The Vaginal Virome—Balancing Female Genital Tract Bacteriome, Mucosal Immunity, and Sexual and Reproductive Health Outcomes?
Abstract
1. Introduction
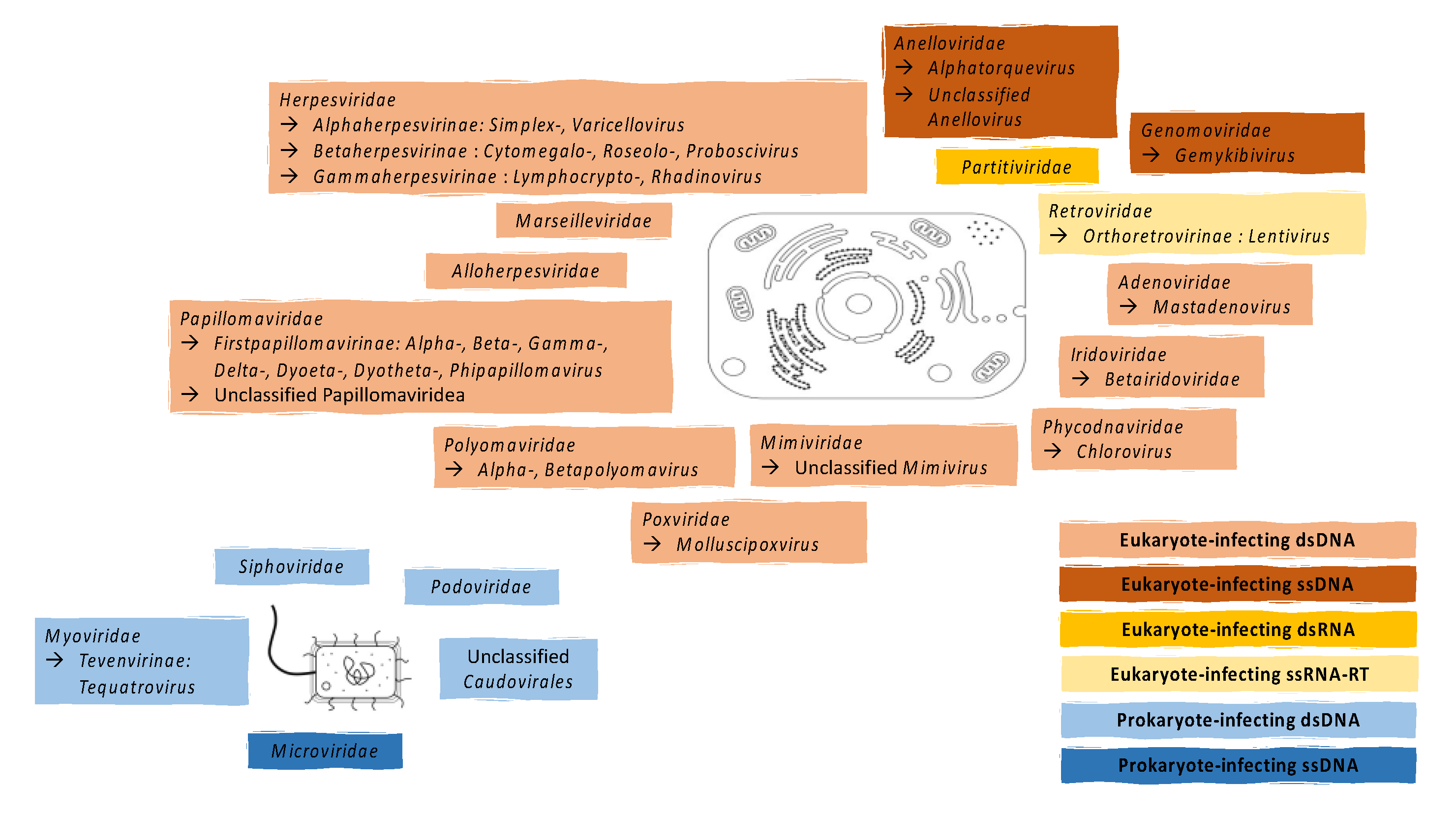
2. Known Components of the Vaginal Virome
2.1. Eukaryote-Infecting Viruses
2.2. Prokaryote-Infecting Viruses
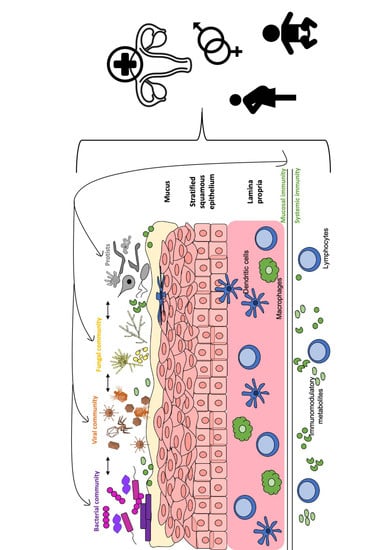
3. Interaction of the Vaginal Virome with Other Components of the Vaginal Microbiota and Human Host
3.1. Interactions between the Viral and Bacterial Microbiota
3.2. Interactions between the Viral and Fungal Microbiota
3.3. Interactions between Virome and Host Immunity
4. The Vaginal Virome and Adverse Sexual and Reproductive Outcomes
4.1. Eukaryotic-Infecting Viruses
4.2. Bacterial Vaginosis
4.3. Infertility
4.4. Adverse Birth Outcomes
5. Limitations of Current Research
6. Conclusions and Future Perspectives
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| ABO | Adverse birth outcome |
| BKPyV | BK polyomavirus |
| BV | Bacterial vaginosis |
| CMV | Cytomegalovirus |
| dsDNA | Double-stranded deoxyribonucleic acid |
| dsRNA | Double-stranded ribonucleic acid |
| FGT | Female genital tract |
| GBS | Group B Streptococcus |
| HHV | Human Herpesvirus |
| HHV | Human Herpesvirus |
| HIV | Human immunodeficiency virus |
| HMP | Human Microbiome Project |
| HPV | Human papillomavirus |
| HSV | Herpes simplex virus |
| IFN | Interferon |
| IP-10 | Interferon Gamma-induced Protein 10 |
| JCPyV | JC polyomavirus |
| MCP1 | Monocyte Chemotactic Protein 1 |
| MCPyV | Merkel cell polyomavirus |
| NK | Natural killer |
| PTB | Preterm birth |
| ssDNA | Single-stranded deoxyribonucleic acid |
| ssRNA-RT | Reverse transcribing single-stranded ribonucleic acid |
| SARS-CoV-2 | Severe acute respiratory syndrome coronavirus 2 |
| SGA | Small-for-gestational age |
| STI | Sexually transmitted infection |
| Th | T helper |
| OTU | Operational taxonomic unit |
References
- Ma, B.; France, M.T.; Crabtree, J.; Holm, J.B.; Humphrys, M.S.; Brotman, R.M.; Ravel, J. A comprehensive non-redundant gene catalog reveals extensive within-community intraspecies diversity in the human vagina. Nat. Commun. 2020, 11, 940. [Google Scholar] [CrossRef]
- Dridi, B.; Raoult, D.; Drancourt, M. Archaea as emerging organisms in complex human microbiomes. Anaerobe 2011, 17, 56–63. [Google Scholar] [CrossRef] [PubMed]
- Belay, N.; Mukhopadhyay, B.; Conway de Macario, E.; Galask, R.; Daniels, L. Methanogenic bacteria in human vaginal samples. J. Clin. Microbiol. 1990, 28, 1666–1668. [Google Scholar] [CrossRef] [PubMed]
- Virgin, H.W. The virome in mammalian physiology and disease. Cell 2014, 157, 142–150. [Google Scholar] [CrossRef] [PubMed]
- Wagner, J.; Maksimovic, J.; Farries, G.; Sim, W.H.; Bishop, R.F.; Cameron, D.J.; Catto-smith, A.G.; Kirkwood, C.D. Bacteriophages in Gut Samples From Pediatric Crohn’s Disease Patients: Metagenomic Analysis Using 454 Pyrosequencing. Inflamm Bowel Dis 2013, 19, 1598–1608. [Google Scholar] [CrossRef] [PubMed]
- Norman, J.M.; Handley, S.A.; Baldridge, M.T.; Droit, L.; Liu, C.Y.; Keller, B.C.; Kambal, A.; Monaco, C.L.; Zhao, G.; Fleshner, P.; et al. Disease-specific alterations in the enteric virome in inflammatory bowel disease. Cell 2015, 160, 447–460. [Google Scholar] [CrossRef] [PubMed]
- Monaco, C.L.; Gootenberg, D.B.; Zhao, G.; Handley, S.A.; Ghebremichael, M.S.; Lim, E.S.; Lankowski, A.; Baldridge, M.T.; Wilen, C.B.; Flagg, M.; et al. Altered Virome and Bacterial Microbiome in Human Immunodeficiency Virus-Associated Acquired Immunodeficiency Syndrome. Cell Host Microbe 2016, 19, 311–322. [Google Scholar] [CrossRef]
- Gogokhia, L.; Buhrke, K.; Bell, R.; Hoffman, B.; Brown, D.G.; Hanke-Gogokhia, C.; Ajami, N.J.; Wong, M.C.; Ghazaryan, A.; Valentine, J.F.; et al. Expansion of Bacteriophages Is Linked to Aggravated Intestinal Inflammation and Colitis. Cell Host Microbe 2019, 25, 285–299.e8. [Google Scholar] [CrossRef]
- Wylie, K.M.; Mihindukulasuriya, K.A.; Zhou, Y.; Sodergren, E.; Storch, G.A.; Weinstock, G.M. Metagenomic analysis of double-stranded DNA viruses in healthy adults. BMC Biol. 2014, 12, 71. [Google Scholar] [CrossRef]
- Schmitt, M.; Depuydt, C.; Benoy, I.; Bogers, J.; Antoine, J.; Arbyn, M.; Pawlita, M.; Group, on behalf of the V.S. Prevalence and viral load of 51 genital human papillomavirus types and three subtypes. Int. J. Cancer 2013, 132, 2395–2403. [Google Scholar] [CrossRef]
- Karolinska-Institutet International Human Papillomavirus Reference Center. Available online: https://www.hpvcenter.se/human_reference_clones/ (accessed on 18 May 2020).
