FLAVi: An Enhanced Annotator for Viral Genomes of Flaviviridae
Abstract
1. Introduction
2. Materials and Methods
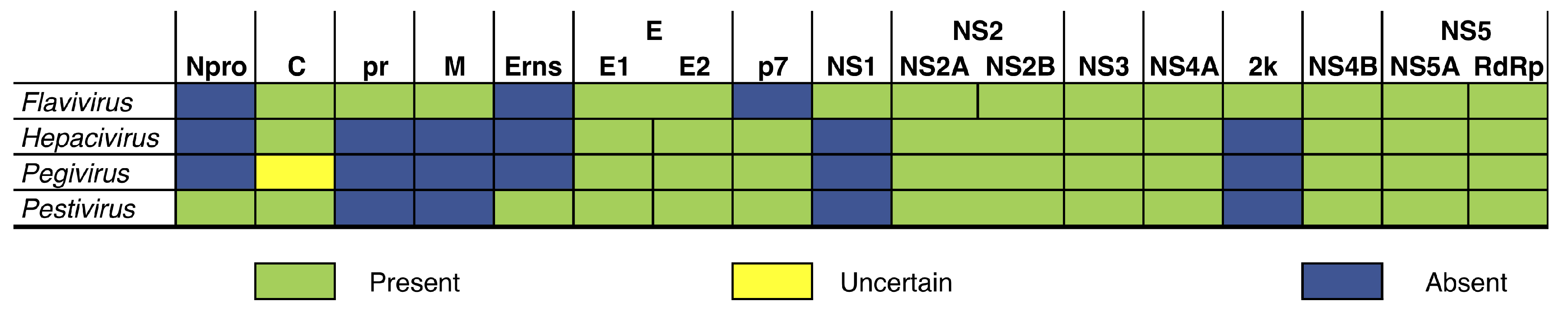
2.1. Taxon Sampling
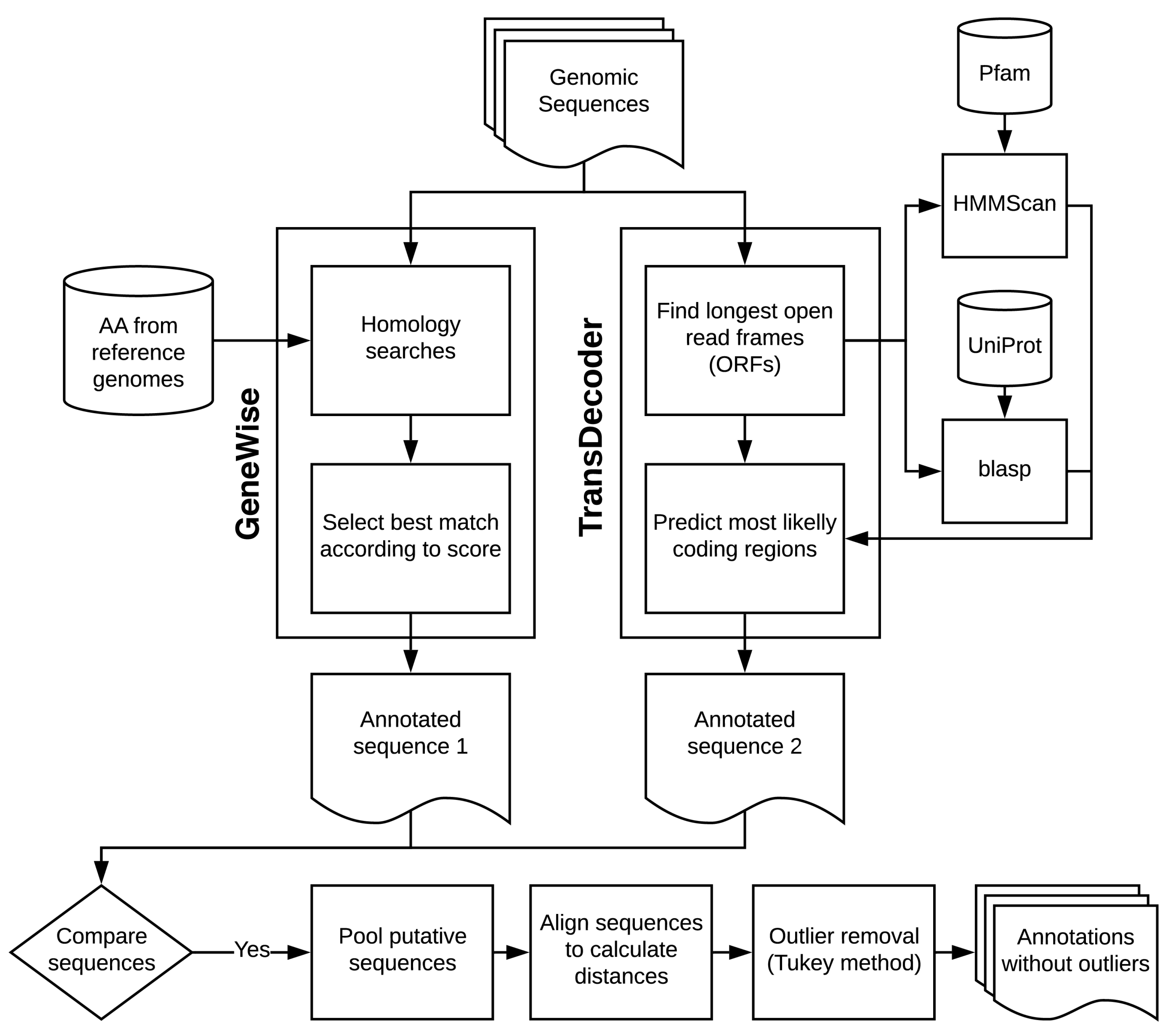
2.2. New Approaches for Gene Prediction in Flaviviruses
2.3. Gene Partitions
2.4. Multiple Sequence Alignment Test
2.5. Tree Search
2.6. Sensitivity Analysis
2.7. Long-Branch Attraction Analysis
2.8. Character Categorization
2.9. Computational Resources
3. Results
3.1. Misalignments Are Frequently Observed When Data Is Not Partitioned
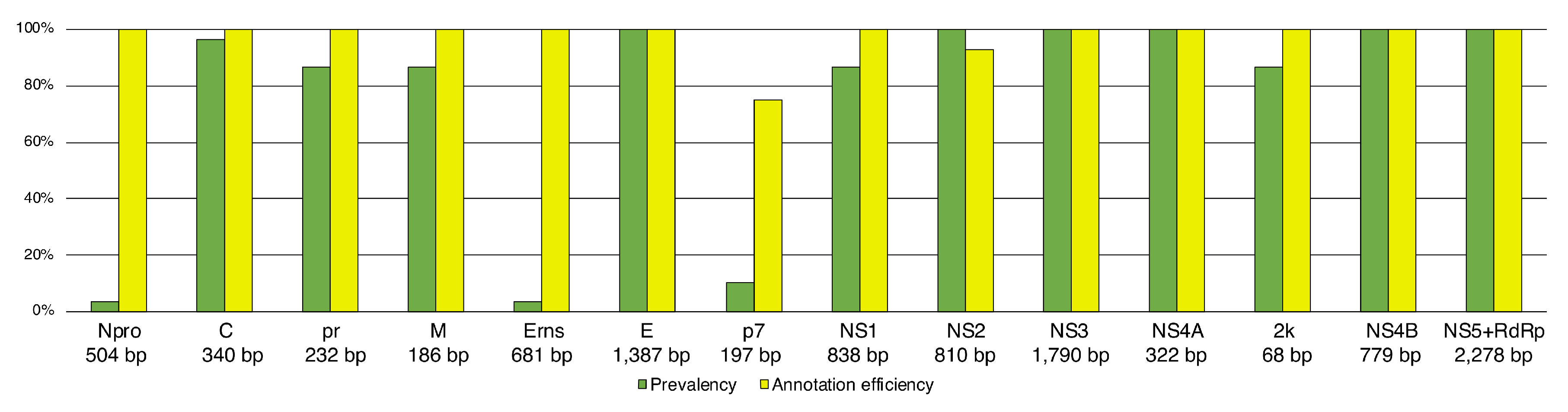
3.2. High Efficiency in Genome Annotation
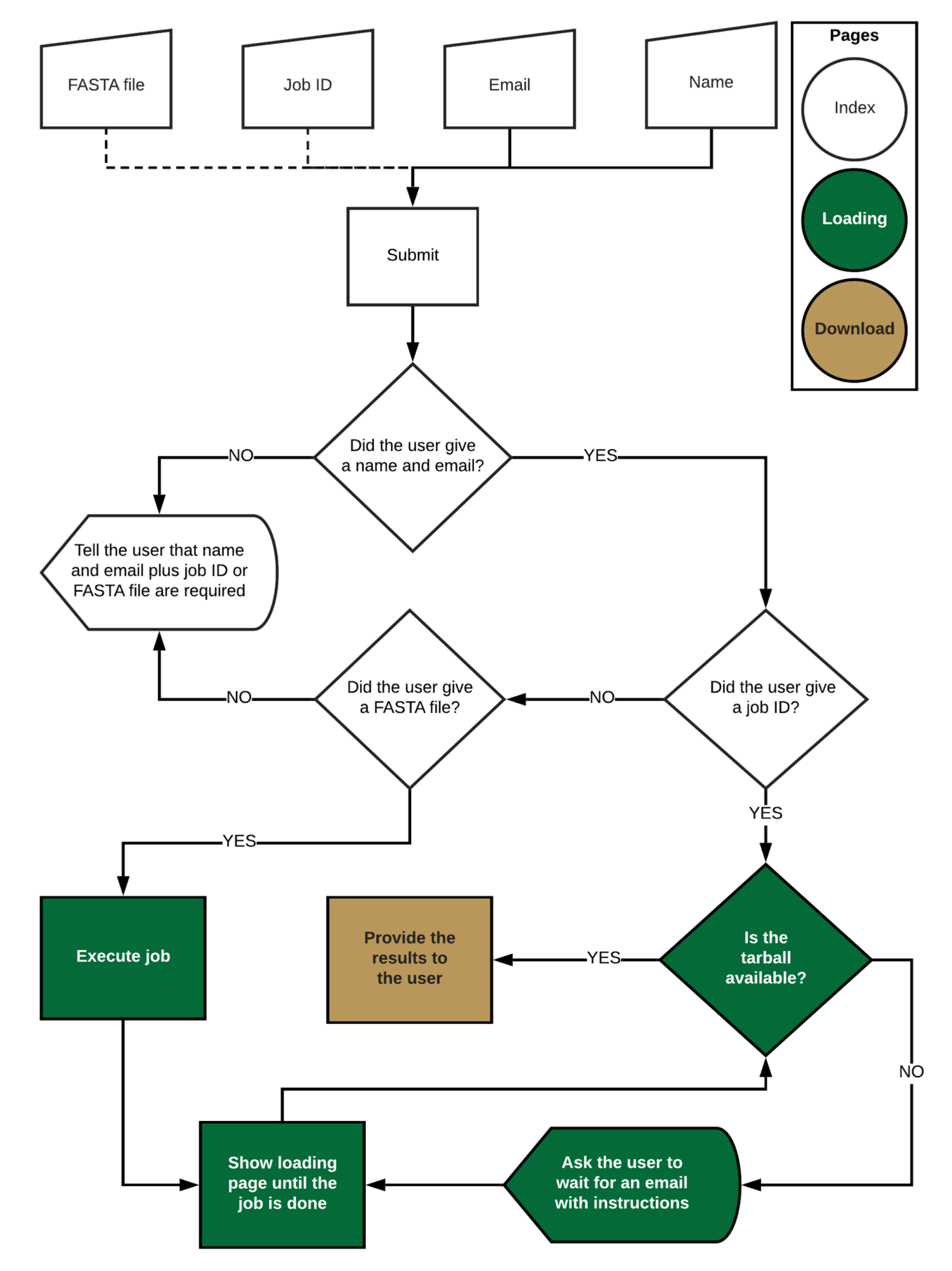
3.3. Web Application
3.4. Topological Distances Formed Two Clusters
3.5. The Scores Were Sensitive to Outgroup Selection and Data Partitioning
3.6. No Long-Branch Distortions Were Observed
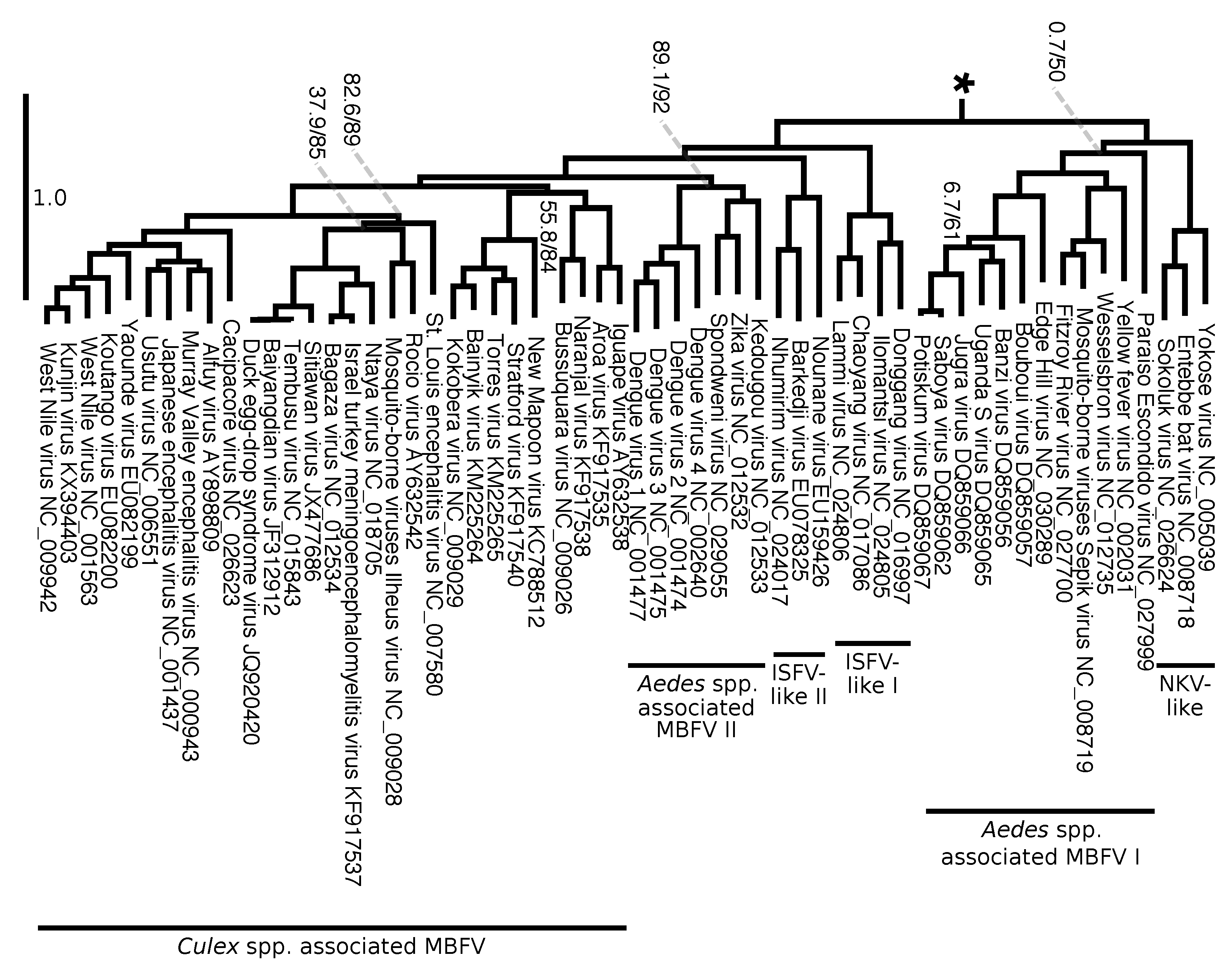
3.7. An Updated Phylogeny of Flaviviridae
4. Discussion
4.1. Msas of Non-Partitioned Genome Sequences Can Be Misleading
4.2. Flexible and Conservative Genome Annotation
4.3. Outgroup Comparison and Lba
4.4. Previous Work on the Phylogeny of Flavivirus
4.5. Phylogenetic Insights
4.6. Final Remarks
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Simmonds, P.; Becher, P.; Bukh, J.; Gould, E.A.; Meyers, G.; Monath, T.; Muerhoff, S.; Pletnev, A.; Rico-Hesse, R.; Smith, D.B.; et al. ICTV virus taxonomy profile: Flaviviridae. J. Gen. Virol. 2017, 98, 2. [Google Scholar] [CrossRef] [PubMed]
- Thomas, D.L.; Astemborski, J.; Rai, R.M.; Anania, F.A.; Schaeffer, M.; Galai, N.; Nolt, K.; Nelson, K.E.; Strathdee, S.A.; Johnson, L.; et al. The natural history of hepatitis C virus infection: Host, viral, and environmental factors. JAMA 2000, 284, 450–456. [Google Scholar] [CrossRef] [PubMed]
- Balcom, E.F.; Doan, M.A.; Branton, W.G.; Jovel, J.; Blevins, G.; Edguer, B.; Hobman, T.C.; Yacyshyn, E.; Emery, D.; Box, A.; et al. Human pegivirus-1 associated leukoencephalitis: Clinical and molecular features. Ann. Neurol. 2018, 84, 781–787. [Google Scholar] [CrossRef]
- Houe, H. Epidemiological features and economical importance of bovine virus diarrhoea virus (BVDV) infections. Vet. Microbiol. 1999, 64, 89–107. [Google Scholar] [CrossRef]
- Holbrook, M. Historical perspectives on Flavivirus research. Viruses 2017, 9, 97. [Google Scholar] [CrossRef]
