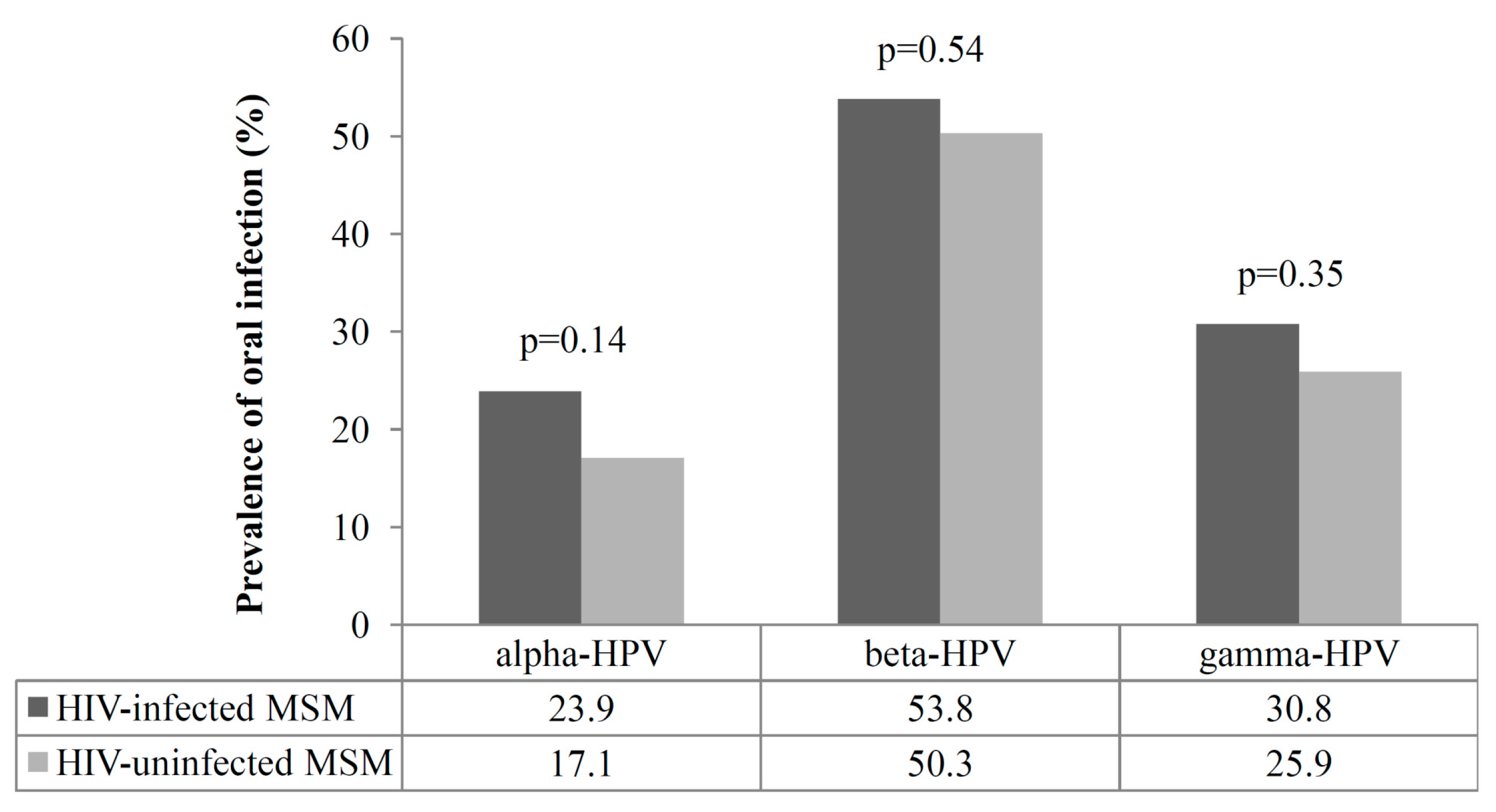
3.2. Prevalence of Oral Infection by Alpha, Beta, and Gamma HPVs
3.3. NGS Data and PV Sequence Analyses
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Appendix A
References
- Mlakar, B.; Kocjan, B.J.; Hošnjak, L.; Fujs Komloš, K.; Milošević, M.; Poljak, M. Betapapillomaviruses in the Anal Canal of HIV Positive and HIV Negative Men Who have Sex with Men. J. Clin. Virol. 2014, 61, 237–241. [Google Scholar] [CrossRef] [PubMed]
- Torres, M.; Gheit, T.; McKay-Chopin, S.; Rodriguez, C.; Romero, J.D.; Filotico, R.; Donà, M.G.; Ortiz, M.; Tommasino, M. Prevalence of Beta and Gamma Human Papillomaviruses in the Anal Canal of Men Who have Sex with Men is Influenced by HIV Status. J. Clin. Virol. 2015, 67, 47–51. [Google Scholar] [CrossRef] [PubMed]
- Donà, M.G.; Gheit, T.; Latini, A.; Benevolo, M.; Torres, M.; Smelov, V.; McKay-Chopin, S.; Giglio, A.; Cristaudo, A.; Zaccarelli, M.; et al. Alpha, Beta and Gamma Human Papillomaviruses in the Anal Canal of HIV-Infected and Uninfected Men Who have Sex with Men. J. Infect. 2015, 71, 74–84. [Google Scholar] [CrossRef] [PubMed]
- Forslund, O.; Johansson, H.; Madsen, K.G.; Kofoed, K. The Nasal Mucosa Contains a Large Spectrum of Human Papillomavirus Types from the Betapapillomavirus and Gammapapillomavirus Genera. J. Infect. Dis. 2013, 208, 1335–1341. [Google Scholar] [CrossRef] [PubMed]
- Wong, M.C.S.; Vlantis, A.C.; Liang, M.; Wong, P.Y.; Ho, W.C.S.; Boon, S.S.; Sze, R.K.H.; Leung, C.; Chan, P.K.S.; Chen, Z. Prevalence and Epidemiologic Profile of Oral Infection with Alpha, Beta, and Gamma Papillomaviruses in an Asian Chinese Population. J. Infect. Dis. 2018, 218, 388–397. [Google Scholar] [CrossRef]
- Nunes, E.M.; Sudenga, S.L.; Gheit, T.; Tommasino, M.; Baggio, M.L.; Ferreira, S.; Galan, L.; Silva, R.C.; Pierce Campbell, C.M.; Lazcano-Ponce, E.; et al. Diversity of Beta-Papillomavirus at Anogenital and Oral Anatomic Sites of Men: The HIM Study. Virology 2016, 495, 33–41. [Google Scholar] [CrossRef] [PubMed]
- Winer, R.L.; Gheit, T.; Feng, Q.; Stern, J.E.; Lin, J.; Cherne, S.; Tommasino, M. Prevalence and Correlates of Beta- and Gamma-Human Papillomavirus Detection in Oral Samples from Mid-Adult Women. J. Infect. Dis. 2019, 219, 1067–1075. [Google Scholar] [CrossRef]
- Nunes, E.M.; Lopez, R.V.M.; Sudenga, S.L.; Gheit, T.; Tommasino, M.; Baggio, M.L.; Ferreira, S.; Galan, L.; Silva, R.C.; Lazcano-Ponce, E.; et al. Concordance of Beta-Papillomavirus Across Anogenital and Oral Anatomic Sites of Men: The HIM Study. Virology 2017, 510, 55–59. [Google Scholar] [CrossRef]
- Bolatti, E.M.; Hosnjak, L.; Chouhy, D.; Casal, P.E.; Re-Louhau, M.F.; Bottai, H.; Komlos, K.F.; Poljak, M.; Giri, A.A. Assessing Gammapapillomavirus Infections of Mucosal Epithelia with Two Broad-Spectrum PCR Protocols. BMC Infect. Dis. 2020, 20, 274. [Google Scholar] [CrossRef]
- Zhang, C.; Liu, F.; Pan, Y.; Deng, Q.; Li, X.; He, Z.; Liu, M.; Ning, T.; Guo, C.; Liang, Y.; et al. Incidence and Clearance of Oral Human Papillomavirus Infection: A Population-Based Cohort Study in Rural China. Oncotarget 2017, 8, 59831–59844. [Google Scholar] [CrossRef][Green Version]