- Wylie, K.M.; Wylie, T.N.; Cahill, A.G.; Macones, G.A.; Tuuli, M.G.; Stout, M.J. The vaginal eukaryotic DNA virome and preterm birth. Am. J. Obstet. Gynecol. 2018, 219, 189.e1–189.e12. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Chen, C.; Wei, W.; Wang, Z.; Dai, J.; Hao, L.; Song, L.; Zhang, X.; Zeng, L.; Du, H.; et al. The metagenome of the female upper reproductive tract. Gigascience 2018, 7. [Google Scholar] [CrossRef] [PubMed]
- Jakobsen, R.R.; Haahr, T.; Humaidan, P.; Jensen, J.S.; Kot, W.; Castro-Mejia, J.; Deng, L.; Leser, T.D.; Nielsen, D.S. Characterization of the vaginal DNA virome in health and dysbiosis: An opening study in patients with non-female factor infertility. bioRxiv 2019, 755710. [Google Scholar] [CrossRef]
- Eskew, A.M.; Stout, M.J.; Bedrick, B.S.; Riley, J.K.; Omurtag, K.R.; Jimenez, P.T.; Odem, R.R.; Ratts, V.S.; Keller, S.L.; Jungheim, E.S.; et al. Association of the eukaryotic vaginal virome with prophylactic antibiotic exposure and reproductive outcomes in a subfertile population undergoing in vitro fertilisation: A prospective exploratory study. BJOG An Int. J. Obstet. Gynaecol. 2020, 127, 208–216. [Google Scholar] [CrossRef] [PubMed]
- Siqueira, J.D.; Curty, G.; Xutao, D.; Hofer, C.B.; Machado, E.S.; Seuánez, H.N.; Soares, M.A.; Delwart, E.; Soares, E.A. Composite analysis of the virome and bacteriome of HIV/HPV co-infected women reveals proxies for immunodeficiency. Viruses 2019, 11, 9–12. [Google Scholar] [CrossRef] [PubMed]
- Ameur, A.; Meiring, T.L.; Bunikis, I.; Häggqvist, S.; Lindau, C.; Lindberg, J.H.; Gustavsson, I.; Mbulawa, Z.Z.A.; Williamson, A.-L.; Gyllensten, U. Comprehensive profiling of the vaginal microbiome in HIV positive women using massive parallel semiconductor sequencing. Sci. Rep. 2014, 4, 4398. [Google Scholar] [CrossRef] [PubMed]
- Grignolo, S.; Bruzzone, B.; Gabbi, L.; Gerbaldo, D.; Gallo, F.; Nigro, N.; Icardi, G.; Viscoli, C.; Di Biagio, A. Vaginal HIV-1 shedding among HIV-1 infected women in the current era of combined antiretroviral therapy: A cross sectional study. Virulence 2017, 8, 101–108. [Google Scholar] [CrossRef] [PubMed]
- Kovacs, A.; Wasserman, S.S.; Burns, D.; Wright, D.J.; Cohn, J.; Landay, A.; Weber, K.; Cohen, M.; Levine, A.; Minkoff, H.; et al. Determinants of HIV-1 shedding in the genital tract of women. Lancet 2001, 358, 1593–1601. [Google Scholar] [CrossRef]
- Bull, M.E.; Legard, J.; Tapia, K.; Sorensen, B.; Cohn, S.E.; Garcia, R.; Holte, S.E.; Coombs, R.W.; Hitti, J.E. HIV-1 shedding from the female genital tract is associated with increased Th1 cytokines/chemokines that maintain tissue homeostasis and proportions of CD8+FOXP3+ T cells. J. Acquir. Immune Defic. Syndr. 2014, 67, 357–364. [Google Scholar] [CrossRef]
- Pfeiffer, J.K.; Virgin, H.W. Viral immunity. Transkingdom control of viral infection and immunity in the mammalian intestine. Science 2016, 351, aad5872. [Google Scholar] [CrossRef]
- Kernbauer, E.; Ding, Y.; Cadwell, K. An enteric virus can replace the beneficial function of commensal bacteria. Nature 2014, 516, 94–98. [Google Scholar] [CrossRef]
- Metzger, R.N.; Krug, A.B.; Eisenächer, K. Enteric Virome Sensing-Its Role in Intestinal Homeostasis and Immunity. Viruses 2018, 10, 146. [Google Scholar] [CrossRef]
- Labrie, S.J.; Samson, J.E.; Moineau, S. Bacteriophage resistance mechanisms. Nat. Rev. Microbiol. 2010, 8, 317. [Google Scholar] [CrossRef] [PubMed]
- Manrique, P.; Bolduc, B.; Walk, S.T.; Van Der Oost, J.; De Vos, W.M.; Young, M.J. Healthy human gut phageome. PNAS 2016, 113, 10400–10405. [Google Scholar] [CrossRef] [PubMed]
- Lepage, P.; Colombet, J.; Marteau, P.; Some-Ngando, T.; Dore, J.; Leclerc, M. Dysbiosis in inflammatory bowel disease: A role for bacteriophages ? Gut Lett. 2008, 57, 424–425. [Google Scholar] [CrossRef]
- Parkes, G.C.; Whelan, K.; Lindsay, J.O. Smoking in inflammatory bowel disease: Impact on disease course and insights into the aetiology of its effect. J. Crohn’s Colitis 2014, 8, 717–725. [Google Scholar] [CrossRef] [PubMed]
- Pride, D.T.; Salzman, J.; Haynes, M.; Rohwer, F.; Davis-long, C.; Iii, R.A.W.; Loomer, P.; Armitage, G.C.; Relman, D.A. Evidence of a robust resident bacteriophage population revealed through analysis of the human salivary virome. ISME J. 2012, 6, 915–926. [Google Scholar] [CrossRef] [PubMed]
- Willner, D.; Furlan, M.; Schmieder, R.; Grasis, J.A.; Pride, D.T.; Relman, D.A.; Angly, F.E.; McDole, T.; Mariella, R.P.; Rohwer, F.; et al. Metagenomic detection of phage-encoded platelet- binding factors in the human oral cavity. PNAS 2011, 108, 4547–4553. [Google Scholar] [CrossRef]