- Hulo, C.; De Castro, E.; Masson, P.; Bougueleret, L.; Bairoch, A.; Xenarios, I.; Le Mercier, P. ViralZone: A knowledge resource to understand virus diversity. Nucleic Acids Res. 2010, 39, D576–D582. [Google Scholar] [CrossRef] [PubMed]
- Clark, K.; Karsch-Mizrachi, I.; Lipman, D.J.; Ostell, J.; Sayers, E.W. GenBank. Nucleic Acids Res. 2015, 44, D67–D72. [Google Scholar] [CrossRef] [PubMed]
- Charles, J.; Tangudu, C.S.; Hurt, S.L.; Tumescheit, C.; Firth, A.E.; Garcia-Rejon, J.E.; Machain-Williams, C.; Blitvich, B.J. Detection of novel and recognized RNA viruses in mosquitoes from the Yucatan Peninsula of Mexico using metagenomics and characterization of their in vitro host ranges. J. Gen. Virol. 2018, 99, 1729–1738. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Wu, Z.; Liu, B.; Du, J.; Zhang, J.; Lu, L.; Zhu, G.; Han, Y.; Su, H.; Yang, L.; Zhang, S.; et al. Discovery of diverse rodent and bat pestiviruses with distinct genomic and phylogenetic characteristics in several Chinese provinces. Front. Microbiol. 2018, 9, 2562. [Google Scholar] [CrossRef]
- El-Gebali, S.; Mistry, J.; Bateman, A.; Eddy, S.R.; Luciani, A.; Potter, S.C.; Qureshi, M.; Richardson, L.J.; Salazar, G.A.; Smart, A.; et al. The Pfam protein families database in 2019. Nucleic Acids Res. 2018, 47, D427–D432. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, A.L.; Attwood, T.K.; Babbitt, P.C.; Blum, M.; Bork, P.; Bridge, A.; Brown, S.D.; Chang, H.Y.; El-Gebali, S.; Fraser, M.I.; et al. InterPro in 2019: Improving coverage, classification and access to protein sequence annotations. Nucleic Acids Res. 2018, 47, D351–D360. [Google Scholar] [CrossRef] [PubMed]
- Marchler-Bauer, A.; Bo, Y.; Han, L.; He, J.; Lanczycki, C.J.; Lu, S.; Chitsaz, F.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R.; et al. CDD/SPARCLE: Functional classification of proteins via subfamily domain architectures. Nucleic Acids Res. 2016, 45, D200–D203. [Google Scholar] [CrossRef] [PubMed]
- Wen, S.; Ma, D.; Lin, Y.; Li, L.; Hong, S.; Li, X.; Wang, X.; Xi, J.; Qiu, L.; Pan, Y.; et al. Complete Genome Characterization of the 2017 Dengue Outbreak in Xishuangbanna, a Border City of China, Burma and Laos. Front. Cell. Infect. Microbiol. 2018, 8, 148. [Google Scholar] [CrossRef] [PubMed]
