Synthesis of Full-Length cDNA Infectious Clones of Soybean Mosaic Virus and Functional Identification of a Key Amino Acid in the Silencing Suppressor Hc-Pro
Abstract
1. Introduction
2. Materials and Methods
2.1. Assembly of Full-Length cDNA Infectious Clones for SMV
2.2. Virus Infection, Determination of Host Ranges, Detection of SMV by ELISA and Western Blots
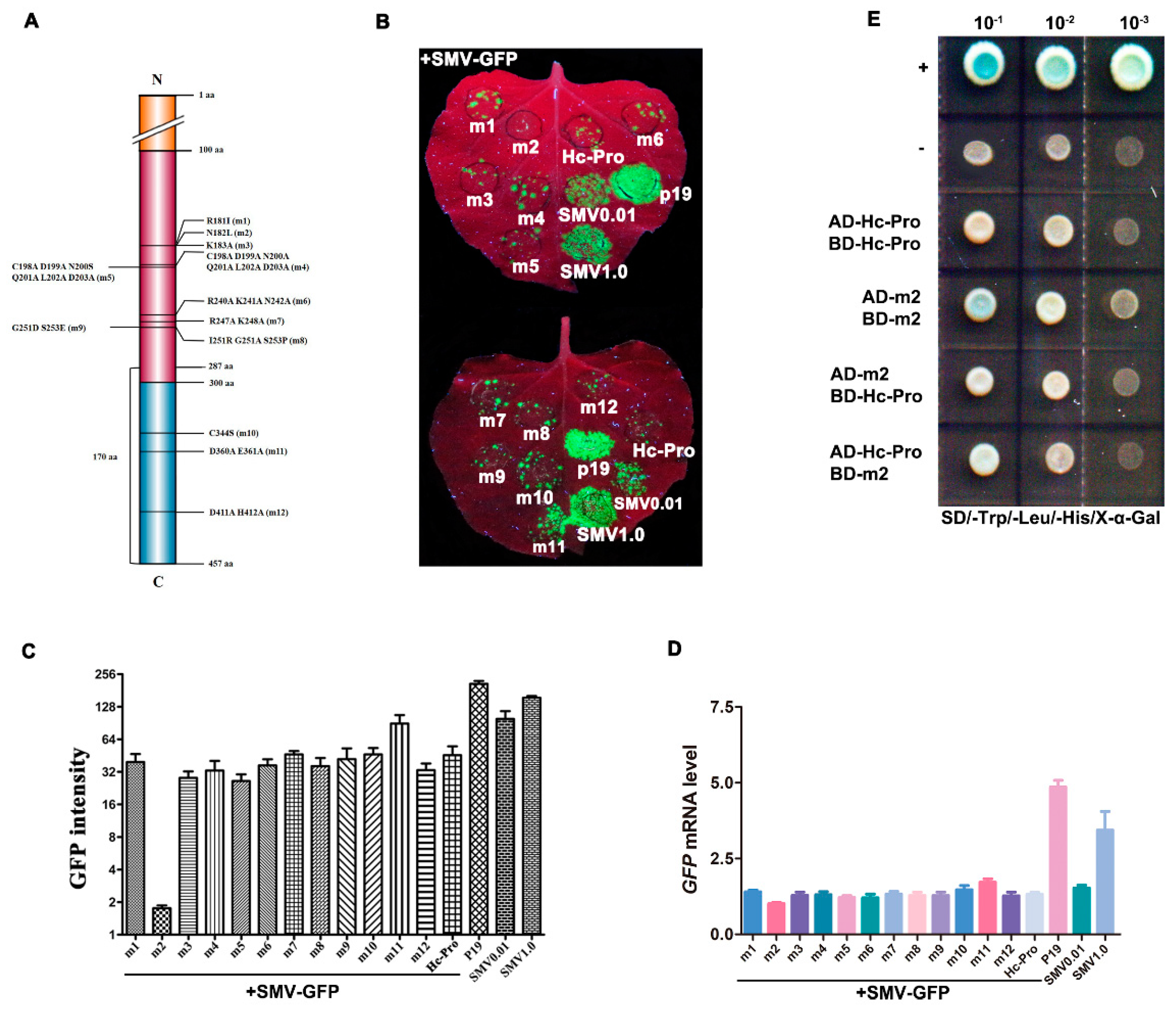
2.3. Recombinant Vector Constructions for Hc-Pro Mutants
2.4. Virus Inoculation and Agroinfiltration
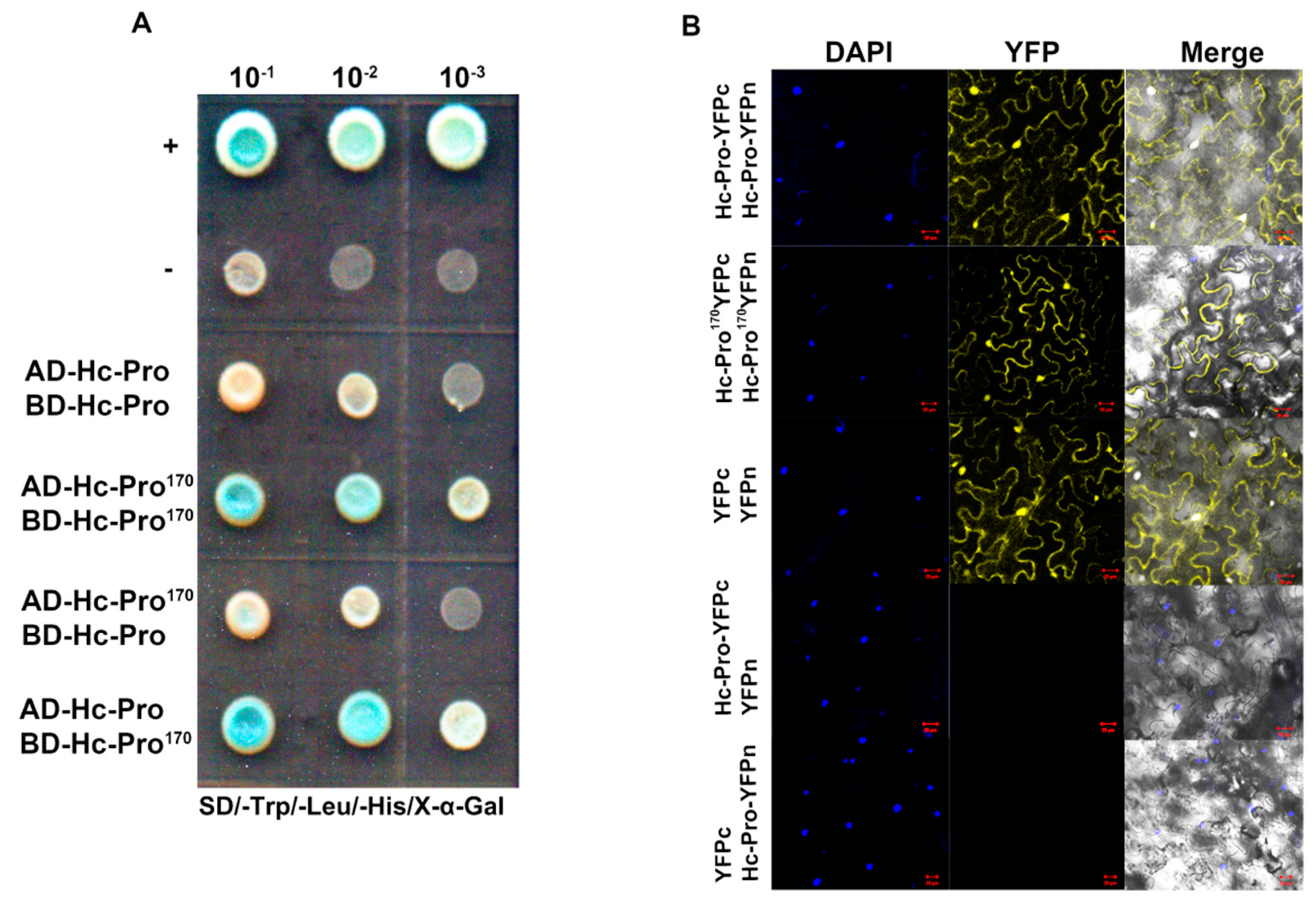
2.5. Yeast Two-Hybrid (Y2H) and Bimolecular Fluorescence Complementation (BiFC) Assays
2.6. RNA Extraction and PCR Analysis
3. Results
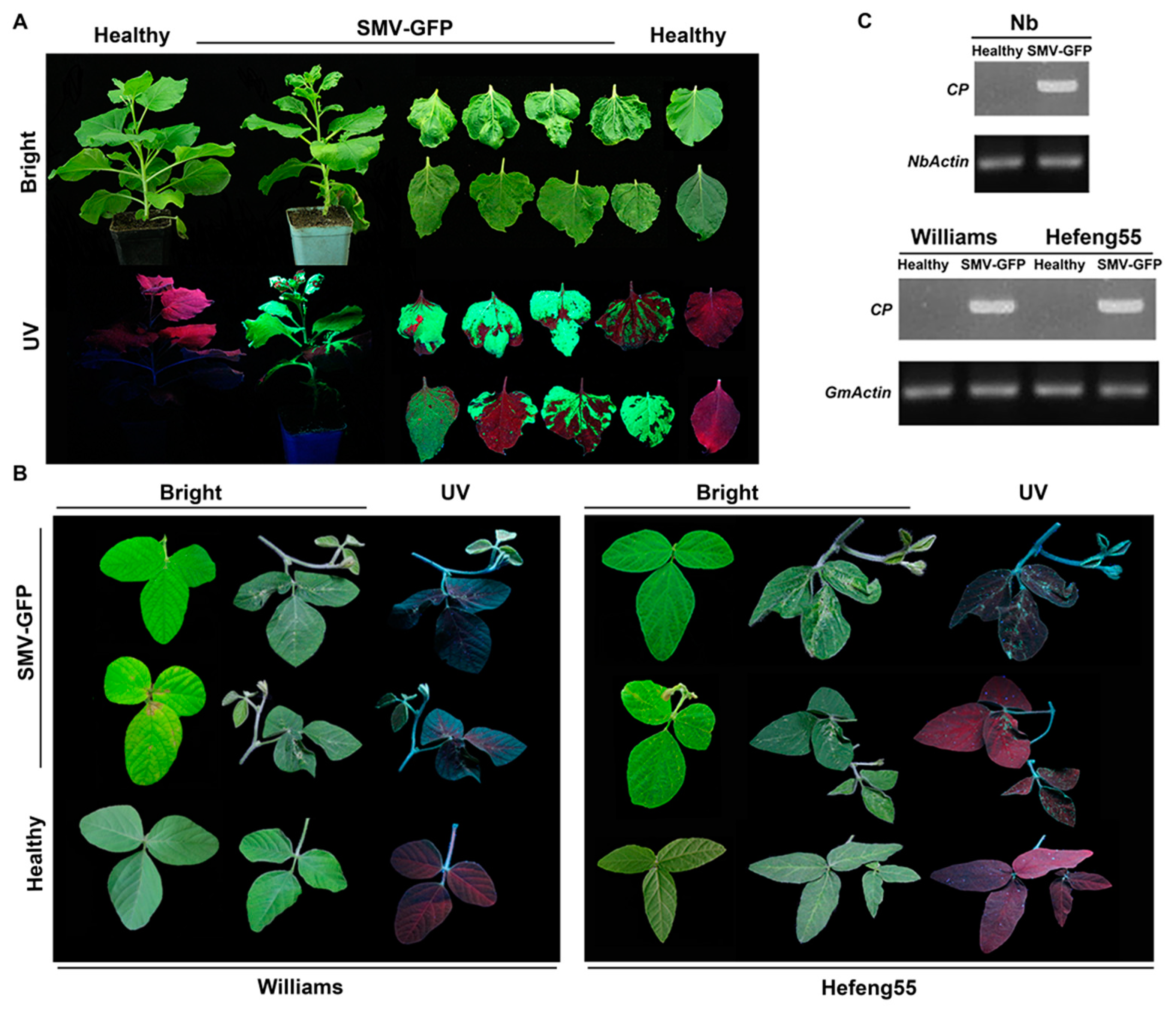
3.1. One-Step Assembly of SMV cDNA Infectious Clone and Its Infectivity
3.2. Construction of GFP-Tagged SMV Infectious Clone and Infection Assays
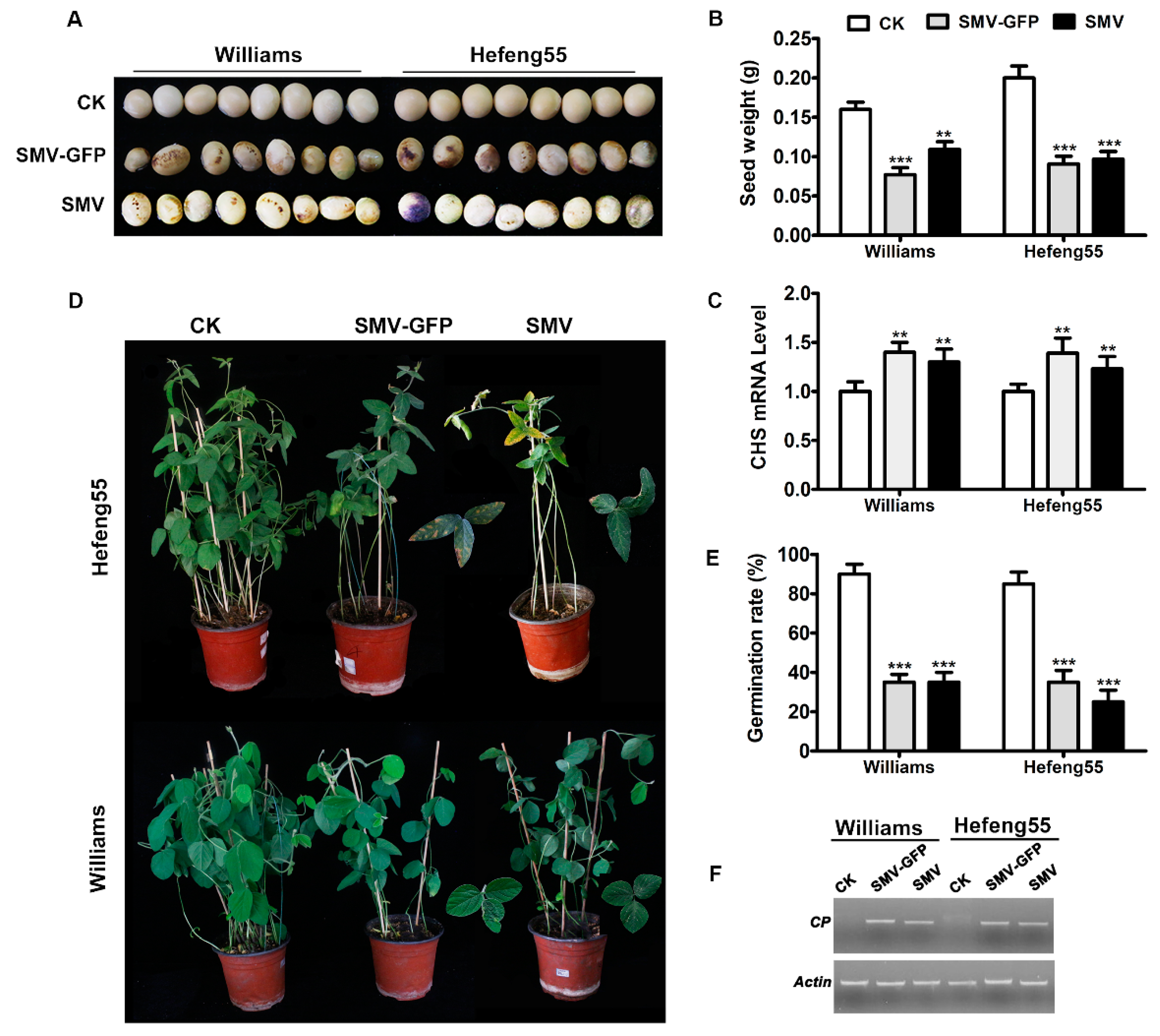
3.3. Seed Transmission of Infectious Clone-Derived SMV in Soybeans
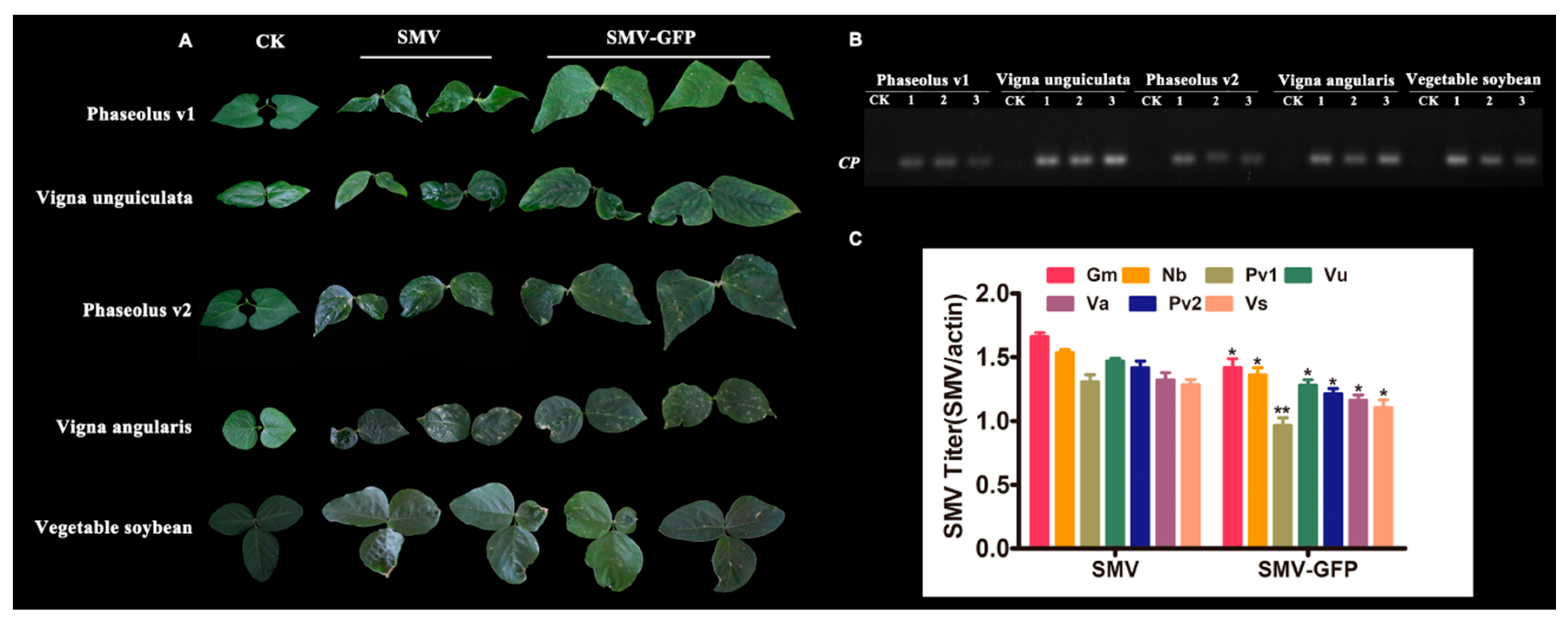
3.4. Host Ranges of SMV Derived from Infectious Clones
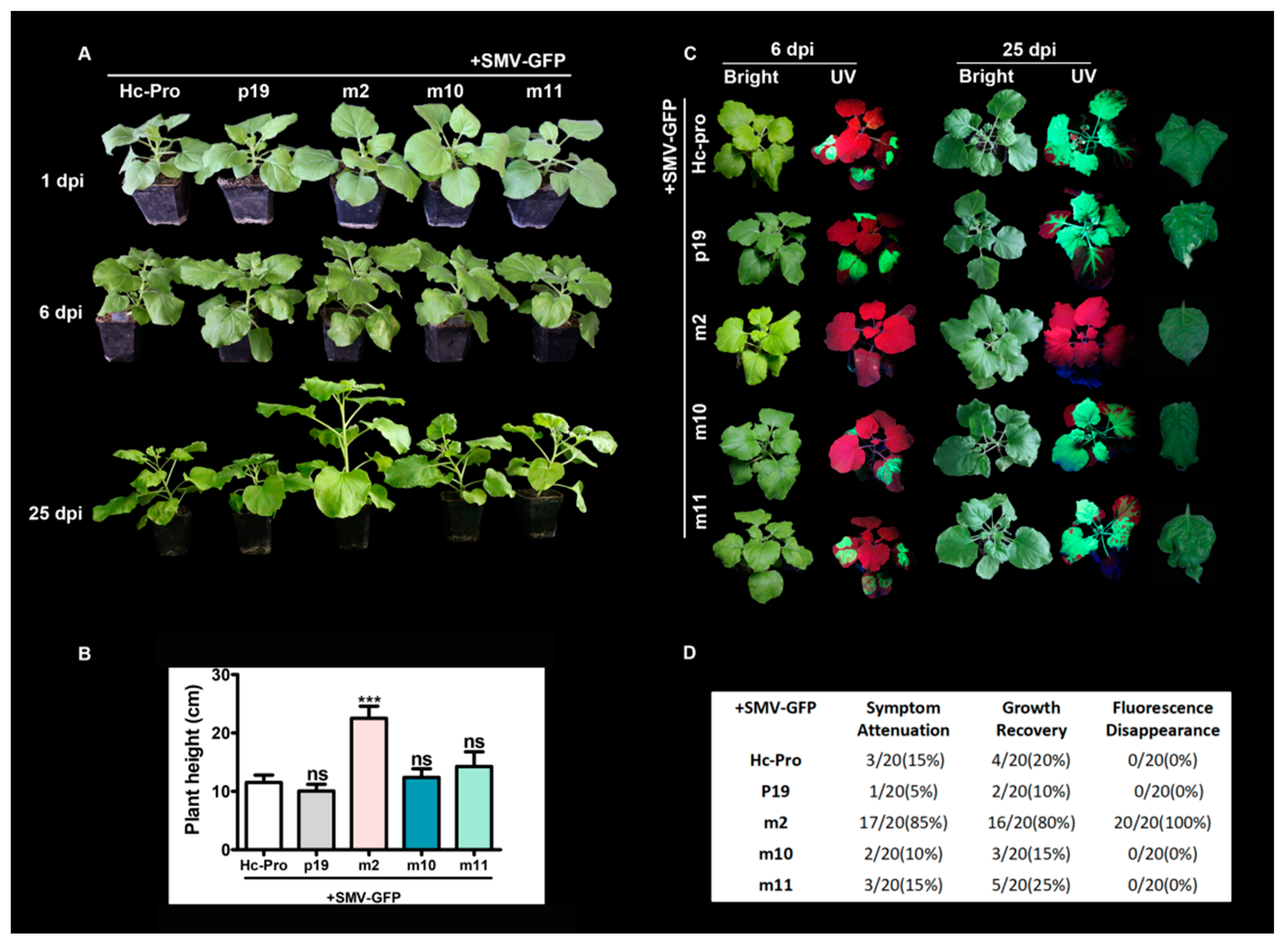
3.5. The Effects of Mutant Hc-Pro on SMV Infectivity and Symptom Development
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Scholthof, K.B.; Adkins, S.; Czosnek, H.; Palukaitis, P.; Jacquot, E.; Hohn, T.; Hohn, B.; Saunders, K.; Candresse, T.; Ahlquist, P.; et al. Top 10 plant viruses in molecular plant pathology. Mol. Plant Pathol. 2011, 12, 938–954. [Google Scholar] [CrossRef]
- Roossinck, M.J. Plant virus metagenomics: Biodiversity and ecology. Annu. Rev. Genet. 2012, 46, 359–369. [Google Scholar] [CrossRef] [PubMed]
- Rever, F.; García, J.A. Molecular biology of potyviruses. Adv. Virus Res. 2015, 92, 101–199. [Google Scholar]
- Carrington, J.C.; Freed, D.D.; Oh, C.S. Expression of potyviral polyproteins in transgenic plants reveals three proteolytic activities required for complete processing. EMBO J. 1990, 9, 1347–1353. [Google Scholar] [CrossRef] [PubMed]
- Wen, R.H.; Hajimorad, M.R. Mutational analysis of the putative pipo of soybean mosaic virus suggests disruption of PIPO protein impedes movement. Virology 2010, 400, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Hill, J.H.; Whitham, S.A. Control of virus diseases in soybeans. Adv. Virus Res. 2014, 90, 355–390. [Google Scholar]
- Widyasari, K.; Alazem, M.; Kim, K.H. Soybean Resistance to Soybean Mosaic Virus. Plants 2020, 9, 219. [Google Scholar] [CrossRef]
- Cho, E.K.; Goodman, R.M. Strains of soybean mosaic virus: Classification based on virulence in resistant soybean cultivars. Phytopathology 1979, 69, 467–470. [Google Scholar] [CrossRef]
- Saruta, M.; Kikuchi, A.; Okabe, A.; Sasaya, T. Molecular characterization of A2 and D strains of Soybean mosaic virus, which caused a recent virus outbreak in soybean cultivar Sachiyutaka in Chugoku and Shikoku regions of Japan. J. Gen. Plant Pathol. 2005, 71, 431–435. [Google Scholar] [CrossRef]