- Marinelli, L.J.; Fitz-gibbon, S.; Hayes, C.; Marinelli, L.J.; Fitz-gibbon, S.; Hayes, C.; Bowman, C.; Inkeles, M.; Loncaric, A.; Russell, D.A. Propionibacterium acnes Bacteriophages Display Limited Genetic Diversity and Broad Killing Activity against Bacterial Skin Isolates. MBio 2012, 3. [Google Scholar] [CrossRef]
- Liu, J.; Yan, R.; Zhong, Q.; Ngo, S.; Bangayan, N.J.; Nguyen, L.; Lui, T.; Liu, M.; Erfe, M.C.; Craft, N.; et al. The diversity and host interactions of Propionibacterium acnes bacteriophages on human skin. ISME J. 2015, 9, 2078–2093. [Google Scholar] [CrossRef]
- Segura-Wang, M.; Gorzer, I.; Jaksch, P.; Puchhammer-Stockl, E. Temporal dynamics of the lung and plasma viromes in lung transplant recipients. PLoS ONE 2018, 13, e0200428. [Google Scholar] [CrossRef] [PubMed]
- Hobbs, Z.; Abedon, S.T. Diversity of phage infection types and associated terminology: The problem with ‘Lytic or lysogenic’ . FEMS Microbiol. Lett. 2016, 363. [Google Scholar] [CrossRef] [PubMed]
- Pavlova, S.I.; Tao, L. Induction of vaginal Lactobacillus phages by the cigarette smoke chemical benzo[a]pyrene diol epoxide. Mutat. Res. 2000, 466, 57–62. [Google Scholar] [CrossRef]
- Kilic, A.O.; Pavlova, S.I.; Alpay, S.; Kilic, S.S.; Tao, L. Comparative Study of Vaginal Lactobacillus Phages Isolated from Women in the United States and Turkey: Prevalence, Morphology, Host Range, and DNA Homology. Clin. Diagn. Lab. Immunol. 2001, 8, 31–39. Available online: http://cvi.asm.org/ (accessed on 13 February 2018). [CrossRef] [PubMed]
- Damelin, L.H.; Paximadis, M.; Mavri-damelin, D.; Birkhead, M.; Lewis, D.A.; Tiemessen, C.T. Identification of predominant culturable vaginal Lactobacillus species and associated bacteriophages from women with and without vaginal discharge syndrome in South Africa. J. Med. Microbiol. 2011, 60, 180–183. [Google Scholar] [CrossRef]
- Malki, K.; Shapiro, J.W.; Price, T.K.; Hilt, E.E.; Thomas-white, K.; Sircar, T.; Rosenfeld, A.B.; Kuffel, G.; Zilliox, M.J.; Wolfe, A.J.; et al. Genomes of Gardnerella Strains Reveal an Abundance of Prophages within the Bladder Microbiome. PLoS ONE 2016, 11, e0166757. [Google Scholar] [CrossRef]
- Martín, R.; Soberón, N.; Escobedo, S.; Suárez, J.E. Bacteriophage induction versus vaginal homeostasis: Role of H2O2 in the selection of Lactobacillus defective prophages. Int. Microbiol. 2009, 12, 131–136. [Google Scholar] [CrossRef]
- van der Mee-Marquet, N.; Diene, S.M.; Barbera, L.; Courtier-Martinez, L.; Lafont, L.; Ouachée, A.; Valentin, A.-S.; Dos Santos, S.; Quentin, R.; François, P. Analysis of the prophages carried by human infecting isolates provides new insight into the evolution of Group B Streptococcus species. Clin. Microbiol. Infect. 2018, 24, 514–521. [Google Scholar] [CrossRef]
- Miller-Ensminger, T.; Garretto, A.; Brenner, J.; Thomas-White, K.; Zambom, A.; Wolfe, A.J.; Putonti, C. Bacteriophages of the Urinary Microbiome. J. Bacteriol. 2018, 200. [Google Scholar] [CrossRef]
- Manrique, P.; Dills, M.; Young, M.J. The Human Gut Phage Community and Its Implications for Health and Disease. Viruses 2017, 9, 141. [Google Scholar] [CrossRef]
- Shannon, B.; Gajer, P.; Yi, T.J.; Ma, B.; Humphrys, M.S.; Thomas-Pavanel, J.; Chieza, L.; Janakiram, P.; Saunders, M.; Tharao, W.; et al. Distinct Effects of the Cervicovaginal Microbiota and Herpes Simplex Type 2 Infection on Female Genital Tract Immunology. J. Infect. Dis. 2017, 215, 1366–1375. [Google Scholar] [CrossRef] [PubMed]
- Torcia, M.G. Interplay among Vaginal Microbiome, Immune Response and Sexually Transmitted Viral Infections. Int. J. Mol. Sci. 2019, 20, 266. [Google Scholar] [CrossRef] [PubMed]
- Gosmann, C.; Anahtar, M.N.; Handley, S.A.; Farcasanu, M.; Abu-Ali, G.; Bowman, B.A.; Padavattan, N.; Desai, C.; Droit, L.; Moodley, A.; et al. Lactobacillus-Deficient Cervicovaginal Bacterial Communities Are Associated with Increased HIV Acquisition in Young South African Women. Immunity 2017, 46, 29–37. [Google Scholar] [CrossRef]
- Brotman, R. S11.3 The vaginal microenvironment prior to incident STI. Sex. Transm. Infect. 2019, 95, A19. [Google Scholar] [CrossRef]
- Jang, S.J.; Lee, K.; Kwon, B.; You, H.J.; Ko, G. Vaginal lactobacilli inhibit growth and hyphae formation of Candida albicans. Sci. Rep. 2019, 9, 8121. [Google Scholar] [CrossRef]
- Tortelli, B.A.; Lewis, W.G.; Allsworth, J.E.; Member-Meneh, N.; Foster, L.R.; Reno, H.E.; Peipert, J.F.; Fay, J.C.; Lewis, A.L. Associations between the vaginal microbiome and Candida colonization in women of reproductive age. Am. J. Obstet. Gynecol. 2020, 222, 471.e1–471.e9. [Google Scholar] [CrossRef]