- Yachdav, G.; Kloppmann, E.; Kajan, L.; Hecht, M.; Goldberg, T.; Hamp, T.; Hönigschmid, P.; Schafferhans, A.; Roos, M.; Bernhofer, M.; et al. PredictProtein: An open resource for online prediction of protein structural and functional features. Nucleic Acids Res. 2014, 42, W337–W343. [Google Scholar] [CrossRef]
- Liu, L.; Xia, H.; Wahlberg, N.; Belák, S.; Baule, C. Phylogeny, classification and evolutionary insights into pestiviruses. Virology 2009, 385, 351–357. [Google Scholar] [CrossRef]
- Thézé, J.; Lowes, S.; Parker, J.; Pybus, O.G. Evolutionary and phylogenetic analysis of the hepaciviruses and pegiviruses. Genome Biol. Evol. 2015, 7, 2996–3008. [Google Scholar] [CrossRef][Green Version]
- Moureau, G.; Cook, S.; Lemey, P.; Nougairede, A.; Forrester, N.L.; Khasnatinov, M.; Charrel, R.N.; Firth, A.E.; Gould, E.A.; De Lamballerie, X. New insights into Flavivirus evolution, taxonomy and biogeographic history, extended by analysis of canonical and alternative coding sequences. PLoS ONE 2015, 10, e0117849. [Google Scholar] [CrossRef]
- Maddison, W.P.; Donoghue, M.J.; Maddison, D.R. Outgroup analysis and parsimony. Syst. Biol. 1984, 33, 83–103. [Google Scholar] [CrossRef]
- Hess, P.N.; De Moraes Russo, C.A. An empirical test of the midpoint rooting method. Biol. J. Linn. Soc. 2007, 92, 669–674. [Google Scholar] [CrossRef]
- Kinene, T.; Wainaina, J.; Maina, S.; Boykin, L. Rooting trees, methods for. Encycl. Evol. Biol. 2016, 489–493. [Google Scholar] [CrossRef]
- Wenzel, J. Origins of SARS-CoV-1 and SARS-CoV-2 are often poorly explored in leading publications. Cladistics 2020. [Google Scholar] [CrossRef]
- Grant, T. Outgroup sampling in phylogenetics: Severity of test and successive outgroup expansion. J. Zool. Syst. Evol. Res. 2019, 57, 748–763. [Google Scholar] [CrossRef]
- de Bernardi Schneider, A.; Malone, R.W.; Guo, J.T.; Homan, J.; Linchangco, G.; Witter, Z.L.; Vinesett, D.; Damodaran, L.; Janies, D.A. Molecular evolution of Zika virus as it crossed the Pacific to the Americas. Cladistics 2017, 33, 1–20. [Google Scholar] [CrossRef]
- Shi, M.; Lin, X.D.; Vasilakis, N.; Tian, J.H.; Li, C.X.; Chen, L.J.; Eastwood, G.; Diao, X.N.; Chen, M.H.; Chen, X.; et al. Divergent viruses discovered in arthropods and vertebrates revise the evolutionary history of the Flaviviridae and related viruses. J. Virol. 2016, 90, 659–669. [Google Scholar] [CrossRef]
