Whole-Genome Sequencing of Human Enteroviruses from Clinical Samples by Nanopore Direct RNA Sequencing
Abstract
1. Introduction
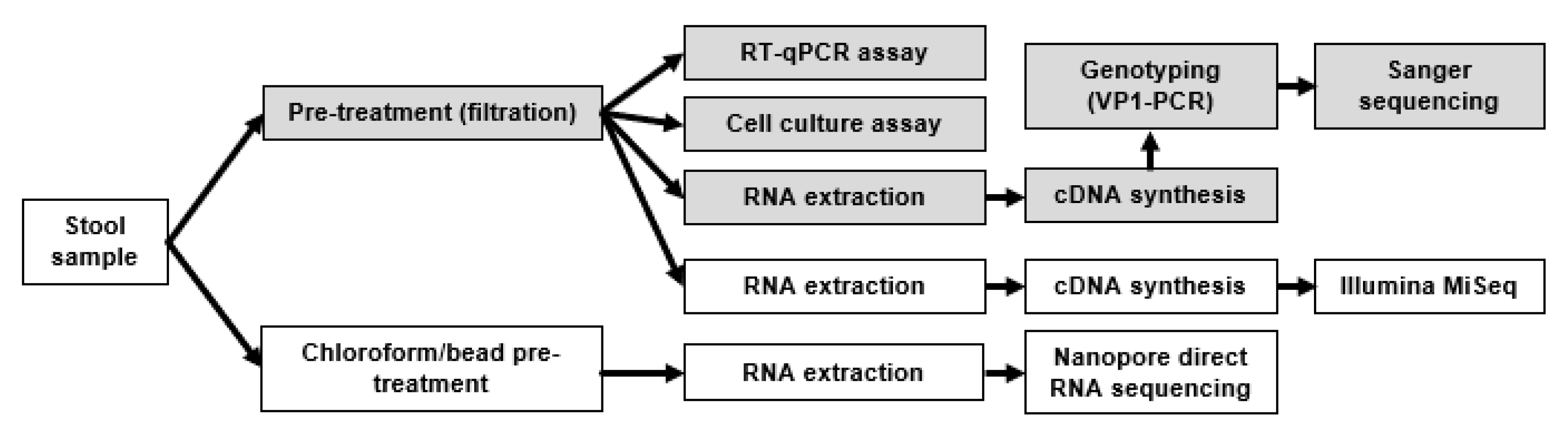
2. Materials and Methods
2.1. Sample Description
2.2. Routine Diagnostic Approach
2.3. RNA Extraction and Purification
2.4. Pre-Treatment (Chloroform/Beads/Centrifugation)
2.5. Real-Time RT PCR
2.6. Enterovirus Genotyping
2.7. Nanopore Sequencing
2.8. Illumina MiSeq. Primer Design for Coxsackievirus A6 cDNA Synthesis
2.9. cDNA Synthesis for Illumina MiSeq
2.10. Illumina MiSeq Sequencing
2.11. Bioinformatic Analysis
2.12. Data Availability
3. Results
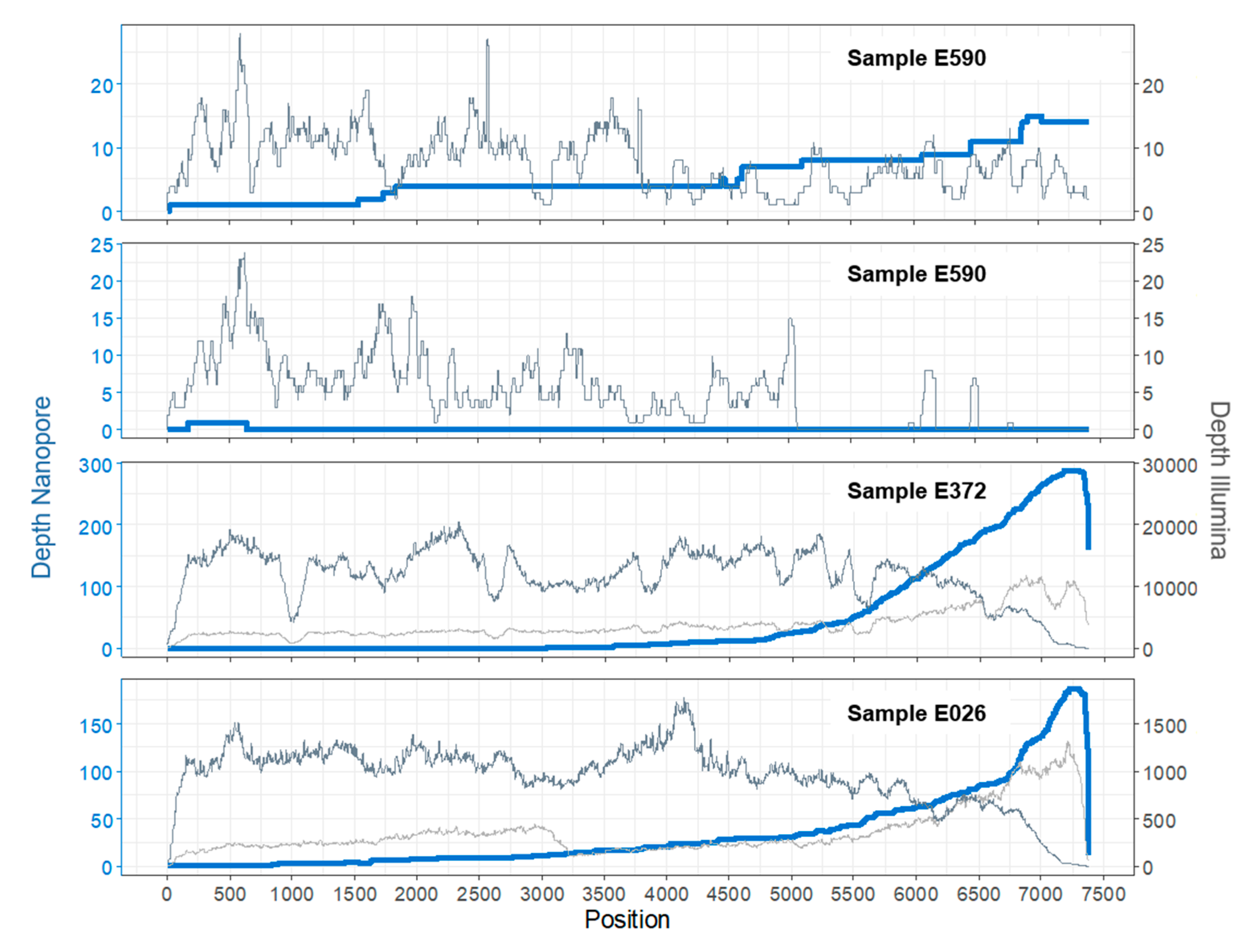
3.1. Sample E590
3.2. Sample E372
3.3. Sample E026
4. Discussion
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Tan, L.V.; Nguyen, T.K.T.; Tran, T.T.; Tran, T.N.; Hoang, M.T.V.; Saraswathy, S.; Tran, T.M.V.; Lei, T.M.T.; Lam, A.N.; Geoghegan, J.L. A generic assay for whole-genome amplification and deep sequencing of enterovirus A71. J. Virol. Methods 2015, 215, 30–36. [Google Scholar] [CrossRef] [PubMed]
- Royston, L.; Tapparel, C. Rhinoviruses and Respiratory Enteroviruses: Not. as Simple as ABC. Viruses 2016, 8, 16. [Google Scholar] [CrossRef] [PubMed]
- Oberste, M.S.; Maher, K.; Williams, A.J.; Dybdahl-Sissoko, N.; Brown, B.A.; Gookin, M.S.; Penaranda, S.; Mishrik, N.; Uddin, M.; Pallansch, M.A. Species-specific RT-PCR amplification of human enteroviruses: A tool for rapid species identification of uncharacterized enteroviruses. J. Gen. Virol. 2006, 87, 119–128. [Google Scholar] [CrossRef] [PubMed]
- Solomon, T.; Lewthwaite, P.; Perera, D.; Cardosa, M.J.; McMinn, P.; Ooi, M.H. Virology, epidemiology, pathogenesis, and control of enterovirus 71. Lancet Infect. Dis. 2010, 10, 778–790. [Google Scholar] [CrossRef]
- Yozwiak, N.L.; Skewes-Cox, P.; Gordon, A.; Saborio, S.; Kuan, G.; Balmaseda, A.; Ganem, D.; Harris, E.; DeRisi, J.L. Human enterovirus 109: A novel interspecies recombinant enterovirus isolated from a case of acute pediatric respiratory illness in Nicaragua. J. Virol. 2010, 84, 9047–9058. [Google Scholar] [CrossRef]