- Donà, M.G.; Pichi, B.; Rollo, F.; Gheit, T.; Laquintana, V.; Covello, R.; Pescarmona, E.; Spriano, G.; Pellini, R.; Giuliani, M.; et al. Mucosal and Cutaneous Human Papillomaviruses in Head and Neck Squamous Cell Papillomas. Head Neck 2017, 39, 254–259. [Google Scholar] [CrossRef] [PubMed]
- Kerge, S.; Vuorinen, J.; Hurme, S.; Soukka, T.; Gheit, T.; Tommasino, M.; Syrjanen, S.; Rautava, J. Benign Proliferative Epithelial Lesions of Oral Mucosa are Infrequently Associated with Alpha-, Beta-, Or Gamma Human Papillomaviruses. Laryngoscope Investig. Otolaryngol. 2018, 4, 43–48. [Google Scholar] [CrossRef] [PubMed]
- Agalliu, I.; Gapstur, S.; Chen, Z.; Wang, T.; Anderson, R.L.; Teras, L.; Kreimer, A.R.; Hayes, R.B.; Freedman, N.D.; Burk, R.D. Associations of Oral Alpha-, Beta-, and Gamma-Human Papillomavirus Types with Risk of Incident Head and Neck Cancer. JAMA Oncol. 2016, 2, 599–606. [Google Scholar] [CrossRef] [PubMed]
- Bouvard, V.; Baan, R.; Straif, K.; Grosse, Y.; Secretan, B.; El Ghissassi, F.; Benbrahim-Tallaa, L.; Guha, N.; Freeman, C.; Galichet, L.; et al. A Review of Human Carcinogens—Part B: Biological Agents. IARC Press 2009, 10, 321–322. [Google Scholar] [CrossRef]
- D’Souza, G.; Wentz, A.; Kluz, N.; Zhang, Y.; Sugar, E.; Youngfellow, R.M.; Guo, Y.; Xiao, W.; Gillison, M.L. Sex Differences in Risk Factors and Natural History of Oral Human Papillomavirus Infection. J. Infect. Dis. 2016, 213, 1893–1896. [Google Scholar] [CrossRef] [PubMed]
- Colon-Lopez, V.; Quinones-Avila, V.; Del Toro-Mejias, L.M.; Reyes, K.; Rivera, M.E.; Nieves, K.; Sanchez-Vazquez, M.M.; Martinez-Ferrer, M.; Ortiz, A.P. Oral HPV Infection in a Clinic-Based Sample of Hispanic Men. BMC Oral Health. 2014, 14, 7. [Google Scholar] [CrossRef] [PubMed]
- Read, T.R.; Hocking, J.S.; Vodstrcil, L.A.; Tabrizi, S.N.; McCullough, M.J.; Grulich, A.E.; Garland, S.M.; Bradshaw, C.S.; Chen, M.Y.; Fairley, C.K. Oral Human Papillomavirus in Men having Sex with Men: Risk-Factors and Sampling. PLoS ONE 2012, 7, e49324. [Google Scholar] [CrossRef]
- Chin-Hong, P.V.; Vittinghoff, E.; Cranston, R.D.; Buchbinder, S.; Cohen, D.; Colfax, G.; Da Costa, M.; Darragh, T.; Hess, E.; Judson, F.; et al. Age-Specific Prevalence of Anal Human Papillomavirus Infection in HIV-Negative Sexually Active Men Who have Sex with Men: The EXPLORE Study. J. Infect. Dis. 2004, 190, 2070–2076. [Google Scholar] [CrossRef]
- Goldstone, S.; Palefsky, J.M.; Giuliano, A.R.; Moreira, E.D., Jr.; Aranda, C.; Jessen, H.; Hillman, R.J.; Ferris, D.G.; Coutlee, F.; Liaw, K.L.; et al. Prevalence of and Risk Factors for Human Papillomavirus (HPV) Infection among HIV-Seronegative Men Who have Sex with Men. J. Infect. Dis. 2011, 203, 66–74. [Google Scholar] [CrossRef]
- Machalek, D.A.; Poynten, M.; Jin, F.; Fairley, C.K.; Farnsworth, A.; Garland, S.M.; Hillman, R.J.; Petoumenos, K.; Roberts, J.; Tabrizi, S.N.; et al. Anal Human Papillomavirus Infection and Associated Neoplastic Lesions in Men Who have Sex with Men: A Systematic Review and Meta-Analysis. Lancet. Oncol. 2012, 13, 487–500. [Google Scholar] [CrossRef]