- Birney, E.; Clamp, M.; Durbin, R. GeneWise and Genomewise. Genome Res. 2004, 14, 988–995. [Google Scholar] [CrossRef]
- Haas, B.J.; Papanicolaou, A.; Yassour, M.; Grabherr, M.; Blood, P.D.; Bowden, J.; Couger, M.B.; Eccles, D.; Li, B.; Lieber, M.; et al. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 2013, 8, 1494. [Google Scholar] [CrossRef]
- Consortium, T.U. UniProt: A worldwide hub of protein knowledge. Nucleic Acids Res. 2018, 47, D506–D515. [Google Scholar] [CrossRef]
- Rice, P.; Longden, I.; Bleasby, A. EMBOSS: The European molecular biology open software suite. Trends Genet. 2000, 16, 276–277. [Google Scholar] [CrossRef]
- Tukey, J.W. Exploratory Data Analysis; Section 2C; Addison-Wesley: Boston, MA, USA, 1977. [Google Scholar]
- Sievers, F.; Wilm, A.; Dineen, D.; Gibson, T.J.; Karplus, K.; Li, W.; Lopez, R.; McWilliam, H.; Remmert, M.; Söding, J.; et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011, 7, 539. [Google Scholar] [CrossRef]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef] [PubMed]
- Goloboff, P.A.; Farris, J.S.; Nixon, K.C. TNT, a free program for phylogenetic analysis. Cladistics 2008, 24, 774–786. [Google Scholar] [CrossRef]
- Nguyen, L.T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2014, 32, 268–274. [Google Scholar] [CrossRef] [PubMed]
- Bouckaert, R.; Heled, J.; Kühnert, D.; Vaughan, T.; Wu, C.H.; Xie, D.; Suchard, M.A.; Rambaut, A.; Drummond, A.J. BEAST 2: A software platform for Bayesian evolutionary analysis. PLoS Comput. Biol. 2014, 10, e1003537. [Google Scholar] [CrossRef]
- Guindon, S.; Dufayard, J.F.; Lefort, V.; Anisimova, M.; Hordijk, W.; Gascuel, O. New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst. Biol. 2010, 59, 307–321. [Google Scholar] [CrossRef]
- Minh, B.Q.; Nguyen, M.A.T.; von Haeseler, A. Ultrafast approximation for phylogenetic bootstrap. Mol. Biol. Evol. 2013, 30, 1188–1195. [Google Scholar] [CrossRef]
- Bogdanowicz, D.; Giaro, K. Matching split distance for unrooted binary phylogenetic trees. IEEE/ACM Trans. Comput. Biol. Bioinform. 2012, 9, 150–160. [Google Scholar] [CrossRef]
- Robinson, D.F.; Foulds, L.R. Comparison of phylogenetic trees. Math. Biosci. 1981, 53, 131–147. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2014; Available online: http://www.R-project.org/ (accessed on 13 August 2020).