- Goodwin, S.; McPherson, J.D.; McCombie, W.R. Coming of age: Ten years of next-generation sequencing technologies. Nat. Rev. Genet. 2016, 17, 333–351. [Google Scholar] [CrossRef]
- Quick, J.; Grubaugh, N.D.; Pullan, S.T.; Claro, I.M.; Smith, A.D.; Gangavarapu, G.O.; Robles-Sikisaka, R.; Rogers, T.F.; Beutler, N.A.; Burton, D.R.; et al. Multiplex PCR method for MinION and Illumina sequencing of Zika and other virus genomes directly from clinical samples. Nat. Protoc. 2017, 12, 1261–1276. [Google Scholar] [CrossRef]
- Moldovan, N.; Balázs, Z.; Tombácz, D.; Csabai, Z.; Szűcs, A.; Snyder, M.; Boldogkői, Z. Multi-platform analysis reveals a complex transcriptome architecture of a circovirus. Virus Res. 2017, 237, 37–46. [Google Scholar] [CrossRef][Green Version]
- Tombacz, D.; Csabai, Z.; Szűcs, A.; Balázs, Z.; Moldován, N.; Sharon, D.; Boldogkői, M.S.Z. Long-Read. Isoform Sequencing Reveals a Hidden Complexity of the Transcriptional Landscape of Herpes Simplex Virus Type 1. Front. Microbiol. 2017, 8, 1079. [Google Scholar] [CrossRef]
- Uchida, Y.; Kouyama, J.; Naiki, K.; Uemura, H.; Tsuji, G.; Sugawara, K.; Nakao, M.; Motoya, D.; Nakayama, N.; Imai, Y. A case of genotype-3b hepatitis C virus in which the whole genome was successfully analyzed using third-generation nanopore sequencing. Hepatol. Res. 2019, 49, 1083–1087. [Google Scholar] [CrossRef]
- Eckert, S.E.; Chan, J.Z.M.; Houniet, D. Enrichment by hybridisation of long DNA fragments for Nanopore sequencing. Microb. Genom. 2016, 2, e000087. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Ke, Y.A.; Zhang, Y.; Huang, K.Q.; Wang, L.; Shen, X.X.; Dong, P.X.; Xu, W.B.; Ma, X.J. Rapid and Accurate Sequencing of Enterovirus Genomes Using MinION Nanopore Sequencer. Biomed. Environ. Sci. 2017, 30, 718–726. [Google Scholar] [PubMed]
- Lewandowski, K.; Pullan, S.T.; Lumley, S.F.; Foster, D.; Sanderson, N.; Vipond, R. Metagenomic Nanopore Sequencing of Influenza Virus Direct from Clinical Respiratory Samples. J. Clin. Microbiol. 2019, 58, e00963-19. [Google Scholar] [CrossRef] [PubMed]