- Rollo, F.; Latini, A.; Pichi, B.; Colafigli, M.; Benevolo, M.; Sinopoli, I.; Sperduti, I.; Laquintana, V.; Fabbri, G.; Frasca, M.; et al. Prevalence and Determinants of Oral Infection by Human Papillomavirus in HIV-Infected and Uninfected Men Who have Sex with Men. PLoS ONE 2017, 12, e0184623. [Google Scholar] [CrossRef] [PubMed]
- Giuliani, M.; Rollo, F.; Vescio, M.F.; Pichi, B.; Latini, A.; Benevolo, M.; Pellini, R.; Cristaudo, A.; Dona’, M.G. Oral Human Papillomavirus Infection in HIV-Infected and HIV-Uninfected MSM: The OHMAR Prospective Cohort Study. Sex. Transm. Infect. 2020. published online ahead of print. [Google Scholar] [CrossRef] [PubMed]
- Iannacone, M.R.; Gheit, T.; Waterboer, T.; Giuliano, A.R.; Messina, J.L.; Fenske, N.A.; Cherpelis, B.S.; Sondak, V.K.; Roetzheim, R.G.; Ferrer-Gil, S.; et al. Case-Control Study of Cutaneous Human Papillomavirus Infection in Basal Cell Carcinoma of the Skin. J. Invest. Dermatol. 2013, 133, 1512–1520. [Google Scholar] [CrossRef] [PubMed]
- Forslund, O.; Antonsson, A.; Nordin, P.; Stenquist, B.; Hansson, B.G. A Broad Range of Human Papillomavirus Types Detected with a General PCR Method Suitable for Analysis of Cutaneous Tumours and Normal Skin. J. Gen. Virol. 1999, 80, 2437–2443. [Google Scholar] [CrossRef] [PubMed]
- Chouhy, D.; Gorosito, M.; Sanchez, A.; Serra, E.C.; Bergero, A.; Fernandez Bussy, R.; Giri, A.A. New Generic Primer System Targeting Mucosal/Genital and Cutaneous Human Papillomaviruses Leads to the Characterization of HPV 115, a Novel Beta-Papillomavirus Species 3. Virology 2010, 397, 205–216. [Google Scholar] [CrossRef]
- Brancaccio, R.N.; Robitaille, A.; Dutta, S.; Cuenin, C.; Santare, D.; Skenders, G.; Leja, M.; Fischer, N.; Giuliano, A.R.; Rollison, D.E.; et al. Generation of a Novel Next-Generation Sequencing-Based Method for the Isolation of New Human Papillomavirus Types. Virology 2018, 520, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Galati, L.; Brancaccio, R.N.; Robitaille, A.; Cuenin, C.; Luzi, F.; Fiorucci, G.; Chiantore, M.V.; Marascio, N.; Matera, G.; Liberto, M.C.; et al. Detection of Human Papillomaviruses in Paired Healthy Skin and Actinic Keratosis by Next Generation Sequencing. Papillomavirus Res. 2020, 9, 100196. [Google Scholar] [CrossRef]
- Berger, S.A.; Stamatakis, A. Aligning Short Reads to Reference Alignments and Trees. Bioinformatics 2011, 27, 2068–2075. [Google Scholar] [CrossRef]
- Stamatakis, A. RAxML Version 8: A Tool for Phylogenetic Analysis and Post-Analysis of Large Phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef]
- Van Doorslaer, K.; Li, Z.; Xirasagar, S.; Maes, P.; Kaminsky, D.; Liou, D.; Sun, Q.; Kaur, R.; Huyen, Y.; McBride, A.A. The Papillomavirus Episteme: A Major Update to the Papillomavirus Sequence Database. Nucleic Acids Res. 2017, 45, D499–D506. [Google Scholar] [CrossRef]
- International HPV Reference Center. Available online: https://ki.se/en/labmed/international-hpv-reference-center (accessed on 1 April 2020).