- Hester, R.A.; Kennedy, S.B. Candida infection as a risk factor for HIV transmission. J. Womens Health (Larchmt) 2003, 12, 487–494. [Google Scholar] [CrossRef]
- van de Wijgert, J.H.H.M.; Morrison, C.S.; Cornelisse, P.G.A.; Munjoma, M.; Moncada, J.; Awio, P.; Wang, J.; Van der Pol, B.; Chipato, T.; Salata, R.A.; et al. Bacterial Vaginosis and Vaginal Yeast, But Not Vaginal Cleansing, Increase HIV-1 Acquisition in African Women. JAIDS J. Acquir. Immune Defic. Syndr. 2008, 48, 203–210. [Google Scholar] [CrossRef]
- Apalata, T.; Carr, W.H.; Sturm, W.A.; Longo-Mbenza, B.; Moodley, P. Determinants of Symptomatic Vulvovaginal Candidiasis among Human Immunodeficiency Virus Type 1 Infected Women in Rural KwaZulu-Natal, South Africa. Infect. Dis. Obstet. Gynecol. 2014, 2014, 387070. [Google Scholar] [CrossRef]
- Plotkin, B.J.; Sigar, I.M.; Tiwari, V.; Halkyard, S. Herpes Simplex Virus (HSV) Modulation of Staphylococcus aureus and Candida albicans Initiation of HeLa 299 Cell-Associated Biofilm. Curr. Microbiol. 2016, 72, 529–537. [Google Scholar] [CrossRef]
- Mazaheritehrani, E.; Sala, A.; Orsi, C.F.; Neglia, R.G.; Morace, G.; Blasi, E.; Cermelli, C. Human pathogenic viruses are retained in and released by Candida albicans biofilm in vitro. Virus Res. 2014, 179, 153–160. [Google Scholar] [CrossRef] [PubMed]
- Nazik, H.; Joubert, L.-M.; Secor, P.R.; Sweere, J.M.; Bollyky, P.L.; Sass, G.; Cegelski, L.; Stevens, D.A. Pseudomonas phage inhibition of Candida albicans. Microbiology 2017, 163, 1568–1577. [Google Scholar] [CrossRef]
- Cermelli, C.; Orsi, C.F.; Ardizzoni, A.; Lugli, E.; Cenacchi, V.; Cossarizza, A.; Blasi, E. Herpes simplex virus type 1 dysregulates anti-fungal defenses preventing monocyte activation and downregulating toll-like receptor-2. Microbiol. Immunol. 2008, 52, 575–584. [Google Scholar] [CrossRef]
- Gruber, A.; Lell, C.P.; Speth, C.; Stoiber, H.; Lass-Flörl, C.; Sonneborn, A.; Ernst, J.F.; Dierich, M.P.; Würzner, R. Human immunodeficiency virus type 1 Tat binds to Candida albicans, inducing hyphae but augmenting phagocytosis in vitro. Immunology 2001, 104, 455–461. [Google Scholar] [CrossRef] [PubMed]
- Würzner, R.; Gruber, A.; Stoiber, H.; Spruth, M.; Chen, Y.-H.; Lukasser-Vogl, E.; Schwendinger, M.G.; Dierich, M.P. Human Immunodeficiency Virus Type 1 gp41 Binds to Candida albicans via Complement C3-like Region. J. Infect. Dis. 1997, 176, 492–498. [Google Scholar] [CrossRef] [PubMed]
- Gruber, A.; Lukasser-Vogl, E.; Zepelin, M.B.; Dierich, M.P.; Würzner, R. Human Immunodeficiency Virus Type 1 gp160 and gp41 Binding to Candida albicans Selectively Enhances Candidal Virulence In Vitro. J. Infect. Dis. 1998, 177, 1057–1063. [Google Scholar] [CrossRef] [PubMed]
- Murphy, K.; Mitchell, C.M.; Hospital, M.G.; Hospital, M.G. The interplay of host immunity and environment on risk of bacterial vaginosisand associated reproductive health outcomes. J. Infect. Dis. 2016, 214 (Suppl. S1), S29–S35. [Google Scholar] [CrossRef]
- Muzny, C.A.; Laniewski, P.; Schwebke, J.R.; Herbst-Kralovetz, M.M. Host-vaginal microbiota interactions in the pathogenesis of bacterial vaginosis. Curr. Opin. Infect. Dis. 2020, 33, 59–65. [Google Scholar] [CrossRef]
- Amjadi, F.; Salehi, E.; Mehdizadeh, M.; Aflatoonian, R. Role of the innate immunity in female reproductive tract. Adv. Biomed. Res. 2014, 3, 1. [Google Scholar] [CrossRef]
- Nguyen, P.V.; Kafka, J.K.; Ferreira, V.H.; Roth, K.; Kaushic, C. Innate and adaptive immune responses in male and female reproductive tracts in homeostasis and following HIV infection. Cell. Mol. Immunol. 2014, 11, 410–427. [Google Scholar] [CrossRef]
- Campos, A.C.C.; Murta, E.F.C.; Michelin, M.A.; Reis, C. Evaluation of Cytokines in Endocervical Secretion and Vaginal pH from Women with Bacterial Vaginosis or Human Papillomavirus. ISRN Obstet. Gynecol. 2012, 2012, 342075. [Google Scholar] [CrossRef] [PubMed]
- Scott, M.E.; Shvetsov, Y.B.; Thompson, P.J.; Hernandez, B.Y.; Zhu, X.; Wilkens, L.R.; Killeen, J.; Vo, D.D.; Moscicki, A.-B.; Goodman, M.T. Cervical cytokines and clearance of incident human papillomavirus infection: Hawaii HPV cohort study. Int. J. Cancer 2013, 133, 1187–1196. [Google Scholar] [CrossRef] [PubMed]
- Liebenberg, L.J.P.; McKinnon, L.R.; Yende-Zuma, N.; Garrett, N.; Baxter, C.; Kharsany, A.B.M.; Archary, D.; Rositch, A.; Samsunder, N.; Mansoor, L.E.; et al. HPV infection and the genital cytokine milieu in women at high risk of HIV acquisition. Nat. Commun. 2019, 10, 5227. [Google Scholar] [CrossRef] [PubMed]