- Garalde, D.R.; Warland, A.; James, P.; Admassu, T.; Pantic, N.; Bruce, M.; Lloyd, J.H.; Sipos, B.; Jachimowicz, D.; Snell, E.A.; et al. Highly parallel direct RNA sequencing on an array of nanopores. Nat. Methods 2018, 15, 201–206. [Google Scholar] [CrossRef] [PubMed]
- Workman, R.E.; Tang, A.D.; Tang, P.S.; Jain, M.; Tyson, J.R.; Zuzarte, P.C.; Gilpatrick, T.; Razaghi, R.; Quick, J.; Sadowski, N.; et al. Nanopore native RNA sequencing of a human poly(A) transcriptome. bioRxiv 2018, 459529. [Google Scholar] [CrossRef]
- Depledge, D.P.; Srinivas, K.P.; Sadaoka, T.; Bready, D.; Mori, Y.; Placantonakis, D.G.; Mohr, I.; Wilson, A.C. Direct RNA sequencing on nanopore arrays redefines the transcriptional complexity of a viral pathogen. Nat. Commun. 2019, 10, 754. [Google Scholar] [CrossRef]
- Moldovan, N.; Tombacz, D.; Szucs, A.; Csabai, Z.; Snyder, M.; Boldogkoi, Z. Multi-Platform Sequencing Approach Reveals a Novel Transcriptome Profile in Pseudorabies Virus. Front. Microbiol. 2017, 8, 2708. [Google Scholar] [CrossRef]
- Keller, M.W.; Rambo-Martin, B.L.; Wilson, M.M.; Ridenour, C.A.; Shepard, S.S.; Stark, T.J.; Neuhaus, E.B.; Dugan, V.G.; Wentworth, D.E.; Barnes, J.R. Direct RNA Sequencing of the Coding Complete Influenza A Virus Genome. Sci. Rep. 2018, 8, 14408. [Google Scholar] [CrossRef]
- Viehweger, A.; Krautwurst, S.; Lamiekiwicz, K.; Madhugiri, R.; Ziebuhr, J.; Holzer, M.; Marz, M. Direct RNA nanopore sequencing of full-length coron-avirus genomes provides novel insights into structural variants and enables modification analysis. bioRxiv 2019, 483693. [Google Scholar]
- Wongsurawat, T.; Jenjaroenpun, P.; Taylor, M.K.; Lee, J.; Tolardo, A.L.; Parvathareddy, J.; Kandel, S.; Wadley, T.D.; Kaewnapan, B.; Athipanyasilp, N. Rapid Sequencing of Multiple RNA Viruses in Their Native Form. Front. Microbiol. 2019, 10, 260. [Google Scholar] [CrossRef]
- Dubuis, O.; Gorgievski-Hrisoho, M.; Germann, D.; Matter, L. Evaluation of 2-SP transport medium for detection of Chlamydia trachomatis and Neisseria gonorrhoeae by two automated amplification systems and culture for chlamydia. J. Clin. Pathol. 1997, 50, 947–950. [Google Scholar] [CrossRef] [PubMed]