- Rollo, F.; Pichi, B.; Benevolo, M.; Giuliani, M.; Latini, A.; Lorenzon, L.; Colafigli, M.; Frasca, M.; Pellini, R.; Cristaudo, A.; et al. Oral Testing for High-Risk Human Papillomavirus DNA and E6/E7 Messenger RNA in Healthy Individuals at Risk for Oral Infection. Cancer 2019, 125, 2587–2593. [Google Scholar] [CrossRef]
- Bottalico, D.; Chen, Z.; Dunne, A.; Ostoloza, J.; McKinney, S.; Sun, C.; Schlecht, N.F.; Fatahzadeh, M.; Herrero, R.; Schiffman, M.; et al. The Oral Cavity Contains Abundant Known and Novel Human Papillomaviruses from the Betapapillomavirus and Gammapapillomavirus Genera. J. Infect. Dis. 2011, 204, 787–792. [Google Scholar] [CrossRef] [PubMed]
- Hampras, S.S.; Rollison, D.E.; Giuliano, A.R.; McKay-Chopin, S.; Minoni, L.; Sereday, K.; Gheit, T.; Tommasino, M. Prevalence and Concordance of Cutaneous Beta Human Papillomavirus Infection at Mucosal and Cutaneous Sites. J. Infect. Dis. 2017, 216, 92–96. [Google Scholar] [CrossRef] [PubMed]
- Kreimer, A.R.; Alberg, A.J.; Daniel, R.; Gravitt, P.E.; Viscidi, R.; Garrett, E.S.; Shah, K.V.; Gillison, M.L. Oral Human Papillomavirus Infection in Adults is Associated with Sexual Behavior and HIV Serostatus. J. Infect. Dis. 2004, 189, 686–698. [Google Scholar] [CrossRef] [PubMed]
- Sias, C.; Salichos, L.; Lapa, D.; Del Nonno, F.; Baiocchini, A.; Capobianchi, M.R.; Garbuglia, A.R. Alpha, Beta, Gamma Human PapillomaViruses (HPV) Detection with a Different Sets of Primers in Oropharyngeal Swabs, Anal and Cervical Samples. Virol. J. 2019, 16, 27. [Google Scholar] [CrossRef] [PubMed]
- Smelov, V.; Muwonge, R.; Sokolova, O.; McKay-Chopin, S.; Eklund, C.; Komyakov, B.; Gheit, T. Beta and Gamma Human Papillomaviruses in Anal and Genital Sites among Men: Prevalence and Determinants. Sci. Rep. 2018, 8, 8241. [Google Scholar] [CrossRef]
- Gheit, T. Mucosal and Cutaneous Human Papillomavirus Infections and Cancer Biology. Front. Oncol. 2019, 9, 355. [Google Scholar] [CrossRef]
- O’Brien, P.M.; Campo, M.S. Papillomaviruses: A Correlation between Immune Evasion and Oncogenicity? Trends Microbiol. 2003, 11, 300–305. [Google Scholar] [CrossRef]
- Cornet, I.; Bouvard, V.; Campo, M.S.; Thomas, M.; Banks, L.; Gissmann, L.; Lamartine, J.; Sylla, B.S.; Accardi, R.; Tommasino, M. Comparative Analysis of Transforming Properties of E6 and E7 from Different Beta Human Papillomavirus Types. J. Virol. 2012, 86, 2366–2370. [Google Scholar] [CrossRef]
- Viarisio, D.; Muller-Decker, K.; Zanna, P.; Kloz, U.; Aengeneyndt, B.; Accardi, R.; Flechtenmacher, C.; Gissmann, L.; Tommasino, M. Novel Ss-HPV49 Transgenic Mouse Model of Upper Digestive Tract Cancer. Cancer Res. 2016, 76, 4216–4225. [Google Scholar] [CrossRef]
| NGS Pool | PCR Protocol | Specimen HIV Status | Total Number * |
|---|---|---|---|
| 1 | CUT | HIV− | 29 |
| 2 | CUT | HIV+ | 30 |
| 3 | FAP | HIV− | 30 |
| 4 | FAP | HIV+ | 28 |
| 5 | FAPM1 | HIV− | 18 |
| 6 | FAPM1 | HIV+ | 14 |
| Alpha Species/Type | HIV-Infected n = 117 n (%) | HIV-Uninfected n = 193 n (%) | |||||
|---|---|---|---|---|---|---|---|
| Single Infection | Multiple Infection | Total | Single Infection | Multiple Infection | Total | p Value a | |