- Sammon, J.W. A nonlinear mapping for data structure analysis. IEEE Trans. Comput. 1969, 100, 401–409. [Google Scholar] [CrossRef]
- Felsenstein, J. Cases in which parsimony or compatibility methods will be positively misleading. Syst. Zool. 1978, 27, 401–410. [Google Scholar] [CrossRef]
- Farris, J.S. Likelihood and inconsistency. Cladistics 1999, 15, 199–204. [Google Scholar]
- Siddall, M.E.; Whiting, M.F. Long-branch abstractions. Cladistics 1999, 15, 9–24. [Google Scholar] [CrossRef]
- Kück, P.; Mayer, C.; Wägele, J.W.; Misof, B. Long branch effects distort maximum likelihood phylogenies in simulations despite selection of the correct model. PLoS ONE 2012, 7, e36593. [Google Scholar] [CrossRef] [PubMed]
- Chang, J.T. Inconsistency of evolutionary tree topology reconstruction methods when substitution rates vary across characters. Math. Biosci. 1996, 134, 189–215. [Google Scholar] [CrossRef]
- Machado, D.J. YBYRÁ facilitates comparison of large phylogenetic trees. BMC Bioinform. 2015, 16, 204. [Google Scholar] [CrossRef] [PubMed]
- Grant, T.; Kluge, A.G. Transformation series as an ideographic character concept. Cladistics 2004, 20, 23–31. [Google Scholar] [CrossRef]
- Nixon, K.C.; Carpenter, J.M. On homology. Cladistics 2012, 28, 160–169. [Google Scholar] [CrossRef]
- Grinberg, M. Flask Web Development: Developing Web Applications with Python, 1st ed.; O’Reilly Media, Inc.: Sebastopol, CA, USA, 2014. [Google Scholar]
- Huelsenbeck, J.P. The robustness of two phylogenetic methods: Four-taxon simulations reveal a slight superiority of maximum likelihood over neighbor joining. Mol. Biol. Evol. 1995, 12, 843–849. [Google Scholar]
- Buckley, T.R.; Simon, C.; Chambers, G.K. Exploring among-site rate variation models in a maximum likelihood framework using empirical data: Effects of model assumptions on estimates of topology, branch lengths, and bootstrap support. Syst. Biol. 2001, 50, 67–86. [Google Scholar] [CrossRef]
- Nylander, J.A.; Ronquist, F.; Huelsenbeck, J.P.; Nieves-Aldrey, J. Bayesian phylogenetic analysis of combined data. Syst. Biol. 2004, 53, 47–67. [Google Scholar] [CrossRef]
- Brown, J.M.; Lemmon, A.R. The importance of data partitioning and the utility of Bayes factors in Bayesian phylogenetics. Syst. Biol. 2007, 56, 643–655. [Google Scholar] [CrossRef]
- Kainer, D.; Lanfear, R. The effects of partitioning on phylogenetic inference. Mol. Biol. Evol. 2015, 32, 1611–1627. [Google Scholar] [CrossRef]
- Bekal, S.; Domier, L.L.; Gonfa, B.; McCoppin, N.K.; Lambert, K.N.; Bhalerao, K. A novel Flavivirus in the soybean cyst nematode. J. Gen. Virol. 2014, 95, 1272–1280. [Google Scholar] [CrossRef] [PubMed]