- Thépaut, M.; Grandjean, T.; Hober, D.; Lobert, P.-E.; Bortolotti, P.; Faure, K.; Dessein, R.; Kipnis, E.; Guery, B. Protective role of murine norovirus against Pseudomonas aeruginosa acute pneumonia. Vet. Res. 2015, 46, 91. [Google Scholar] [CrossRef][Green Version]
- Lee, L.-Y.; Garland, S.M. Human papillomavirus vaccination: The population impact. F1000Research 2017, 6, 866. [Google Scholar] [CrossRef]
- Spicknall, I.H.; Looker, K.J.; Gottlieb, S.L.; Chesson, H.W.; Schiffer, J.T.; Elmes, J.; Boily, M.-C. Review of mathematical models of HSV-2 vaccination: Implications for vaccine development. Vaccine 2019, 37, 7396–7407. [Google Scholar] [CrossRef]
- Looker, K.J.; Elmes, J.A.R.; Gottlieb, S.L.; Schiffer, J.T.; Vickerman, P.; Turner, K.M.E.; Boily, M.-C. Effect of HSV-2 infection on subsequent HIV acquisition: An updated systematic review and meta-analysis. Lancet Infect. Dis. 2017, 17, 1303–1316. [Google Scholar] [CrossRef]
- Si, J.; Yu, C.; Guo, Y.; Bian, Z.; Meng, R.; Yang, L.; Chen, Y.; Jin, J.; Liu, J.; Guo, Z.; et al. Chronic hepatitis B virus infection and total and cause-specific mortality: A prospective cohort study of 0.5 million people. BMJ Open 2019, 9, e027696. [Google Scholar] [CrossRef]
- Sangkomkamhang, U.S.; Lumbiganon, P.; Laopaiboon, M. Hepatitis B vaccination during pregnancy for preventing infant infection. Cochrane Database Syst. Rev. 2014. [Google Scholar] [CrossRef]
- Lanzieri, T.M.; Dollard, S.C.; Bialek, S.R.; Grosse, S.D. Systematic review of the birth prevalence of congenital cytomegalovirus infection in developing countries. Int. J. Infect. Dis. 2014, 22, 44–48. [Google Scholar] [CrossRef]
- Clark, P.; Kurtzer, T.; Duff, P. Role of Bacterial Vaginosis in Peripartum Infections. Infect. Dis. Obstet. Gynecol. 1994, 2, 179–183. [Google Scholar] [CrossRef] [PubMed]
- Nelson, D.B.; Hanlon, A.; Nachamkin, I.; Haggerty, C.; Mastrogiannis, D.S.; Liu, C.; Fredricks, D.N. Early pregnancy changes in bacterial vaginosis-associated bacteria and preterm delivery. Paediatr. Perinat. Epidemiol. 2014, 28, 88–96. [Google Scholar] [CrossRef] [PubMed]
- Haggerty, C.L.; Hillier, S.L.; Bass, D.C.; Ness, R.B.; Evaluation, P.I.D.; study investigators, C.H. Bacterial vaginosis and anaerobic bacteria are associated with endometritis. Clin. Infect. Dis. 2004, 39, 990–995. [Google Scholar] [CrossRef] [PubMed]
- Cohen, C.R.; Lingappa, J.R.; Baeten, J.M.; Ngayo, M.O.; Spiegel, C.A.; Hong, T.; Donnell, D.; Celum, C.; Kapiga, S.; Delany, S.; et al. Bacterial vaginosis associated with increased risk of female-to-male HIV-1 transmission: A prospective cohort analysis among african couples. PLoS Med. 2012, 9, 18. [Google Scholar] [CrossRef] [PubMed]
- Anahtar, M.N.; Byrne, E.H.; Doherty, K.E.; Bowman, B.A.; Yamamoto, S.; Soumillon, M.; Padavattan, N.; Ismail, N.; Moodley, A.; Sabatini, M.E.; et al. Inflammatory Responses in the Female Genital Tract. Immunity 2016, 42, 965–976. [Google Scholar] [CrossRef]
- Blackwell, A.L. Vaginal bacterial phaginosis ? Sex Trans Infect 1999, 75, 352–353. [Google Scholar] [CrossRef]
- Pavlova, S.I.; Kilic, A.O.; Mou, S.M.; Tao, L. Phage Infection in Vaginal Lactobacilli: An In Vitro Study. Infect. Dis. Obstet. Gynecol. 1997, 5, 36–44. [Google Scholar]
- Vander Borght, M.; Wyns, C. Fertility and infertility: Definition and epidemiology. Clin. Biochem. 2018, 62, 2–10. [Google Scholar] [CrossRef]
- Africa, C.W.J.; Nel, J.; Stemmet, M. Anaerobes and Bacterial Vaginosis in Pregnancy: Virulence Factors Contributing to Vaginal Colonisation. Int. J. Environ. Res. Public Heal. 2014, 11, 6979–7000. [Google Scholar] [CrossRef]
- van Oostrum, N.; De Sutter, P.; Meys, J.; Verstraelen, H. Risks associated with bacterial vaginosis in infertility patients: A systematic review and meta-analysis. Hum. Reprod. 2013, 28, 1809–1815. [Google Scholar] [CrossRef]
- Salah, R.M.; Allam, A.M.; Magdy, A.M.; Mohamed, A.S. Bacterial vaginosis and infertility: Cause or association? Eur. J. Obstet. Gynecol. Reprod. Biol. 2013, 167, 59–63. [Google Scholar] [CrossRef]
- Ruggeri, M.; Cannas, S.; Cubeddu, M.; Molicotti, P.; Piras, G.L.; Dessole, S.; Zanetti, S. Bacterial agents as a cause of infertility in humans. New Microbiol. 2016, 39, 206–209. [Google Scholar]
- Cattani, P.; Cerimele, F.; Porta, D.; Graffeo, R.; Ranno, S.; Marchetti, S.; Ricci, R.; Capodicasa, N.; Fuga, L.; Amico, R.; et al. Age-specific seroprevalence of Human Herpesvirus 8 in Mediterranean regions. Clin. Microbiol. Infect. 2003, 9, 274–279. [Google Scholar] [CrossRef] [PubMed]