- WHO. Enterovirus Surveillance Guidelines: Guidelines for Enterovirus Surveillance in Support of the Polio Eradication Initiative; World Health Organization (WHO) Regional Office for Europe: Geneva, Switzerland, 2015; Available online: http://www.euro.who.int/__data/assets/pdf_file/0020/272810/EnterovirusSurveillanceGuidelines.pdf (accessed on 25 February 2018).
- Nix, A.W.; Oberste, M.S.; Pallansch, M.A. Sensitive, seminested PCR amplification of VP1 sequences for direct identification of all enterovirus serotypes from original clinical specimens. J. Clin. Microbiol. 2006, 44, 2698–2704. [Google Scholar] [CrossRef] [PubMed]
- Dierssen, U.; Rehren, F.; Henke-Gendo, C.; Harste, G.; Heim, A. Rapid routine detection of enterovirus RNA in cerebrospinal fluid by a one-step real-time RT-PCR assay. J. Clin. Virol. 2008, 42, 58–64. [Google Scholar] [CrossRef] [PubMed]
- Grädel, C.; Miani, T.A.M.; Barbani, T.M.; Leib, L.S.; Suter-Riniker, F.; Ramette, A. Rapid and Cost-Efficient Enterovirus Genotyping from Clinical Samples Using Flongle Flow Cells. Genes 2019, 10, 659. [Google Scholar] [CrossRef]
- Oberste, M.S.; Nix, A.W.; Maher, K.; Pallansch, A.M. Improved molecular identification of enteroviruses by RT-PCR and amplicon sequencing. J. Clin. Virol. 2003, 26, 375–377. [Google Scholar] [CrossRef]
- Bessaud, M.; Jegouic, S.; Joffret, L.M.; Barge, C.; Balanant, J.; Gouandjika-Vasilache, I.; Delpeyroux, F. Characterization of the genome of human enteroviruses: Design of generic primers for amplification and sequencing of different regions of the viral genome. J. Virol. Methods 2008, 149, 277–284. [Google Scholar] [CrossRef]
- Huson, D.H.; Auch, A.F.; Qi, J.; Schuster, S.C. MEGAN analysis of metagenomic data. Genome Res. 2007, 17, 377–386. [Google Scholar] [CrossRef]
- Li, H. Minimap2: Pairwise alignment for nucleotide sequences. Bioinformatics 2018, 34, 3094–3100. [Google Scholar] [CrossRef]
- Bushnell, B. BBMap: A Fast, Accurate, Splice-Aware Aligner. 2014. Available online: https:/sourceforge.net/projects/bbmap/ (accessed on 11 January 2018).