- Wheeler, W.C. Nucleic acid sequence phylogeny and random outgroups. Cladistics 1990, 6, 363–367. [Google Scholar] [CrossRef]
- Bergsten, J. A review of long-branch attraction. Cladistics 2005, 21, 163–193. [Google Scholar] [CrossRef]
- Pol, D.; Siddall, M.E. Biases in maximum likelihood and parsimony: A simulation approach to a 10-taxon case. Cladistics 2001, 17, 266–281. [Google Scholar] [CrossRef]
- Farris, J.S. Estimating phylogenetic trees from distance matrices. Am. Nat. 1972, 106, 645–668. [Google Scholar] [CrossRef]
- Farris, J.S. Outgroups and parsimony. Syst. Zool. 1982, 31, 328–334. [Google Scholar] [CrossRef]
- Lundberg, J.G. Wagner networks and ancestors. Syst. Biol. 1972, 21, 398–413. [Google Scholar] [CrossRef]
- Kluge, A.G.; Grant, T. From conviction to anti-superfluity: Old and new justifications of parsimony in phylogenetic inference. Cladistics 2006, 22, 276–288. [Google Scholar] [CrossRef]
- Grant, T.; Kluge, A.G. Perspective: Parsimony, explanatory power, and dynamic homology testing. Syst. Biodivers. 2009, 7, 357–363. [Google Scholar] [CrossRef]
- Grant, T.; Rada, M.; Anganoy-Criollo, M.; Batista, A.; Dias, P.H.; Jeckel, A.M.; Machado, D.J.; Rueda-Almonacid, J.V. Phylogenetic systematics of dart-poison frogs and their relatives revisited (Anura: Dendrobatoidea). South Am. J. Herpetol. 2017, 12. [Google Scholar] [CrossRef]
- Huelsenbeck, J.P. Is the Felsenstein zone a fly trap? Syst. Biol. 1997, 46, 69–74. [Google Scholar] [CrossRef] [PubMed]
- Wheeler, W.C. Systematics: A Course of Lectures; John Wiley & Sons: Hoboken, NJ, USA, 2012. [Google Scholar]
- Wheeler, W. Homology and the optimization of DNA sequence data. Cladistics 2001, 17, S3–S11. [Google Scholar] [CrossRef] [PubMed]
- Kuno, G.; Chang, G.J.J.; Tsuchiya, K.R.; Karabatsos, N.; Cropp, C.B. Phylogeny of the genus Flavivirus. J. Virol. 1998, 72, 73–83. [Google Scholar] [CrossRef] [PubMed]
- Jenkins, G.M.; Pagel, M.; Gould, E.A.; Paolo, M.d.A.; Holmes, E.C. Evolution of base composition and codon usage bias in the genus Flavivirus. J. Mol. Evol. 2001, 52, 383–390. [Google Scholar] [CrossRef]
- Billoir, F.; de Chesse, R.; Tolou, H.; de Micco, P.; Gould, E.A.; de Lamballerie, X. Phylogeny of the genus Flavivirus using complete coding sequences of arthropod-borne viruses and viruses with no known vector. J. Gen. Virol. 2000, 81, 781–790. [Google Scholar] [CrossRef]
- Schubert, A.M.; Putonti, C. Evolution of the sequence composition of flaviviruses. Infect. Genet. Evol. 2010, 10, 129–136. [Google Scholar] [CrossRef][Green Version]