- Marci, R.; Gentili, V.; Bortolotti, D.; Lo Monte, G.; Caselli, E.; Bolzani, S.; Rotola, A.; Di Luca, D.; Rizzo, R. Presence of HHV-6A in Endometrial Epithelial Cells from Women with Primary Unexplained Infertility. PLoS ONE 2016, 11, e0158304. [Google Scholar] [CrossRef] [PubMed]
- Caselli, E.; Bortolotti, D.; Marci, R.; Rotola, A.; Gentili, V.; Soffritti, I.; D’Accolti, M.; Lo Monte, G.; Sicolo, M.; Barao, I.; et al. HHV-6A Infection of Endometrial Epithelial Cells Induces Increased Endometrial NK Cell-Mediated Cytotoxicity. Front. Microbiol. 2017, 8, 2525. [Google Scholar] [CrossRef] [PubMed]
- Doody, K.J.; Doody, K.M.; Pratt, P.F., Jr.; Knox, K. Prospective evaluation of human herpesvirus 6 (HHV-6) in endometrial tissues of women with repeat implantation failure. Fertil. Steril. 2019, 112, e334. [Google Scholar] [CrossRef]
- Teixeira, F.M.E.; Pietrobon, A.J.; de Oliveira, L.M.; da Oliveira, L.M.S.; Sato, M.N. Maternal-Fetal Interplay in Zika Virus Infection and Adverse Perinatal Outcomes. Front. Immunol. 2020, 11, 175. [Google Scholar] [CrossRef]
- Dagnew, M.; Belachew, A.; Tiruneh, M.; Moges, F. Hepatitis E virus infection among pregnant women in Africa: Systematic review and meta-analysis. BMC Infect. Dis. 2019, 19, 519. [Google Scholar] [CrossRef]
- Endalamaw, A.; Demsie, A.; Eshetie, S.; Habtewold, T.D. A systematic review and meta-analysis of vertical transmission route of HIV in Ethiopia. BMC Infect. Dis. 2018, 18, 283. [Google Scholar] [CrossRef]
- Bhatta, A.K.; Keyal, U.; Liu, Y.; Gellen, E. Vertical transmission of herpes simplex virus: An update. J. Dtsch. Dermatol. Ges. 2018, 16, 685–692. [Google Scholar] [CrossRef]
- Kagan, K.O.; Hamprecht, K. Cytomegalovirus infection in pregnancy. Arch. Gynecol. Obstet. 2017, 296, 15–26. [Google Scholar] [CrossRef] [PubMed]
- Bascietto, F.; Liberati, M.; Murgano, D.; Buca, D.; Iacovelli, A.; Flacco, M.E.; Manzoli, L.; Familiari, A.; Scambia, G.; D’Antonio, F. Outcome of fetuses with congenital parvovirus B19 infection: Systematic review and meta-analysis. Ultrasound Obstet. Gynecol. 2018, 52, 569–576. [Google Scholar] [CrossRef] [PubMed]
- Mandelbrot, L. Fetal varicella—Diagnosis, management, and outcome. Prenat. Diagn. 2012, 32, 511–518. [Google Scholar] [CrossRef] [PubMed]
- Fettweis, J.M.; Serrano, M.G.; Brooks, J.P.; Edwards, D.J.; Girerd, P.H.; Parikh, H.I.; Huang, B.; Arodz, T.J.; Edupuganti, L.; Glascock, A.L.; et al. The vaginal microbiome and preterm birth. Nat. Med. 2019, 25, 1012–1021. [Google Scholar] [CrossRef] [PubMed]
- Hočevar, K.; Maver, A.; Vidmar Šimic, M.; Hodžić, A.; Haslberger, A.; Premru Seršen, T.; Peterlin, B. Vaginal Microbiome Signature Is Associated With Spontaneous Preterm Delivery. Front. Med. 2019, 6, 201. [Google Scholar] [CrossRef] [PubMed]
- Brown, R.G.; Al-Memar, M.; Marchesi, J.R.; Lee, Y.S.; Smith, A.; Chan, D.; Lewis, H.; Kindinger, L.; Terzidou, V.; Bourne, T.; et al. Establishment of vaginal microbiota composition in early pregnancy and its association with subsequent preterm prelabor rupture of the fetal membranes. Transl. Res. 2019, 207, 30–43. [Google Scholar] [CrossRef]
- Amabebe, E.; Chapman, D.R.; Stern, V.L.; Stafford, G.; Anumba, D.O.C. Mid-gestational changes in cervicovaginal fluid cytokine levels in asymptomatic pregnant women are predictive markers of inflammation-associated spontaneous preterm birth. J. Reprod. Immunol. 2018, 126, 1–10. [Google Scholar] [CrossRef]
- Ford, J.H.; Li, M.; Scheil, W.; Roder, D. Human papillomavirus infection and intrauterine growth restriction: A data-linkage study. J. Matern. Neonatal Med. 2019, 32, 279–285. [Google Scholar] [CrossRef]
- Ford, J.H. Preconception risk factors and SGA babies: Papilloma virus, omega 3 and fat soluble vitamin deficiencies. Early Hum. Dev. 2011, 87, 785–789. [Google Scholar] [CrossRef]
- Van den Veyver, I.B.; Ni, J.; Bowles, N.; Carpenter, R.J.; Weiner, C.P.; Yankowitz, J.; Moise, K.J.; Henderson, J.; Towbin, J.A. Detection of Intrauterine Viral Infection Using the Polymerase Chain Reaction. Mol. Genet. Metab. 1998, 63, 85–95. [Google Scholar] [CrossRef]
- DiGiulio, D.B.; Romero, R.; Kusanovic, J.P.; Gomez, R.; Kim, C.J.; Seok, K.S.; Gotsch, F.; Mazaki-Tovi, S.; Vaisbuch, E.; Sanders, K.; et al. Prevalence and diversity of microbes in the amniotic fluid, the fetal inflammatory response, and pregnancy outcome in women with preterm pre-labor rupture of membranes. Am. J. Reprod. Immunol. 2010, 64, 38–57. [Google Scholar] [CrossRef] [PubMed]
- DiGiulio, D.B.; Gervasi, M.T.; Romero, R.; Vaisbuch, E.; Mazaki-Tovi, S.; Kusanovic, J.P.; Seok, K.S.; Gomez, R.; Mittal, P.; Gotsch, F.; et al. Microbial invasion of the amniotic cavity in pregnancies with small-for-gestational-age fetuses. J. Perinat. Med. 2010, 38, 495–502. [Google Scholar] [CrossRef] [PubMed]