| Any alpha-3 | 6 (5.1) | 6 (5.1) | 12 (10.2) | 6 (3.1) | 4 (2.1) | 10 (5.2) | |
| 61 | 1 (0.9) | 1 (0.9) | 2 (1.7) | 0 (0.0) | 0 (0.0) | 0 (0.0) | |
| 62 | 2 (1.7) | 0 (0.0) | 2 (1.7) | 1 (0.5) | 0 (0.0) | 1 (0.5) | |
| 72 | 2 (1.7) | 2 (1.7) | 4 (3.4) | 0 (0.0) | 1 (0.5) | 1 (0.5) | 0.049 |
| 81 | 0 (0.0) | 1 (0.9) | 1 (0.9) | 0 (0.0) | 0 (0.0) | 0 (0.0) | |
| 83 | 0 (0.0) | 0 (0.0) | 0 (0.0) | 1 (0.5) | 1 (0.5) | 2 (1.0) | |
| 84 | 1 (0.9) | 3 (2.6) | 4 (3.4) | 0 (0.0) | 2 (1.0) | 2 (1.0) | |
| 89 (CP6108) | 0 (0.0) | 3 (2.6) | 3 (2.6) | 4 (2.1) | 1 (0.5) | 5 (2.6) | |
| Any alpha-5 | 2 (1.7) | 2 (1.7) | 4 (3.4) | 1 (0.5) | 1 (0.5) | 2 (1.0) | |
| 26 | 0 (0.0) | 0 (0.0) | 0 (0.0) | 1 (0.5) | 0 (0.0) | 1 (0.5) | |
| 51 | 0 (0.0) | 2 (1.7) | 2 (1.7) | 0 (0.0) | 0 (0.0) | 0 (0.0) | |
| 69 | 1 (0.9) | 0 (0.0) | 1 (0.9) | 0 (0.0) | 0 (0.0) | 0 (0.0) | |
| 82 | 1 (0.9) | 1 (0.9) | 2 (1.7) | 0 (0.0) | 1 (0.5) | 1 (0.5) | |
| Any alpha-6 | 0 (0.0) | 3 (2.6) | 3 (2.6) | 6 (3.1) | 4 (2.1) | 10 (5.2) | |
| 53 | 0 (0.0) | 1 (0.9) | 1 (0.9) | 1 (0.5) | 2 (1.0) | 3 (1.5) | |
| 56 | 0 (0.0) | 0 (0.0) | 0 (0.0) | 2 (1.0) | 2 (1.0) | 4 (2.1) | |
| 66 | 0 (0.0) | 2 (1.7) | 2 (1.7) | 3 (1.5) | 0 (0.0) | 3 (1.5) | |
| Any alpha-7 | 5 (4.3) | 4 (3.4) | 9 (7.7) | 2 (1.0) | 6 (3.1) | 8 (4.1) | |
| 18 | 1 (0.9) | 2 (1.7) | 3 (2.6) | 1 (0.5) | 0 (0.0) | 1 (0.5) | |
| 39 | 1 (0.9) | 1 (0.9) | 2 (1.7) | 0 (0.0) | 0 (0.0) | 0 (0.0) | |
| 45 | 0 (0.0) | 1 (0.9) | 1 (0.9) | 0 (0.0) | 3 (1.5) | 3 (1.5) | |
| 59 | 0 (0.0) | 2 (1.7) | 2 (1.7) | 1 (0.5) | 0 (0.0) | 1 (0.5) | |
| 68 | 2 (1.7) | 0 (0.0) | 2 (1.7) | 0 (0.0) | 2 (1.0) | 2 (1.0) | |
| 70 | 1 (0.9) | 0 (0.0) | 1 (0.9) | 0 (0.0) | 1 (0.5) | 1 (0.5) | |
| Any alpha-9 | 0 (0.0) | 3 (2.6) | 3 (2.6) | 6 (3.1) | 6 (3.1) | 12 (6.2) | |
| 16 | 0 (0.0) | 3 (2.6) | 3 (2.6) | 2 (1.0) | 6 (3.1) | 8 (4.1) | |
| 33 | 0 (0.0) | 1 (0.9) | 1 (0.9) | 2 (1.0) | 0 (0.0) | 2 (1.0) | |
| 35 | 0 (0.0) | 0 (0.0) | 0 (0.0) | 1 (0.5) | 0 (0.0) | 1 (0.5) | |
| 58 | 0 (0.0) | 0 (0.0) | 0 (0.0) | 1 (0.5) | 0 (0.0) | 1 (0.5) | |
| Any alpha-10 | 0 (0.0) | 3 (2.6) | 3 (2.6) | 2 (1.0) | 0 (0.0) | 2 (1.0) | |
| 6 | 0 (0.0) | 1 (0.9) | 1 (0.9) | 1 (0.5) | 0 (0.0) | 1 (0.5) | |
| 11 | 0 (0.0) | 2 (1.7) | 2 (1.7) | 1 (0.5) | 0 (0.0) | 1 (0.5) | |
| Any alpha-11 | 0 (0.0) | 1 (0.9) | 1 (0.9) | 1 (0.5) | 0 (0.0) | 1 (0.5) | |
| 73 | 0 (0.0) | 1 (0.9) | 1 (0.9) | 1 (0.5) | 0 (0.0) | 1 (0.5) | |
| Any alpha-13 | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 1 (0.5) | 1 (0.5) | |
| 54 | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 1 (0.5) | 1 (0.5) | |
| Any alpha-14 | 0 (0.0) | 1 (0.9) | 1 (0.9) | 0 (0.0) | 0 (0.0) | 0 (0.0) | |
| 71 | 0 (0.0) | 1 (0.9) | 1 (0.9) | 0 (0.0) | 0 (0.0) | 0 (0.0) | |
| Other | 5 (4.3) | 3 (2.6) | 8 (6.8) | 0 (0.0) | 2 (1.0) | 2 (1.0) | |
| 55 (44 subtype) | 3 (2.6) | 1 (0.9) | 4 (3.4) | 0 (0.0) | 2 (1.0) | 2 (1.0) | |
| IS39 (82 subtype) | 2 (1.7) | 2 (1.7) | 4 (3.4) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0.01 |
| Beta Species/Type | HIV-Infected n = 117 n (%) | HIV-Uninfected n = 193 n (%) | |||||
|---|---|---|---|---|---|---|---|