- Seo, J.K.; Lee, H.G.; Choi, H.S.; Lee, S.H.; Kim, K.H. Infectious in vivo Transcripts from a Full-length Clone of Soybean mosaic virus Strain G5H. Plant Pathol. J. 2009, 25, 54–61. [Google Scholar] [CrossRef][Green Version]
- Li, K.; Yang, Q.H.; Zhi, H.J.; Gai, J.Y. Identification and Distribution of Soybean mosaic virus Strains in Southern China. Plant Dis. 2010, 94, 351–357. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.G.; Li, H.W.; Zhi, H.J.; Tian, Z.; Hu, C.; Hu, G.Y.; Huang, Z.P.; Zhang, L. Identification of strains and screening of resistance resources to soybean mosaic virus in Anhui Province. Chin. J. Oil Crop Sci. 2014, 36, 374–379. [Google Scholar]
- Wang, Y.; Zhi, H.; Guo, D.; Gai, J.; Chen, Q.; Li, K.; Li, H. Classification and distribution of strain groups of soybean mosaic virus in Northern China spring planting soybean region. Soybean Sci. 2005, 24, 263–268. [Google Scholar]
- Jossey, S.; Hobbs, H.A.; Domier, L.L. Role of Soybean mosaic virus-Encoded Proteins in Seed and Aphid Transmission in Soybean. Phytopathology 2013, 103, 941–948. [Google Scholar] [CrossRef]
- Bowers, G.R.; Goodman, R.M. Strain Specificity of Soybean Mosaic Virus Seed Transmission in Soybean. Crop Sci. 1991, 31, 1171–1174. [Google Scholar] [CrossRef]
- Tuteja, J.H.; Zabala, G.; Varala, K.; Hudson, M.; Vodkin, L.O. Endogenous, Tissue-Specific Short Interfering RNAs Silence the Chalcone Synthase Gene Family in Glycine max Seed Coats. Plant Cell 2009, 21, 3063–3077. [Google Scholar] [CrossRef]
- Senda, M. Patterning of Virus-Infected Glycine max Seed Coat Is Associated with Suppression of Endogenous Silencing of Chalcone Synthase Genes. Plant Cell 2004, 16, 807–818. [Google Scholar] [CrossRef]
- Hartman, G.L.; Sinclair, J.B.; Rupe, J.C. Compendium of Soybean Diseases, 4th ed.; Soybean Disease Compendium: New York, NY, USA, 1999; pp. 37–39. [Google Scholar]
- Laviolette, F.A. Longevity of Tobacco Ringspot Virus in Soybean Seed. Phytopathology 1971, 6, 61. [Google Scholar] [CrossRef]
- Boyer, J.C.; Haenni, A.L. Infectious transcripts and cDNA clones of RNA viruses. Virology 1994, 198, 415–426. [Google Scholar] [CrossRef] [PubMed]
- Mori, M.; Mise, K.; Kobayashi, K.; Okuno, T.; Furusawa, I. Infectivity of plasmids containing brome mosaic virus cDNA linked to the cauliflower mosaic virus 35S RNA promoter. J. Gen. Virol. 1991, 243–246. [Google Scholar] [CrossRef]
- Sun, K.; Zhao, D.; Liu, Y.; Huang, C.; Zhang, W.; Li, Z. Rapid Construction of Complex Plant RNA Virus Infectious cDNA Clones for Agroinfection Using a Yeast-E. coli-Agrobacterium Shuttle Vector. Viruses 2017, 9, 332. [Google Scholar] [CrossRef] [PubMed]
- Jakab, G.; Droz, E.; Brigneti, G.; Baulcombe, D.; Malnoë, P. Infectious in vivo and in vitro transcripts from a full-length cDNA clone of PVY-N605, a Swiss necrotic isolate of potato virus Y. J. Gen. Virol. 1997, 3141–3145. [Google Scholar] [CrossRef] [PubMed]