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef]
- Kroneman, A.; Vennema, H.; Deforche, K.; Avoort, H.v.D.; Penaranda, S.; Oberste, M.S.; Vinje, J.; Koopmans, M. An automated genotyping tool for enteroviruses and noroviruses. J. Clin. Virol. 2011, 51, 121–125. [Google Scholar] [CrossRef]
- Tan, S.; Dvorak, C.M.T.; Murtaugh, M.P. Rapid, Unbiased PRRSV Strain Detection Using MinION Direct RNA Sequencing and Bioinformatics Tools. Viruses 2019, 11, 1132. [Google Scholar] [CrossRef] [PubMed]
- Oberste, M.S.; Maher, K.; Kilpatrick, D.R.; Flemister, M.R.; Brown, B.A.; Pallansch, M.A. Typing of human enteroviruses by partial sequencing of VP1. J. Clin. Microbiol. 1999, 37, 1288–1293. [Google Scholar] [CrossRef] [PubMed]
- Brown, B.A.; Maher, K.; Flemister, M.R.; Naraghi-Arani, P.; Uddin, M.; Oberste, M.S.; Pallansch, M.A. Resolving ambiguities in genetic typing of human enterovirus species C clinical isolates and identification of enterovirus 96, 99 and 102. J. Gen. Virol. 2009, 90, 1713–1723. [Google Scholar] [CrossRef] [PubMed]
- Smith, M.A.; Ersavas, T.; Ferguson, J.M.; Liu, H.; Lucas, M.C.; Begik, O.; Bojarski, L.; Barton, K.; Novoa, E.M. Barcoding and demultiplexing Oxford Nanopore native RNA sequencing reads with deep residual learning. bioRxiv 2019, 864322. [Google Scholar]
| Sample E590 | Sample E372 | Sample E026 | ||||||
|---|---|---|---|---|---|---|---|---|
| Domain | Nanopore (reads) | Illumina MiSeq (contigs) | Nanopore (reads) | Illumina MiSeq (contigs) | Nanopore (reads) | Illumina MiSeq (contigs) | ||
| DRS | Genotype specific primers | DRS | Random hexamers | Oligo-dT primer | DRS | Random hexamers | Oligo-dT primer | |
| Eukaryota | 8850 | 1 | 10 | 19 | 28 | 1,349,407 | 227 | 56 |
| Bacteria | 199 | 38 | 12,967 | 12,411 | 14,125 | 65,408 | 45,096 | 11,004 |
| Archaea | 0 | 0 | 5 | 1 | 0 | 6 | 7 | 3 |
| Viruses | 16 | 17 | 315 | 64 | 40 | 1579 | 398 | 63 |
| Total | 9065 | 56 | 13,297 | 12,495 | 14,193 | 1,350,992 | 45,728 | 11,126 |
| Sample E590 | Sample E372 | Sample E026 | ||||||
|---|---|---|---|---|---|---|---|---|
| Run ID | E590-DRS | E590-MiSeq | E372-DRS | E372-MiSeqR6 | E372-MiSeqOdT | E026-DRS | E026-MiSeqR6 | E026-MiSeqOdT |
| Technology | DRS | MiSeq (custom primers) | DRS | MiSeq (random hexamers) | MiSeq (oligo dT) | DRS | MiSeq (random hexamers) | MiSeq (oligo dT) |
| Coxsackievirus A6 | 16* | 8 | − | − | − | − | − | − |
| Coxsackievirus A9 | − | − | − | − | − | 1 | − | − |
| Coxsackievirus B1 | − | − | − | − | − | 4 | − | − |
| Coxsackievirus B2 | − | − | 1 | − | − | − | − | − |
| Coxsackievirus B3 | − | 1 | 1 | − | − | 5 | − | − |
| Coxsackievirus B5 | − | − | 1 | − | − | 1 | − | − |
| Echovirus 18 | − | 4 | − | − | − | − | − | − |
| Echovirus 25 | − | − | − | − | − | 12 * | 1 * | 1 * |
| Echovirus 30 | − | − | 124 | 28 | 1 * | − | 9 | 2 |