| Single Infection | Multiple Infection | Total | Single Infection | Multiple Infection | Total | p Value a | |
| Any beta-1 | 17 (14.5) | 29 (24.8) | 46 (39.3) | 21 (10.9) | 45 (23.3) | 66 (34.2) | |
| 5 | 5 (4.3) | 8 (6.8) | 13 (11.1) | 5 (2.6) | 19 (9.8) | 24 (12.4) | |
| 8 | 1 (0.9) | 1 (0.9) | 2 (1.7) | 4 (2.1) | 12 (6.2) | 16 (8.3) | 0.01 |
| 12 | 3 (2.6) | 4 (3.4) | 7 (6.0) | 1 (0.5) | 11 (5.7) | 12 (6.2) | |
| 14 | 2 (1.7) | 1 (0.9) | 3 (2.6) | 0 (0.0) | 3 (1.5) | 3 (1.5) | |
| 19 | 0 (0.0) | 4 (3.4) | 4 (3.4) | 2 (1.0) | 11 (5.7) | 13 (6.7) | |
| 20 | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 1 (0.5) | 1 (0.5) | |
| 21 | 0 (0.0) | 3 (2.6) | 3 (2.6) | 2 (1.0) | 14 (7.3) | 16 (8.3) | 0.04 |
| 24 | 0 (0.0) | 6 (5.1) | 6 (5.1) | 2 (1.0) | 10 (5.2) | 12 (6.2) | |
| 25 | 1 (0.9) | 3 (2.6) | 4 (3.4) | 0 (0.0) | 2 (1.0) | 2 (1.0) | |
| 36 | 1 (0.9) | 1 (0.9) | 2 (1.7) | 2 (1.0) | 3 (1.5) | 5 (2.6) | |
| 47 | 0 (0.0) | 9 (7.7) | 9 (7.7) | 1 (0.5) | 9 (4.7) | 10 (5.2) | |
| 98 | 0 (0.0) | 1 (0.9) | 1 (0.9) | 0 (0.0) | 6 (3.1) | 6 (3.1) | |
| 99 | 0 (0.0) | 3 (2.6) | 3 (2.6) | 0 (0.0) | 4 (2.1) | 4 (2.1) | |
| 105 | 2 (1.7) | 7 (6.0) | 9 (7.7) | 2 (1.0) | 12 (6.2) | 14 (7.3) | |
| 124 | 2 (1.7) | 5 (4.3) | 7 (6.0) | 0 (0.0) | 6 (3.1) | 6 (3.1) | |
| 143 | 0 (0.0) | 1 (0.9) | 1 (0.9) | 0 (0.0) | 1 (0.5) | 1 (0.5) | |
| Any beta-2 | 11 (9.4) | 27 (23.1) | 38 (32.5) | 24 (12.4) | 44 (22.8) | 68 (35.2) | |
| 9 | 3 (2.6) | 5 (4.3) | 8 (6.8) | 0 (0.0) | 8 (4.1) | 8 (4.1) | |
| 15 | 0 (0.0) | 3 (2.6) | 3 (2.6) | 1 (0.5) | 4 (2.1) | 5 (2.6) | |
| 17 | 0 (0.0) | 3 (2.6) | 3 (2.6) | 0 (0.0) | 5 (2.6) | 5 (2.6) | |
| 22 | 0 (0.0) | 1 (0.9) | 1 (0.9) | 0 (0.0) | 3 (1.5) | 3 (1.5) | |
| 23 | 0 (0.0) | 1 (0.9) | 1 (0.9) | 0 (0.0) | 3 (1.5) | 3 (1.5) | |
| 38 | 2 (1.7) | 8 (6.8) | 10 (8.5) | 7 (3.6) | 13 (6.7) | 20 (10.4) | |
| 80 | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 1 (0.5) | 1 (0.5) | |
| 100 | 0 (0.0) | 3 (2.6) | 3 (2.6) | 2 (1.0) | 10 (5.2) | 12 (6.2) | |
| 107 | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 1 (0.5) | 1 (0.5) | |
| 110 | 0 (0.0) | 6 (5.1) | 6 (5.1) | 2 (1.0) | 10 (5.2) | 12 (6.2) | |
| 111 | 0 (0.0) | 4 (3.4) | 4 (3.4) | 1 (0.5) | 10 (5.2) | 11 (5.7) | |
| 120 | 1 (0.9) | 5 (4.3) | 6 (5.1) | 2 (1.0) | 4 (2.1) | 6 (3.1) | |
| 122 | 1 (0.9) | 4 (3.4) | 5 (4.3) | 3 (1.5) | 14 (7.3) | 17 (8.8) | |
| 145 | 1 (0.9) | 3 (2.6) | 4 (3.4) | 0 (0.0) | 7 (3.6) | 7 (3.6) | |
| 151 | 1 (0.9) | 2 (1.7) | 3 (2.6) | 1 (0.5) | 8 (4.1) | 9 (4.7) | |
| 159 | 1 (0.9) | 1 (0.9) | 2 (1.7) | 0 (0.0) | 3 (1.5) | 3 (1.5) | |
| 174 | 1 (0.9) | 4 (3.4) | 5 (4.3) | 5 (2.6) | 6 (3.1) | 11 (5.7) | |
| Any beta-3 | 2 (1.7) | 10 (8.5) | 12 (10.3) | 3 (1.5) | 12 (6.2) | 15 (7.7) | |
| 49 | 0 (0.0) | 3 (2.6) | 3 (2.6) | 0 (0.0) | 4 (2.1) | 4 (2.1) | |
| 75 | 0 (0.0) | 2 (1.7) | 2 (1.7) | 1 (0.5) | 3 (1.5) | 4 (2.1) | |
| 76 | 1 (0.9) | 6 (5.1) | 7 (6.0) | 2 (1.0) | 6 (3.1) | 8 (4.1) | |
| 115 | 1 (0.9) | 0 (0.0) | 1 (0.9) | 0 (0.0) | 0 (0.0) | 0 (0.0) | |
| Any beta-5 | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 2 (1.0) | 2 (1.0) | |