- Tuo, D.; Shen, W.; Yan, P.; Li, X.; Zhou, P. Rapid Construction of Stable Infectious Full-Length cDNA Clone of Papaya Leaf Distortion Mosaic Virus Using In-Fusion Cloning. Viruses 2015, 7, 6241–6250. [Google Scholar] [CrossRef] [PubMed]
- Johansen, I.E. Intron insertion facilitates amplification of cloned virus cDNA in Escherichia coli while biological activity is reestablished after transcription in vivo. Proc. Natl. Acad. Sci. USA 1996, 93, 12400–12405. [Google Scholar] [CrossRef] [PubMed]
- Larionov, V.; Kouprina, N.; Graves, J.; Chen, X.N.; Korenberg, J.R.; Resnick, M.A. Specific cloning of human DNA as yeast artificial chromosomes by transformation-associated recombination. Proc. Natl. Acad. Sci. USA 1996, 93, 491–496. [Google Scholar] [CrossRef] [PubMed]
- Oldenburg, K.R.; Vo, K.T.; Michaelis, S.; Paddon, C. Recombination-mediated PCR-directed plasmid construction in vivo in yeast. Nucleic Acids Res. 1997, 25, 451–452. [Google Scholar] [CrossRef]
- Ding, S.W. RNA-based antiviral immunity. Nat. Rev. Immunol. 2010, 10, 632–644. [Google Scholar] [CrossRef]
- Bologna, N.G.; Voinnet, O. The diversity, biogenesis, and activities of endogenous silencing small RNAs in Arabidopsis. Annu. Rev. Plant Biol. 2014, 65, 473–503. [Google Scholar] [CrossRef]
- Kasschau, K.D.; Carrington, J.C. A Counterdefensive Strategy of Plant Viruses: Suppression of Posttranscriptional Gene Silencing. Cell 1998, 95, 461–470. [Google Scholar] [CrossRef]
- Anandalakshmi, R.; Pruss, G.J.; Ge, X.; Marathe, R.; Mallory, A.C.; Smith, T.H.; Vance, V.B. A viral suppressor of gene silencing in plants. Proc. Natl. Acad. Sci. USA 1998, 95, 13079–13084. [Google Scholar] [CrossRef]
- Lakatos, L.; Csorba, T.; Pantaleo, V.; Chapman, E.J.; Carrington, J.C.; Liu, Y.P.; Dolja, V.V.; Calvino, L.F.; López-Moya, J.J.; Burgyán, J. Small RNA binding is a common strategy to suppress RNA silencing by several viral suppressors. EMBO J. 2006, 25, 2768–2780. [Google Scholar] [CrossRef] [PubMed]
- Ivanov, K.I.; Eskelin, K.; Bašić, M.; De, S.; Lõhmus, A.; Varjosalo, M.; Mäkinen, K. Molecular insights into the function of the viral RNA silencing suppressor HCPro. Plant J. 2016, 85, 30–45. [Google Scholar] [CrossRef] [PubMed]
- Jamous, R.M.; Boonrod, K.; Fuellgrabe, M.W.; Ali-Shtayeh, M.S.; Krczal, G.; Wassenegger, M. The helper component-proteinase of the Zucchini yellow mosaic virus inhibits the Hua Enhancer 1 methyltransferase activity in vitro. J. Gen. Virol. 2011, 92, 2222–2226. [Google Scholar] [CrossRef] [PubMed]
- Hasiów-Jaroszewska, B.; Fares, M.A.; Elena, S.F. Molecular evolution of viral multifunctional proteins: The case of potyvirus HC-Pro. J. Mol. Evol. 2014, 78, 75–86. [Google Scholar] [CrossRef] [PubMed]
- Urcuqui-Inchima, S.; Maia, I.G.; Arruda, P.; Haenni, A.L.; Bernardi, F. Deletion mapping of the potyviral helper component-proteinase reveals two regions involved in RNA binding. Virology 2000, 268, 104–111. [Google Scholar] [CrossRef]