- DiGiulio, D.B.; Gervasi, M.; Romero, R.; Mazaki-Tovi, S.; Vaisbuch, E.; Kusanovic, J.P.; Seok, K.S.; Gomez, R.; Mittal, P.; Gotsch, F.; et al. Microbial invasion of the amniotic cavity in preeclampsia as assessed by cultivation and sequence-based methods. J. Perinat. Med. 2010, 38, 503–513. [Google Scholar] [CrossRef]
- Collado, M.C.; Rautava, S.; Aakko, J.; Isolauri, E.; Salminen, S. Human gut colonisation may be initiated in utero by distinct microbial communities in the placenta and amniotic fluid. Sci. Rep. 2016, 6, 23129. [Google Scholar] [CrossRef] [PubMed]
- Lim, E.S.; Rodriguez, C.; Holtz, L.R. Amniotic fluid from healthy term pregnancies does not harbor a detectable microbial community. Microbiome 2018, 6, 87. [Google Scholar] [CrossRef]
- Payne, M.S.; Keelan, J.A.; Stinson, L.F. Re: “Amniotic fluid from healthy term pregnancies does not harbor a detectable microbial community”. Microbiome 2019, 7, 20. [Google Scholar] [CrossRef]
- Lim, E.S.; Rodriguez, C.; Holtz, L.R. Reply Re: “Amniotic fluid from healthy term pregnancies does not harbor a detectable microbial community”. Microbiome 2019, 7, 21. [Google Scholar] [CrossRef]
- Koi, H.; Zhang, J.; Makrigiannakis, A.; Getsios, S.; MacCalman, C.D.; Kopf, G.S.; Strauss III, J.F.; Parry, S. Differential Expression of the Coxsackievirus and Adenovirus Receptor Regulates Adenovirus Infection of the Placenta1. Biol. Reprod. 2001, 64, 1001–1009. [Google Scholar] [CrossRef]
- Cardenas, I.; Means, R.E.; Aldo, P.; Koga, K.; Lang, S.M.; Booth, C.; Manzur, A.; Oyarzun, E.; Romero, R.; Mor, G. Viral Infection of the Placenta Leads to Fetal Inflammation and Sensitization to Bacterial Products Predisposing to Preterm Labor. J. Immunol. 2010, 185, 1248–1257. [Google Scholar] [CrossRef]
- Racicot, K.; Cardenas, I.; Wünsche, V.; Aldo, P.; Guller, S.; Means, R.E.; Romero, R.; Mor, G. Viral Infection of the Pregnant Cervix Predisposes to Ascending Bacterial Infection. J. Immunol. 2013, 191, 934–941. [Google Scholar] [CrossRef]
- Racicot, K.; Kwon, J.Y.; Aldo, P.; Abrahams, V.; El-Guindy, A.; Romero, R.; Mor, G. Type I Interferon Regulates the Placental Inflammatory Response to Bacteria and is Targeted by Virus: Mechanism of Polymicrobial Infection-Induced Preterm Birth. Am. J. Reprod. Immunol. 2016, 75, 451–460. [Google Scholar] [CrossRef] [PubMed]
- Di Mascio, D.; Khalil, A.; Saccone, G.; Rizzo, G.; Buca, D.; Liberati, M.; Vecchiet, J.; Nappi, L.; Scambia, G.; Berghella, V.; et al. Outcome of Coronavirus spectrum infections (SARS, MERS, COVID 1-19) during pregnancy: A systematic review and meta-analysis. Am. J. Obstet. Gynecol. MFM 2020, 2, 100107. [Google Scholar] [CrossRef] [PubMed]
- Qiu, L.; Liu, X.; Xiao, M.; Xie, J.; Cao, W.; Liu, Z.; Morse, A.; Xie, Y.; Li, T.; Zhu, L. SARS-CoV-2 is not detectable in the vaginal fluid of women with severe COVID-19 infection [published online ahead of print]. Clin. Infect. Dis. 2020, ciaa375. [Google Scholar] [CrossRef]
- Chen, H.; Guo, J.; Wang, C.; Luo, F.; Yu, X.; Zhang, W.; Li, J.; Zhao, D.; Xu, D.; Gong, Q.; et al. Clinical characteristics and intrauterine vertical transmission potential of COVID-19 infection in nine pregnant women: A retrospective review of medical records. Lancet 2020, 395, 809–815. [Google Scholar] [CrossRef]
- Vivanti, A.J.; Vauloup-Fellous, C.; Prevot, S.; Zupan, V.; Suffee, C.; Do Cao, J.; Benachi, A.; De Luca, D. Transplacental transmission of SARS-CoV-2 infection. Nat. Commun. 2020, 11, 3572. [Google Scholar] [CrossRef]
- Groß, R.; Conzelmann, C.; Müller, J.A.; Stenger, S.; Steinhart, K.; Kirchhoff, F.; Münch, J. Detection of SARS-CoV-2 in human breastmilk. Lancet (London, England) 2020, 395, 1757–1758. [Google Scholar] [CrossRef]
- Algarroba, G.N.; Rekawek, P.; Vahanian, S.A.; Khullar, P.; Palaia, T.; Peltier, M.R.; Chavez, M.R.; Vintzileos, A.M. Visualization of SARS-CoV-2 virus invading the human placenta using electron microscopy. Am. J. Obstet. Gynecol. 2020. [Google Scholar] [CrossRef]
- Chinchar, V.G.; Hick, P.; Ince, I.A.; Jancovich, J.K.; Marschang, R.; Qin, Q.; Subramaniam, K.; Waltzek, T.B.; Whittington, R.; Williams, T.; et al. ICTV Virus Taxonomy Profile: Iridoviridae. J. Gen. Virol. 2017, 98, 890–891. [Google Scholar] [CrossRef]
- Wilson, W.; Etten, J.; Schroeder, D.S.; Nagasaki, K.; Brussaard, C.; Delaroque, N.; Bratbak, G.; Suttle, C. ICTV 9th Report 2011 Phycodnaviridae. 2016. Available online: https://www.researchgate.net/publication/303988473_ICTV_9th_Report_2011_Phycodnaviridae (accessed on 27 July 2020).