| 96 | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 2 (1.0) | 2 (1.0) | |
| Gamma Species/Type | HIV-Infected n = 117 n (%) | HIV-Uninfected n = 193 n (%) | |||||
|---|---|---|---|---|---|---|---|
| Single Infection | Multiple Infection | Total | Single Infection | Multiple Infection | Total | p Value a | |
| Any gamma-1 | 2 (1.7) | 2 (1.7) | 4 (3.4) | 0 (0.0) | 1 (0.5) | 1 (0.5) | 0.049 |
| 4 | 1 (0.9) | 1 (0.9) | 2 (1.7) | 0 (0.0) | 1 (0.5) | 1 (0.5) | |
| 65 | 1 (0.9) | 1 (0.9) | 2 (1.7) | 0 (0.0) | 0 (0.0) | 0 (0.0) | |
| Any gamma-2 | 4 (3.4) | 1 (0.9) | 5 (4.3) | 1 (0.5) | 0 (0.0) | 1 (0.5) | 0.02 |
| 200 | 4 (3.4) | 1 (0.9) | 5 (4.3) | 1 (0.5) | 0 (0.0) | 1 (0.5) | 0.02 |
| Any gamma-6 | 2 (1.7) | 0 (0.0) | 2 (1.7) | 0 (0.0) | 1 (0.5) | 1 (0.5) | |
| 108 | 2 (1.7) | 0 (0.0) | 2 (1.7) | 0 (0.0) | 1 (0.5) | 1 (0.5) | |
| Any gamma-7 | 1 (0.9) | 3 (2.6) | 4 (3.4) | 2 (1.0) | 7 (3.6) | 9 (4.7) | |
| 123 | 0 (0.0) | 1 (0.9) | 1 (0.9) | 0 (0.0) | 1 (0.5) | 1 (0.5) | |
| 134 | 0 (0.0) | 1 (0.9) | 1 (0.9) | 1 (0.5) | 0 (0.0) | 1 (0.5) | |
| 149 | 1 (0.9) | 2 (1.7) | 3 (2.6) | 0 (0.0) | 5 (2.6) | 5 (2.6) | |
| 170 | 0 (0.0) | 0 (0.0) | 0 (0.0) | 1 (0.5) | 1 (0.5) | 2 (1.0) | |
| Any gamma-8 | 0 (0.0) | 1 (0.9) | 1 (0.9) | 0 (0.0) | 3 (1.5) | 3 (1.5) | |
| 112 | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 1 (0.5) | 1 (0.5) | |
| 164 | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 1 (0.5) | 1 (0.5) | |
| 168 | 0 (0.0) | 1 (0.9) | 1 (0.9) | 0 (0.0) | 1 (0.5) | 1 (0.5) | |
| Any gamma-10 | 5 (4.3) | 8 (6.8) | 13 (11.1) | 5 (2.6) | 12 (6.2) | 17 (8.8) | |
| 121 | 0 (0.0) | 1 (0.9) | 1 (0.9) | 1 (0.5) | 4 (2.1) | 5 (2.6) | |
| 130 | 3 (2.6) | 3 (2.6) | 6 (5.1) | 1 (0.5) | 2 (1.0) | 3 (1.5) | |
| 133 | 2 (1.7) | 4 (3.4) | 6 (5.1) | 2 (1.0) | 7 (3.6) | 9 (4.7) | |
| 180 | 0 (0.0) | 2 (1.7) | 2 (1.7) | 1 (0.5) | 7 (3.6) | 8 (4.1) | |
| Any gamma-11 | 0 (0.0) | 2 (1.7) | 2 (1.7) | 1 (0.5) | 0 (0.0) | 1 (0.5) | |
| 169 | 0 (0.0) | 2 (1.7) | 2 (1.7) | 1 (0.5) | 0 (0.0) | 1 (0.5) | |
| Any gamma-12 | 1 (0.9) | 2 (1.7) | 3 (2.6) | 4 (2.1) | 7 (3.6) | 11 (5.7) | |
| 127 | 0 (0.0) | 0 (0.0) | 0 (0.0) | 1 (0.5) | 0 (0.0) | 1 (0.5) | |
| 132 | 1 (0.9) | 2 (1.7) | 3 (2.6) | 1 (0.5) | 6 (3.1) | 7 (3.6) | |
| 148 | 0 (0.0) | 1 (0.9) | 1 (0.9) | 2 (1.0) | 1 (0.5) | 3 (1.5) | |
| Any gamma-13 | 0 (0.0) | 2 (1.7) | 2 (1.7) | 4 (2.1) | 3 (1.5) | 7 (3.6) | |
| 128 | 0 (0.0) | 2 (1.7) | 2 (1.7) | 4 (2.1) | 3 (1.5) | 7 (3.6) | |
| Any gamma-15 | 1 (0.9) | 2 (1.7) | 3 (2.6) | 1 (0.5) | 4 (2.1) | 5 (2.6) | |
| 179 | 1 (0.9) | 2 (1.7) | 3 (2.6) | 1 (0.5) | 4 (2.1) | 5 (2.6) | |
| Any gamma-18 | 1 (0.9) | 3 (2.6) | 4 (3.4) | 2 (1.0) | 2 (1.0) | 4 (2.1) | |
| 156 | 1 (0.9) | 3 (2.6) | 4 (3.4) | 2 (1.0) | 2 (1.0) | 4 (2.1) | |
| Any gamma-19 | 1 (0.9) | 0 (0.0) | 1 (0.9) | 0 (0.0) | 1 (0.5) | 1 (0.5) | |
| 161 | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 1 (0.5) | 1 (0.5) | |
| 162 | 1 (0.9) | 0 (0.0) | 1 (0.9) | 0 (0.0) | 0 (0.0) | 0 (0.0) | |
| Any gamma-22 | 3 (2.6) | 1 (0.9) | 4 (3.4) | 1 (0.5) | 2 (1.0) | 3 (1.5) | |
| 172 | 3 (2.6) | 1 (0.9) | 4 (3.4) | 1 (0.5) | 2 (1.0) | 3 (1.5) | |