- Atreya, C.D.; Pirone, T.P. Mutational analysis of the helper component-proteinase gene of a potyvirus: Effects of amino acid substitutions, deletions, and gene replacement on virulence and aphid transmissibility. Proc. Natl. Acad. Sci. USA 1993, 90, 11919–11923. [Google Scholar] [CrossRef]
- Shiboleth, Y.M.; Haronsky, E.; Leibman, D.; Arazi, T.; Wassenegger, M.; Whitham, S.A.; Gaba, V.; Gal-On, A. The conserved FRNK box in HC-Pro, a plant viral suppressor of gene silencing, is required for small RNA binding and mediates symptom development. J. Virol. 2007, 81, 13135–13148. [Google Scholar] [CrossRef]
- Peng, Y.H.; Kadoury, D.; Gal-On, A.; Huet, H.; Wang, Y.; Raccah, B. Mutations in the HC-Pro gene of zucchini yellow mosaic potyvirus: Effects on aphid transmission and binding to purified virions. J. Gen. Virol. 1998, 897–904. [Google Scholar] [CrossRef]
- Guo, B.; Lin, J.; Ye, K. Structure of the autocatalytic cysteine protease domain of potyvirus helper-component proteinase. J. Biol. Chem. 2011, 286, 21937–21943. [Google Scholar] [CrossRef]
- Gal-On, A. A Point Mutation in the FRNK Motif of the Potyvirus Helper Component-Protease Gene Alters Symptom Expression in Cucurbits and Elicits Protection against the Severe Homologous Virus. Phytopathology 2000, 90, 467–473. [Google Scholar] [CrossRef]
- Anandalakshmi, R.; Marathe, R.; Ge, X.; Herr, J.M.; Mau, C.; Mallory, A.; Pruss, G.; Bowman, L.; Vance, V.B. A calmodulin-related protein that suppresses posttranscriptional gene silencing in plants. Science 2000, 290, 142–144. [Google Scholar] [CrossRef] [PubMed]
- Tena, F.; González, I.; Doblas, P.; Rodríguez, C.; Sahana, N.; Kaur, H.; Tenllado, F.; Praveen, S.; Canto, T. The influence of cis-acting P1 protein and translational elements on the expression of Potato virus Y helper-component proteinase (HCPro) in heterologous systems and its suppression of silencing activity. Mol. Plant Pathol. 2013, 14, 530–541. [Google Scholar] [CrossRef] [PubMed]
- Thornbury, D.W.; Hellmann, G.M.; Rhoads, R.E.; Pirone, T.P. Purification and characterization of potyvirus helper component. Virology 1985, 144, 260–267. [Google Scholar] [CrossRef]
- Guo, D.; Merits, A.; Saarma, M. Self-association and mapping of interaction domains of helper component-proteinase of potato A potyvirus. J. Gen. Virol. 1999, 1127–1131. [Google Scholar] [CrossRef]
- Urcuqui-Inchima, S.; Walter, J.; Drugeon, G.; German-Retana, S.; Haenni, A.L.; Candresse, T.; Bernardi, F.; Le, G.O. Potyvirus helper component-proteinase self-interaction in the yeast two-hybrid system and delineation of the interaction domain involved. Virology 1999, 258, 95–99. [Google Scholar] [CrossRef]
- Plisson, C.; Drucker, M.; Blanc, S.; German-Retana, S.; Le, G.O.; Thomas, D.; Bron, P. Structural characterization of HC-Pro, a plant virus multifunctional protein. J. Biol. Chem. 2003, 278, 23753–23761. [Google Scholar] [CrossRef]