- Cook, S.; Moureau, G.; Kitchen, A.; Gould, E.A.; de Lamballerie, X.; Holmes, E.C.; Harbach, R.E. Molecular evolution of the insect-specific flaviviruses. J. Gen. Virol. 2012, 93, 223. [Google Scholar] [CrossRef] [PubMed]
- Zanotto, P.d.; Gould, E.A.; Gao, G.F.; Harvey, P.H.; Holmes, E.C. Population dynamics of flaviviruses revealed by molecular phylogenies. Proc. Natl. Acad. Sci. USA 1996, 93, 548–553. [Google Scholar] [CrossRef]
- Twiddy, S.S.; Pybus, O.G.; Holmes, E.C. Comparative population dynamics of mosquito-borne flaviviruses. Infect. Genet. Evol. 2003, 3, 87–95. [Google Scholar] [CrossRef]
- Lobo, F.P.; Mota, B.E.; Pena, S.D.; Azevedo, V.; Macedo, A.M.; Tauch, A.; Machado, C.R.; Franco, G.R. Virus-host coevolution: Common patterns of nucleotide motif usage in Flaviviridae and their hosts. PLoS ONE 2009, 4, e6282. [Google Scholar] [CrossRef] [PubMed]
- Huhtamo, E.; Cook, S.; Moureau, G.; Uzcátegui, N.Y.; Sironen, T.; Kuivanen, S.; Putkuri, N.; Kurkela, S.; Harbach, R.E.; Firth, A.E.; et al. Novel flaviviruses from mosquitoes: Mosquito-specific evolutionary lineages within the phylogenetic group of mosquito-borne flaviviruses. Virology 2014, 464, 320–329. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; He, L.; He, R.L.; Yau, S.S.T. Zika and flaviviruses phylogeny based on the alignment-free natural vector method. DNA Cell Biol. 2017, 36, 109–116. [Google Scholar] [CrossRef]
- Springer, M.S.; Gatesy, J. On the importance of homology in the age of phylogenomics. Syst. Biodivers. 2018, 16, 210–228. [Google Scholar] [CrossRef]
- De Lamballerie, X.; Crochu, S.; Billoir, F.; Neyts, J.; De Micco, P.; Holmes, E.; Gould, E. Genome sequence analysis of Tamana bat virus and its relationship with the genus Flavivirus. J. Gen. Virol. 2002, 83, 2443–2454. [Google Scholar] [CrossRef]
- Gupta, S.K.; Singh, S.; Nischal, A.; Pant, K.K.; Seth, P.K. Molecular-based identification and phylogeny of genomic and proteomic sequences of mosquito-borne Flavivirus. Genes Genom. 2014, 36, 31–43. [Google Scholar] [CrossRef]
- Alkan, C.; Zapata, S.; Bichaud, L.; Moureau, G.; Lemey, P.; Firth, A.E.; Gritsun, T.S.; Gould, E.A.; de Lamballerie, X.; Depaquit, J.; et al. Ecuador Paraiso Escondido virus, a new Flavivirus isolated from New World sand flies in Ecuador, is the first representative of a novel clade in the genus Flavivirus. J. Virol. 2015, 89, 11773–11785. [Google Scholar] [CrossRef]
- Walker, P.J.; Siddell, S.G.; Lefkowitz, E.J.; Mushegian, A.R.; Dempsey, D.M.; Dutilh, B.E.; Harrach, B.; Harrison, R.L.; Hendrickson, R.C.; Junglen, S.; et al. Changes to virus taxonomy and the International Code of Virus Classification and Nomenclature ratified by the International Committee on Taxonomy of Viruses (2019). Arch. Virol. 2019, 164, 2417–2429. [Google Scholar] [CrossRef]