| Any gamma-23 | 1 (0.9) | 1 (0.9) | 2 (1.7) | 4 (2.1) | 5 (2.6) | 9 (4.7) | |
| 175 | 1 (0.9) | 1 (0.9) | 2 (1.7) | 4 (2.1) | 5 (2.6) | 9 (4.7) | |
| Any gamma-25 | 1 (0.9) | 3 (2.6) | 4 (3.4) | 3 (1.5) | 1 (0.5) | 4 (2.1) | |
| 184 | 1 (0.9) | 3 (2.6) | 4 (3.4) | 3 (1.5) | 1 (0.5) | 4 (2.1) | |
| Any gamma-27 | 0 (0.0) | 2 (1.7) | 2 (1.7) | 0 (0.0) | 2 (1.0) | 2 (1.0) | |
| 201 | 0 (0.0) | 2 (1.7) | 2 (1.7) | 0 (0.0) | 2 (1.0) | 2 (1.0) | |
| Other | 0 (0.0) | 3 (2.6) | 3 (2.6) | 3 (1.5) | 2 (1.0) | 5 (2.6) | |
| SD2 b | 0 (0.0) | 3 (2.6) | 3 (2.6) | 3 (1.5) | 2 (1.0) | 5 (2.6) | |
| HIV-Infected n (reads) | HIV-Uninfected n (reads) | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| PV Genus | Unique PV | FAP | FAPM1 | CUT | Unique PV | FAP | FAPM1 | CUT | Total Unique |
| alpha | 12 (932,336) | 2 (40,556) | 4 (860,360) | 11 (31,420) | 10 (28,995) | 1 (4) | 4 (16,371) | 7 (12,620) | 16 (961,331) |
| beta | 38 (87,932) | 7 (9543) | 13 (2416) | 35 (75,973) | 46 (689,080) | 16 (5828) | 23 (585,069) | 39 (98,183) | 53 (777,012) |
| gamma | 56 (77,009) | 14 (6888) | 20 (44,186) | 41 (25,939) | 45 (803,955) | 8 (34,910) | 17 (739,439) | 35 (29,606) | 76 (880,964) |
| unclassified | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 1 (1230) | 0 (0) | 0 (0) | 1 (1230) | 1 (1230) |
| Sub-total | 106 (1,097,277) | 23 (56,983) | 37 (906,962) | 87 (133,332) | 101 (1,522,260) | 25 (40,742) | 44 (1,340,879) | 82 (141,639) | 146 (2,620,537) |
| Putative new beta | 2 (9) | 0 (0) | 1 (2) | 1 (7) | 6 (179) | 2 (43) | 4 (125) | 1 (11) | 8 (188) |
| Total | 108 (1,097,286) | 23 (56,983) | 38 (906,964) | 88 (133,339) | 107 (1,522,439) | 27 (40,785) | 48 (1,341,004) | 83 (141,650) | 154 (2,620,725) |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gheit, T.; Rollo, F.; Brancaccio, R.N.; Robitaille, A.; Galati, L.; Giuliani, M.; Latini, A.; Pichi, B.; Benevolo, M.; Cuenin, C.; et al. Oral Infection by Mucosal and Cutaneous Human Papillomaviruses in the Men Who Have Sex with Men from the OHMAR Study. Viruses 2020, 12, 899. https://doi.org/10.3390/v12080899
Gheit T, Rollo F, Brancaccio RN, Robitaille A, Galati L, Giuliani M, Latini A, Pichi B, Benevolo M, Cuenin C, et al. Oral Infection by Mucosal and Cutaneous Human Papillomaviruses in the Men Who Have Sex with Men from the OHMAR Study. Viruses. 2020; 12(8):899. https://doi.org/10.3390/v12080899
Chicago/Turabian StyleGheit, Tarik, Francesca Rollo, Rosario N Brancaccio, Alexis Robitaille, Luisa Galati, Massimo Giuliani, Alessandra Latini, Barbara Pichi, Maria Benevolo, Cyrille Cuenin, and et al. 2020. "Oral Infection by Mucosal and Cutaneous Human Papillomaviruses in the Men Who Have Sex with Men from the OHMAR Study" Viruses 12, no. 8: 899. https://doi.org/10.3390/v12080899
APA StyleGheit, T., Rollo, F., Brancaccio, R. N., Robitaille, A., Galati, L., Giuliani, M., Latini, A., Pichi, B., Benevolo, M., Cuenin, C., McKay-Chopin, S., Pellini, R., Cristaudo, A., Morrone, A., Tommasino, M., & Donà, M. G. (2020). Oral Infection by Mucosal and Cutaneous Human Papillomaviruses in the Men Who Have Sex with Men from the OHMAR Study. Viruses, 12(8), 899. https://doi.org/10.3390/v12080899