- Zheng, H.; Yan, F.; Lu, Y.; Sun, L.; Lin, L.; Cai, L.; Hou, M.; Chen, J. Mapping the self-interacting domains of TuMV HC-Pro and the subcellular localization of the protein. Virus Genes 2011, 42, 110–116. [Google Scholar] [CrossRef]
- Lindbo, J.A. TRBO: A High-Efficiency Tobacco Mosaic Virus RNA-Based Overexpression Vector. Plant Physiol. 2007, 145, 1232–1240. [Google Scholar] [CrossRef]
- Diao, P.; Zhang, Q.; Sun, H.; Ma, W.; Cao, A.; Yu, R.; Wang, J.; Niu, Y.; Wuriyanghan, H. NbAGO2 miR403a and SA Are Involved in Mediated Antiviral Defenses Against TMV Infection in. Genes 2019, 10, 526. [Google Scholar] [CrossRef]
- Bao, D.; Ganbaatar, O.; Cui, X.; Yu, R.; Bao, W.; Falk, B.W.; Wuriyanghan, H. Down-regulation of genes coding for core RNAi components and disease resistance proteins via corresponding microRNAs might be correlated with successful Soybean mosaic virus infection in soybean. Mol. Plant Pathol. 2018, 19, 948–960. [Google Scholar] [CrossRef]
- Zhao, Q.; Li, H.; Sun, H.; Li, A.; Liu, S.; Yu, R.; Cui, X.; Zhang, D.; Wuriyanghan, H. Salicylic acid and broad spectrum of NBS-LRR family genes are involved in SMV-soybean interactions. Plant Physiol. Biochem. 2018, 123, 132–140. [Google Scholar] [CrossRef] [PubMed]
- Pasin, F.; Bedoya, L.C.; Bernabé-Orts, J.M.; Gallo, A.; Simón-Mateo, C.; Orzaez, D.; García, J.A. Multiple T-DNA Delivery to Plants Using Novel Mini Binary Vectors with Compatible Replication Origins. ACS Synth. Biol. 2017, 6, 1962–1968. [Google Scholar] [CrossRef] [PubMed]
- Brewer, H.C.; Hird, D.L.; Bailey, A.M.; Seal, S.E.; Foster, G.D. A guide to the contained use of plant virus infectious clones. Plant Biotechnol. J. 2018, 16, 832–843. [Google Scholar] [CrossRef] [PubMed]
- Gao, R.; Tian, Y.P.; Wang, J.; Yin, X.; Li, X.D.; Valkonen, J.P. Construction of an infectious cDNA clone and gene expression vector of Tobacco vein banding mosaic virus (genus Potyvirus). Virus Res. 2012, 169, 276–281. [Google Scholar] [CrossRef]
- López-Moya, J.J.; García, J.A. Construction of a stable and highly infectious intron-containing cDNA clone of plum pox potyvirus and its use to infect plants by particle bombardment. Virus Res. 2000, 68, 99–107. [Google Scholar] [CrossRef]
- Yamshchikov, V.; Mishin, V.; Cominelli, F. A new strategy in design of +RNA virus infectious clones enabling their stable propagation in E. coli. Virology 2001, 281, 272–280. [Google Scholar] [CrossRef]
- Fakhfakh, H.; Vilaine, F.; Makni, M.; Robaglia, C. Cell-free cloning and biolistic inoculation of an infectious cDNA of potato virus Y. J. Gen. Virol. 1996, 519–523. [Google Scholar] [CrossRef]
- Flasinski, S.; Gunasinghe, U.B.; Gonzales, R.A.; Cassidy, B.G. The cDNA sequence and infectious transcripts of peanut stripe virus. Gene 1996, 171, 299–300. [Google Scholar] [CrossRef]
- Hurrelbrink, R.J.; Nestorowicz, A.; McMinn, P.C. Characterization of infectious Murray Valley encephalitis virus derived from a stably cloned genome-length cDNA. J. Gen. Virol. 1999, 3115–3125. [Google Scholar] [CrossRef]