| Echovirus 7 | − | − | 6 | − | − | − | − | − |
| Echovirus E12 | − | − | − | − | − | 5 | − | − |
| Echovirus E18 | − | − | − | − | − | 1 | − | − |
| Echovirus E9 | − | − | − | − | − | 1 | − | − |
| Enterovirus B106 | − | − | − | − | − | 2 | − | − |
| Enterovirus B80 | − | − | − | − | − | 1 | − | − |
| Enterovirus B81 | − | − | − | − | − | 5 | − | − |
| Enterovirus B85 | − | − | − | − | − | 2 | − | − |
| Enterovirus B86 | − | − | − | − | − | 1 | − | − |
| Enterovirus B88 | − | − | − | − | − | 4 | − | − |
| Rhinovirus A | − | − | − | 7 | 8 | − | − | − |
| Bovine arvovirus 3 | − | − | − | − | 1 | − | − | − |
| Cactus virus X | − | − | − | − | − | 21 | 12 * | 5 |
| Caprine arthritis encephalitis virus | − | − | − | − | − | − | 1 | − |
| Carrot cryptic virus | − | − | − | − | − | − | 4 | 1 |
| Caudovirales | − | − | − | 3 | 2 | − | 22 | 5 |
| Escherichia virus FI | − | − | − | − | − | − | 1 * | 2 |
| Escherichia virus phiX174 | − | − | − | 1 * | 1 | − | 2 | 1 |
| Gokushovirus WZ−2015a | − | − | − | − | − | − | − | 1 |
| Human gut gokushovirus | − | − | − | − | − | − | − | 1 |
| Melon necrotic spot virus | − | − | − | 1 | − | 8 | 1 * | 1 * |
| Pitaya virus X | − | − | − | − | − | − | 4 | − |
| Schlumbergera virus X | − | − | − | − | − | − | 12 | − |
| Tomato mosaic virus | − | − | − | 2 | 5 | 1274 * | 248 | 1 * |
| Tomato mottle mosaic virus | − | − | − | − | − | − | 5 | 1 |
| Unclass. bacterial viruses | − | − | − | 18 | 21 | − | 61 | 37 |
| Uncultured Microviridae | − | − | − | − | − | − | 1 | 1 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Grädel, C.; Terrazos Miani, M.A.; Baumann, C.; Barbani, M.T.; Neuenschwander, S.; Leib, S.L.; Suter-Riniker, F.; Ramette, A. Whole-Genome Sequencing of Human Enteroviruses from Clinical Samples by Nanopore Direct RNA Sequencing. Viruses 2020, 12, 841. https://doi.org/10.3390/v12080841
Grädel C, Terrazos Miani MA, Baumann C, Barbani MT, Neuenschwander S, Leib SL, Suter-Riniker F, Ramette A. Whole-Genome Sequencing of Human Enteroviruses from Clinical Samples by Nanopore Direct RNA Sequencing. Viruses. 2020; 12(8):841. https://doi.org/10.3390/v12080841
Chicago/Turabian StyleGrädel, Carole, Miguel A. Terrazos Miani, Christian Baumann, Maria Teresa Barbani, Stefan Neuenschwander, Stephen L. Leib, Franziska Suter-Riniker, and Alban Ramette. 2020. "Whole-Genome Sequencing of Human Enteroviruses from Clinical Samples by Nanopore Direct RNA Sequencing" Viruses 12, no. 8: 841. https://doi.org/10.3390/v12080841
APA StyleGrädel, C., Terrazos Miani, M. A., Baumann, C., Barbani, M. T., Neuenschwander, S., Leib, S. L., Suter-Riniker, F., & Ramette, A. (2020). Whole-Genome Sequencing of Human Enteroviruses from Clinical Samples by Nanopore Direct RNA Sequencing. Viruses, 12(8), 841. https://doi.org/10.3390/v12080841