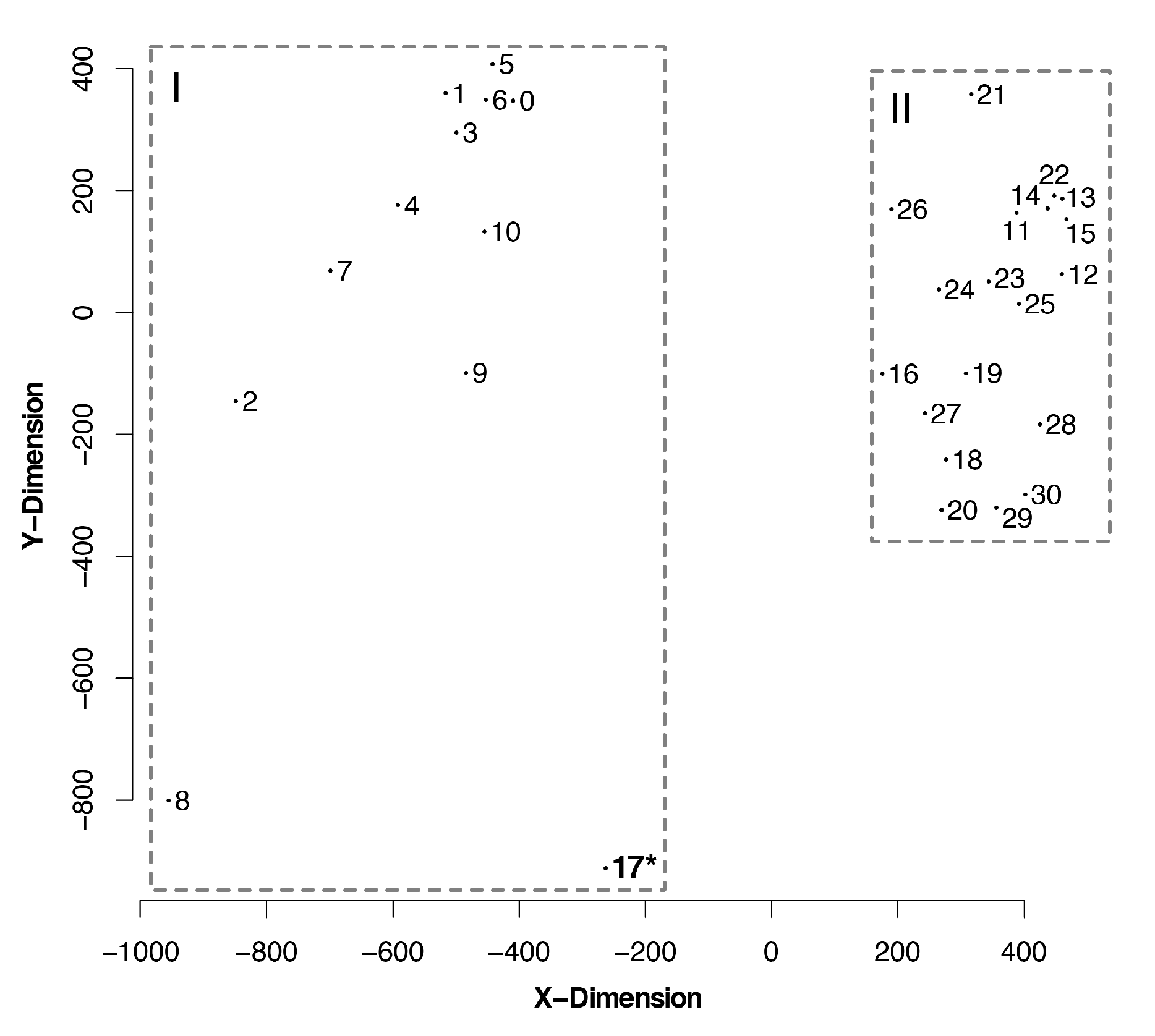
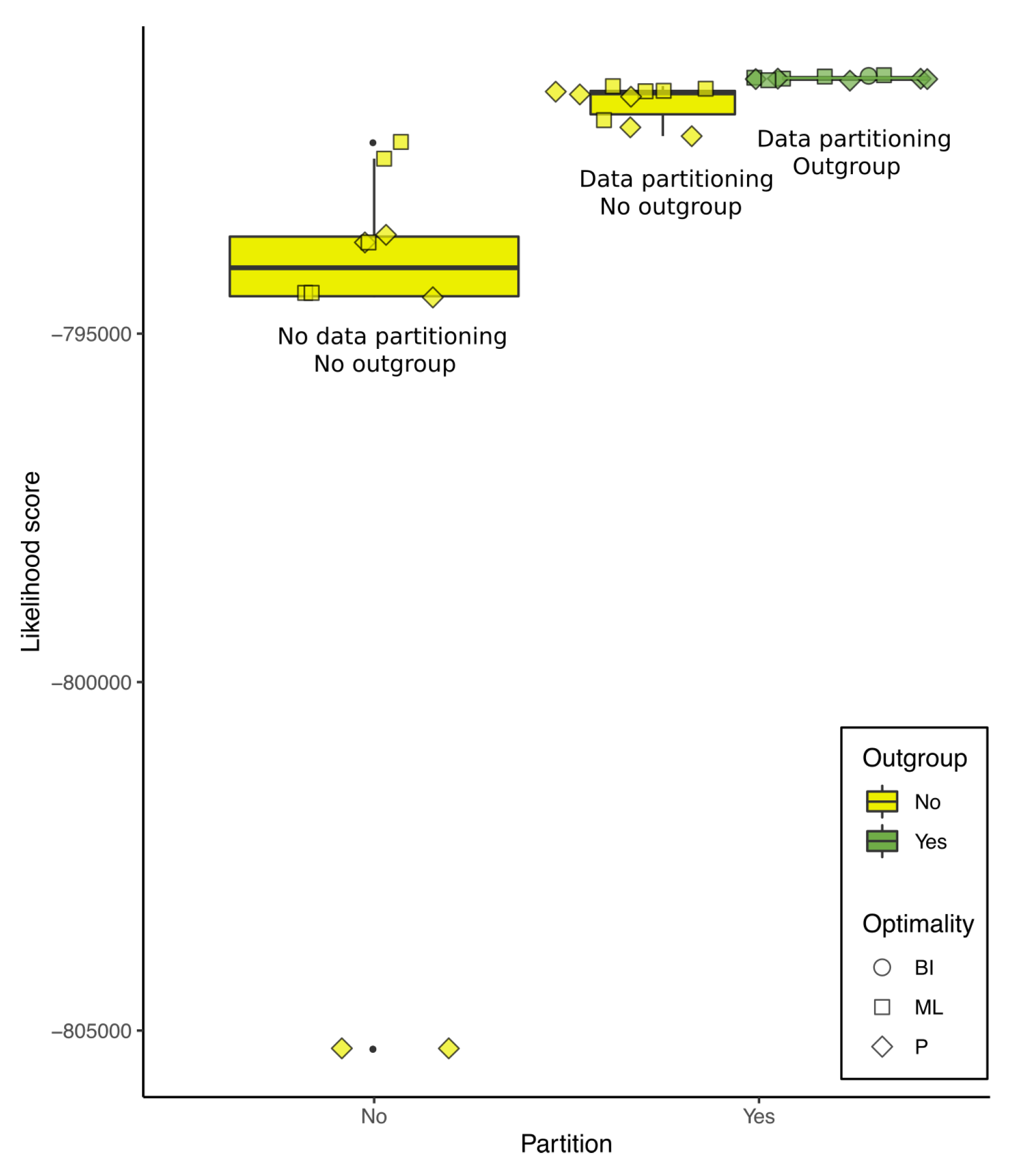
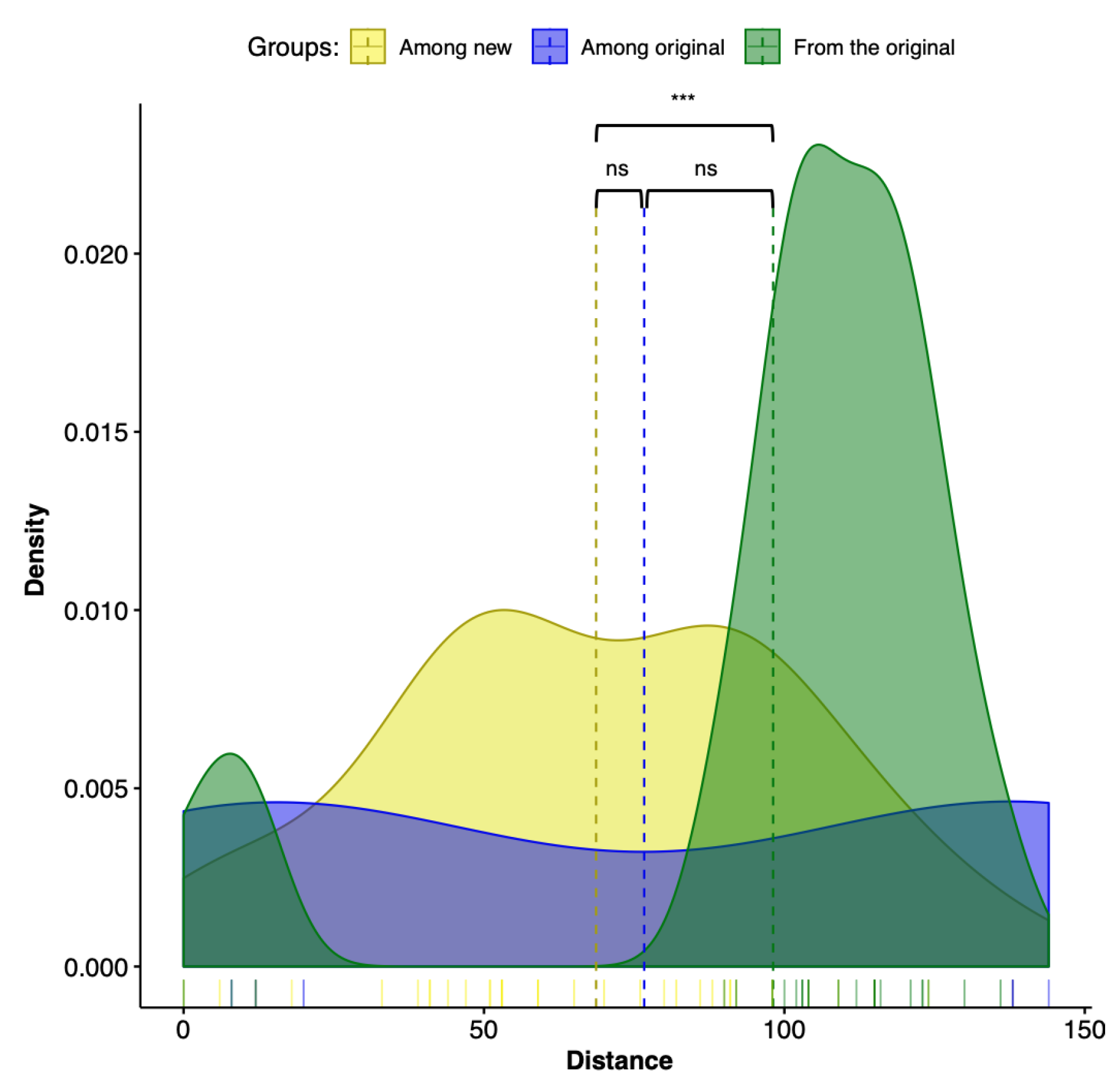
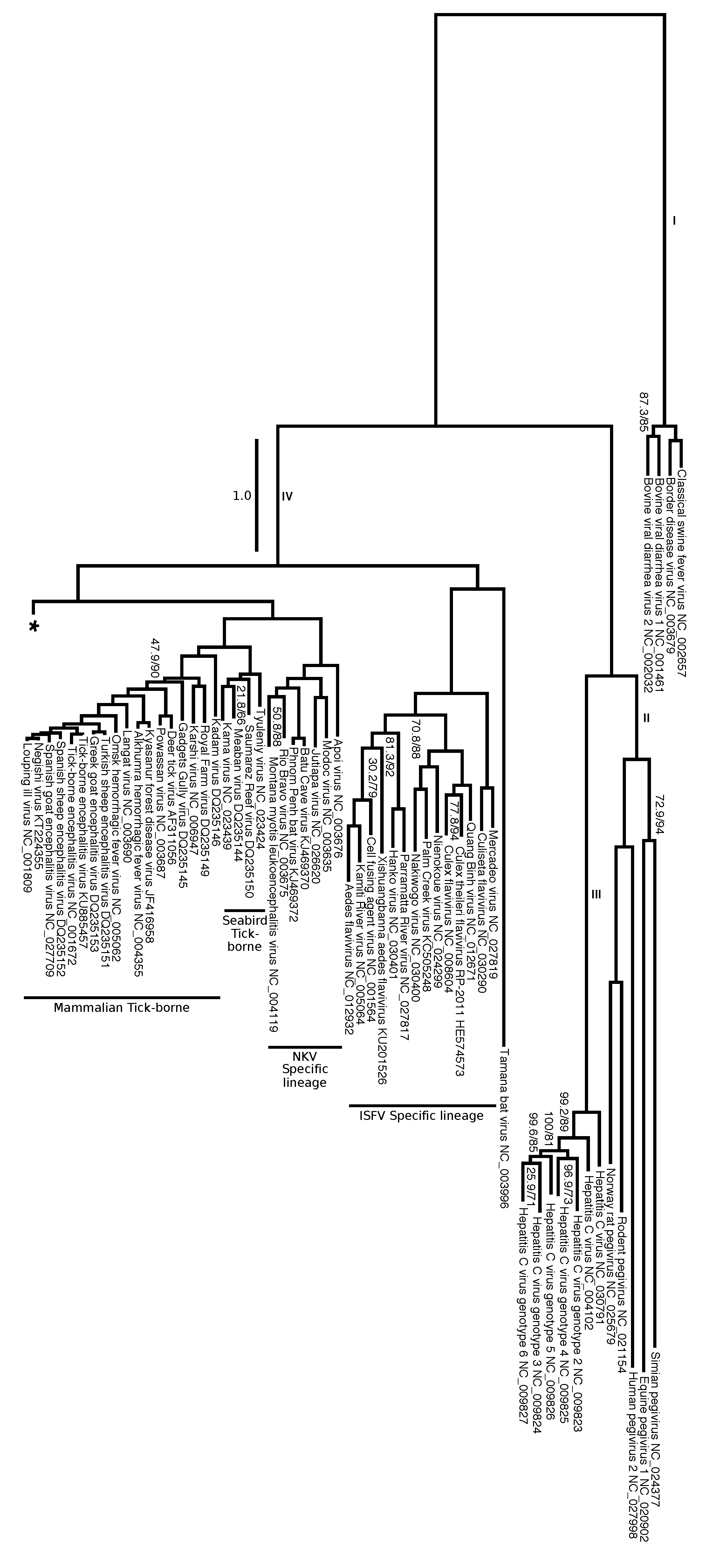
| Tree No. | Partition | Group | Optimality Criteria | Tree Search Program | Alignment Strategy |
|---|---|---|---|---|---|
| 0 | Yes | Flaviviridae | Maximum likelihood | IQ-Tree | Translation (PAM250) |
| 1 | " | " | Bayesian inference | BEAST | Translation (PAM250) |
| 2 | " | " | Parsimony | TNT | Translation (PAM250) |
| 3 | " | " | Maximum likelihood | IQ-tree | Clustal |
| 4 | " | " | " | " | Geneious MSA |
| 5 | " | " | " | " | Mafft |
| 6 | " | " | " | " | MUSCLE |
| 7 | " | " | Parsimony | TNT | Clustal |
| 8 | " | " | " | " | Geneious MSA |
| 9 | " | " | " | " | Mafft |
| 10 | " | " | " | " | MUSCLE |
| 11 | " | Flavivirus | Maximum likelihood | IQ-Tree | Clustal |
| 12 | " | " | " | " | Geneious MSA |
| 13 | " | " | " | " | Mafft |
| 14 | " | " | " | " | MUSCLE |
| 15 | " | " | " | " | Translation (PAM250) |
| 16 | " | " | Parsimony | TNT | Clustal |
| 17 | " | " | " | " | Geneious MSA |
| 18 | " | " | " | " | Mafft |
| 19 | " | " | " | " | MUSCLE |
| 20 | " | " | " | " | Translation (PAM250) |
| 21 | No | " | Maximum likelihood | IQ-Tree | Clustal |
| 22 | " | " | " | " | Mafft |
| 23 | " | " | " | " | Translation (PAM100) |
| 24 | " | " | " | " | Translation (PAM200) |
| 25 | " | " | " | " | Translation (PAM250) |
| 26 | " | " | Parsimony | TNT | Clustal |
| 27 | " | " | " | " | Mafft |
| 28 | " | " | " | " | Translation (PAM100) |
| 29 | " | " | " | " | Translation (PAM200) |
| 30 | " | " | " | " | Translation (PAM250) |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
de Bernadi Schneider, A.; Jacob Machado, D.; Guirales, S.; Janies, D.A. FLAVi: An Enhanced Annotator for Viral Genomes of Flaviviridae. Viruses 2020, 12, 892. https://doi.org/10.3390/v12080892
de Bernadi Schneider A, Jacob Machado D, Guirales S, Janies DA. FLAVi: An Enhanced Annotator for Viral Genomes of Flaviviridae. Viruses. 2020; 12(8):892. https://doi.org/10.3390/v12080892
Chicago/Turabian Stylede Bernadi Schneider, Adriano, Denis Jacob Machado, Sayal Guirales, and Daniel A. Janies. 2020. "FLAVi: An Enhanced Annotator for Viral Genomes of Flaviviridae" Viruses 12, no. 8: 892. https://doi.org/10.3390/v12080892
APA Stylede Bernadi Schneider, A., Jacob Machado, D., Guirales, S., & Janies, D. A. (2020). FLAVi: An Enhanced Annotator for Viral Genomes of Flaviviridae. Viruses, 12(8), 892. https://doi.org/10.3390/v12080892