- Desbiez, C.; Chandeysson, C.; Lecoq, H.; Moury, B. A simple, rapid and efficient way to obtain infectious clones of potyviruses. J. Virol. Methods 2012, 183, 94–97. [Google Scholar] [CrossRef]
- Fernandez-Delmond, I.; Pierrugues, O.; Wispelaere, M.; Guilbaud, L.; Gaubert, S.; Divéki, Z.; Godon, C.; Tepfer, M.; Jacquemond, M. A novel strategy for creating recombinant infectious RNA virus genomes. J. Virol. Methods 2004, 121, 247–257. [Google Scholar] [CrossRef] [PubMed]
- Youssef, F.; Marais, A.; Faure, C.; Gentit, P.; Candresse, T. Strategies to facilitate the development of uncloned or cloned infectious full-length viral cDNAs: Apple chlorotic leaf spot virus as a case study. Virol. J. 2011, 8, 488. [Google Scholar] [CrossRef] [PubMed]
- Polo, S.; Ketner, G.; Levis, R.; Falgout, B. Infectious RNA transcripts from full-length dengue virus type 2 cDNA clones made in yeast. J. Virol. 1997, 71, 5366–5374. [Google Scholar] [CrossRef] [PubMed]
- Bordat, A.; Houvenaghel, M.C.; German-Retana, S. Gibson assembly: An easy way to clone potyviral full-length infectious cDNA clones expressing an ectopic VPg. Virol. J. 2015, 12, 89. [Google Scholar] [CrossRef] [PubMed]
- Tuo, D.; Fu, L.; Shen, W.; Li, X.; Zhou, P.; Yan, P. Generation of stable infectious clones of plant viruses by using Rhizobium radiobacter for both cloning and inoculation. Virology 2017, 510, 99–103. [Google Scholar] [CrossRef]
- Blawid, R.; Nagata, T. Construction of an infectious clone of a plant RNA virus in a binary vector using one-step Gibson Assembly. J. Virol. Methods 2015, 222, 11–15. [Google Scholar] [CrossRef]
- Seo, J.K.; Lee, H.G.; Kim, K.H. Systemic gene delivery into soybean by simple rub-inoculation with plasmid DNA of a Soybean mosaic virus-based vector. Arch. Virol. 2009, 154, 87–99. [Google Scholar] [CrossRef]
- Zhang, L.; Wang, D.; Pei, Y.; Xian, S.; Niu, Y. Construction and characterization of an infectious clone of Soybean mosaic virus isolate from Pinellia ternata. Chin. J. Biotechnol. 2020, 36, 949–958. [Google Scholar]
- Yin, J.; Liu, H.; Xiang, W.; Jin, T.; Guo, D.; Wang, L.; Zhi, H. Discovery of the Agrobacterium growth inhibition sequence in virus and its application to recombinant clone screening. AMB Express 2019, 9, 116. [Google Scholar] [CrossRef]
- Domier, L.L.; Steinlage, T.A.; Hobbs, H.A.; Wang, Y.; Herrera-Rodriguez, G.; Haudenshield, J.S.; McCoppin, N.K.; Hartman, G.L. Similarities in Seed and Aphid Transmission Among Soybean mosaic virus Isolates. Plant Dis. 2007, 91, 546–550. [Google Scholar] [CrossRef]
- Hobbs, H.A.; Hartman, G.L.; Wang, Y.; Hill, C.B.; Bernard, R.L.; Pedersen, W.L.; Domier, L.L. Occurrence of Seed Coat Mottling in Soybean Plants Inoculated with Bean pod mottle virus and Soybean mosaic virus. Plant Dis. 2003, 87, 1333–1336. [Google Scholar] [CrossRef] [PubMed]