- de la Higuera, I.; Kasun, G.W.; Torrance, E.L.; Pratt, A.A.; Maluenda, A.; Colombet, J.; Bisseux, M.; Ravet, V.; Dayaram, A.; Stainton, D.; et al. Unveiling Crucivirus Diversity by Mining Metagenomic Data. bioRxiv 2020. [Google Scholar] [CrossRef]
| Eukaryote-Infecting DNA Viruses | |||||
|---|---|---|---|---|---|
| dsDNA | |||||
| References | Family | Subfamily | Genus | Species | Type |
| [9,12,13,14,15,16] | Herpesviridae | Alphaherpesvirinae | Simplexvirus | HSV-2 | |
| Varicellovirus | Varicella-zoster virus (HHV-3) | ||||
| Betaherpesvirinae | Cytomegalovirus | ||||
| Roseolovirus | HHV-6, HHV-7 | ||||
| Proboscivirus | |||||
| Gammaherpesvirinae | Lymphocryptovirus | Epstein–Barr virus (HHV-4) | |||
| Rhadinovirus | |||||
| [14] | Alloherpesviridae | Cyprinivirus | |||
| [9,10,11,12,13,15,16,17] | Papillomaviridae * | Firstpapillomavirinae | Alphapapillomavirus | Alphapapillomavirus 1 | e.g., HPV32, 42 |
| Alphapapillomavirus 2 | e.g., HPV77 | ||||
| Alphapapillomavirus 3 | e.g., HPV61, 62, 72, 81, 83, 84, 86, 87, 89, 102, 114 | ||||
| Alphapapillomavirus 5 | e.g., HPV51, 26, 82, 69 | ||||
| Alphapapillomavirus 6 | e.g., HPV56, 66, 53, 30 | ||||
| Alphapapillomavirus 7 | e.g., HPV18, 39, 45, 59, 68, 70, 97 | ||||
| Alphapapillomavirus 8 | e.g., HPV7, 40, 43, 91, | ||||
| Alphapapillomavirus 9 | e.g., HPV16, 31, 33, 35, 52, 58, 67, | ||||
| Alphapapillomavirus 10 | e.g., HPV6, 11, 13, 44, 74 | ||||
| Alphapapillomavirus 11 | e.g., HPV73, 34 | ||||
| Alphapapillomavirus 13 | e.g., HPV54 | ||||
| Alphapapillomavirus 14 | e.g., HPV7, 90, 106 | ||||
| Betapapillomavirus | |||||
| Gammapapillomavirus | |||||
| Deltapapillomavirus | |||||
| Dyoetapapillomavirus | |||||
| Dyothetapapillomavirus | |||||
| Phipapillomavirus | |||||
| Unclassified Papillomaviridea | e.g., HPV-85 | ||||
| [9,12,15,17] | Polyomaviridae | Alphapolyomavirus | Human polyomavirus-5 (MCPyV) | ||
| Betapolyomavirus | Human polyomavirus-1 (BKPyV), -2 (JCPyV), | ||||
| [9,12] | Adenoviridae | Mastadenovirus | Human adenovirus B and D | ||
| [12,14] | Poxviridae | Molluscipoxvirus | Molluscum contagiosum virus-1 and -2 | ||
| [14] | Phycodnaviridae | Chlorovirus | |||
| [14] | Mimiviridae | Unclassified Mimivirus | |||
| [14] | Iridoviridae | Betairidoviridae | Iridovirus | ||
| [14] | Marseilleviridae | Marseillevirus | |||
| ssDNA | |||||
| [9,12,15,16,17] | Anelloviridae | Alphatorquevirus | |||
| Unclassified Anellovirus | Torque teno virus, SEN virus | ||||
| [16] | Genomoviridae | Gemykibivirus | |||
| Eukaryote-Infecting RNA Viruses | |||||
| dsRNA | |||||
| [13] | Partitiviridae | ||||
| ssRNA-RT | |||||
| [18,19,20] | Retroviridae | Orthoretrovirinae | Lentivirus | HIV-1 | |
| Prokaryote-Infecting DNA Viruses | |||||
| dsDNA | |||||
| [14] | Podoviridae | ||||
| [14] | Siphoviridae | ||||
| [14] | Myoviridae | Tevenvirinae | Tequatrovirus | T4 virus | |
| ssDNA | |||||
| [14] | Microviridae | ||||
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Happel, A.-U.; Varsani, A.; Balle, C.; Passmore, J.-A.; Jaspan, H. The Vaginal Virome—Balancing Female Genital Tract Bacteriome, Mucosal Immunity, and Sexual and Reproductive Health Outcomes? Viruses 2020, 12, 832. https://doi.org/10.3390/v12080832
Happel A-U, Varsani A, Balle C, Passmore J-A, Jaspan H. The Vaginal Virome—Balancing Female Genital Tract Bacteriome, Mucosal Immunity, and Sexual and Reproductive Health Outcomes? Viruses. 2020; 12(8):832. https://doi.org/10.3390/v12080832
Chicago/Turabian StyleHappel, Anna-Ursula, Arvind Varsani, Christina Balle, Jo-Ann Passmore, and Heather Jaspan. 2020. "The Vaginal Virome—Balancing Female Genital Tract Bacteriome, Mucosal Immunity, and Sexual and Reproductive Health Outcomes?" Viruses 12, no. 8: 832. https://doi.org/10.3390/v12080832
APA StyleHappel, A.-U., Varsani, A., Balle, C., Passmore, J.-A., & Jaspan, H. (2020). The Vaginal Virome—Balancing Female Genital Tract Bacteriome, Mucosal Immunity, and Sexual and Reproductive Health Outcomes? Viruses, 12(8), 832. https://doi.org/10.3390/v12080832