- Porto, M.D.M. Seed Transmission of a Brazilian Isolate of Soybean Mosaic Virus. Phytopathology 1975, 65, 713–716. [Google Scholar] [CrossRef]
- To, J.C. Effect of Different Strains of Soybean Mosaic Virus on Growth, Maturity, Yield, Seed Mottling and Seed Transmission in Several Soybean Cultivars. J. Phytopathol. 1989, 126, 231–236. [Google Scholar] [CrossRef]
- Ren, Q.; Pfeiffer, T.W.; Ghabrial, S.A. Soybean Mosaic Virus Incidence Level and Infection Time: Interaction Effects on Soybean. Crop Sci. 1997, 37, 1706–1711. [Google Scholar] [CrossRef]
- Seo, J.K.; Kang, S.H.; Seo, B.Y.; Jung, J.K.; Kim, K.H. Mutational analysis of interaction between coat protein and helper component-proteinase o Johansen, E.; Edwards, M.C.; Hampton, R.O. Seed Transmission of Viruses: Current Perspectives. Annu. Rev. Phytopathol. 1994, 32, 363–386. [Google Scholar]
- Seo, J.K.; Kang, S.H.; Seo, B.Y.; Jung, J.K.; KIM, K.H. Mutational analysis of interaction between coat protein and helper component-proteinase of Soybean mosaic virus involved in aphid transmission. Mol. Plant Pathol. 2010, 11, 265–276. [Google Scholar] [CrossRef]
- Simmons, H.E.; Dunham, J.P.; Zinn, K.E.; Munkvold, G.P.; Holmes, E.C.; Stephenson, A.G. Zucchini yellow mosaic virus (ZYMV, Potyvirus): Vertical transmission, seed infection and cryptic infections. Virus Res. 2013, 176, 259–264. [Google Scholar] [CrossRef]
- Groves, C.; German, T.; Dasgupta, R.; Mueller, D.; Smith, D.L. Seed Transmission of Soybean vein necrosis virus: The First Tospovirus Implicated in Seed Transmission. PLoS ONE 2016, 11, e0147342. [Google Scholar] [CrossRef]
- Benscher, D.; Pappu, S.S.; Niblett, C.L.; Agudelo, F.V.D.; Lee, R.F. A Strain of Soybean Mosaic Virus Infecting Passiflora spp. in Colombia. Plant Dis. 1996, 80, 258–262. [Google Scholar] [CrossRef]
- Chen, J.; Zheng, H.Y.; Lin, L.; Adams, M.J.; Antoniw, J.F.; Zhao, M.F.; Shang, Y.F.; Chen, J.P. A virus related to soybean mosaic virus from Pinellia ternata in China and its comparison with local soybean SMV isolates. Arch. Virol. 2004, 149, 349–363. [Google Scholar] [CrossRef]
- Sun, H.; ShenTu, S.; Xue, F.; Duns, G.; Chen, J. Molecular characterization and evolutionary analysis of soybean mosaic virus infecting Pinellia ternata in China. Virus Genes 2008, 36, 177–190. [Google Scholar] [CrossRef]
- Yoon, Y.; Lim, S.; Jang, Y.; Kim, B.-S.; Bae, D.; Maharjan, R.; Yi, H.; Bae, S.; Lee, Y.; Lee, B.; et al. First Report of Soybean mosaic virus and Soybean yellow mottle mosaic virus in Vigna angularis. Plant Dis. 2017. [Google Scholar] [CrossRef]
- Gao, L.; Ding, X.; Li, K.; Liao, W.; Zhong, Y.; Ren, R.; Liu, Z.; Adhimoolam, K.; Zhi, H. Characterization of Soybean mosaic virus resistance derived from inverted repeat-SMV-HC-Pro genes in multiple soybean cultivars. Theor. Appl. Genet. 2015, 128, 1489–1505. [Google Scholar] [CrossRef] [PubMed]
- Hunst, P.L.; Tolin, S.A. Isolation and comparison of two strains of soybean mosaic virus. Phytopathology 1982, 72, 710–713. [Google Scholar] [CrossRef]
- Ross, J.P. Pathogenic variation among isolates of soybean mosaic virus. Phytopathology 1969, 59, 829–832. [Google Scholar]
- Govier, D.A.; Kassanis, B.; Pirone, T.P. Partial purification and characterization of the potato virus Y helper component. Virology 1977, 78, 306–314. [Google Scholar] [CrossRef]
- Ruiz-Ferrer, V.; Boskovic, J.; Alfonso, C.; Rivas, G.; Llorca, O.; Lopez-Abella, D.; Lopez-Moya, J.J. Structural Analysis of Tobacco Etch Potyvirus HC-Pro Oligomers Involved in Aphid Transmission†. J. Virol. 2005, 79, 3758–3765. [Google Scholar] [CrossRef]
- Kasschau, K.D.; Cronin, S.; Carrington, J.C. Genome amplification and long-distance movement functions associated with the central domain of tobacco etch potyvirus helper component-proteinase. Virology 1997, 228, 251–262. [Google Scholar] [CrossRef]
- Wang, R.Y.; Pirone, T.P. Purification and Characterization of Turnip Mosaic Virus Helper Component Protein. Phytopathology 1999, 89, 564–567. [Google Scholar] [CrossRef]
- Desbiez, C.; Girard, M.; Lecoq, H. A novel natural mutation in HC-Pro responsible for mild symptomatology of Zucchini yellow mosaic virus (ZYMV, Potyvirus) in cucurbits. Arch. Virol. 2010, 155, 397–401. [Google Scholar] [CrossRef]

| Host | Symptoms Caused by SMV | Symptoms Caused by SMV-GFP |
|---|---|---|
| Vegetable soybean | M, SN, LC | NLL, SN, LC |
| Vigna unguiculata | SN, LC | M, SN, LC |
| Vigna angularis | M, NLL, LC, SN | M, NLL, LC, SN, |
| Vigna radiata | N | N |
| Phaseolus v1 | M, LC, SN | M, NLL, LC |
| Phaseolus v2 | M, LC, SN | M, NLL, LC, SN |
| Vicia faba | N | N |
| Astragalus sinicus | N | N |
| Medicago falcata | N | N |
| Glycine max v1 | N | N |
| Pisum sativum | N | N |
| Phaseolus v3 | N | N |
| Lablab purpureus | N | N |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bao, W.; Yan, T.; Deng, X.; Wuriyanghan, H. Synthesis of Full-Length cDNA Infectious Clones of Soybean Mosaic Virus and Functional Identification of a Key Amino Acid in the Silencing Suppressor Hc-Pro. Viruses 2020, 12, 886. https://doi.org/10.3390/v12080886
Bao W, Yan T, Deng X, Wuriyanghan H. Synthesis of Full-Length cDNA Infectious Clones of Soybean Mosaic Virus and Functional Identification of a Key Amino Acid in the Silencing Suppressor Hc-Pro. Viruses. 2020; 12(8):886. https://doi.org/10.3390/v12080886
Chicago/Turabian StyleBao, Wenhua, Ting Yan, Xiaoyi Deng, and Hada Wuriyanghan. 2020. "Synthesis of Full-Length cDNA Infectious Clones of Soybean Mosaic Virus and Functional Identification of a Key Amino Acid in the Silencing Suppressor Hc-Pro" Viruses 12, no. 8: 886. https://doi.org/10.3390/v12080886
APA StyleBao, W., Yan, T., Deng, X., & Wuriyanghan, H. (2020). Synthesis of Full-Length cDNA Infectious Clones of Soybean Mosaic Virus and Functional Identification of a Key Amino Acid in the Silencing Suppressor Hc-Pro. Viruses, 12(8), 886. https://doi.org/10.3390/v12080886




