A Novel Inducible Prophage from Burkholderia vietnamiensis G4 Is Widely Distributed across the Species and Has Lytic Activity against Pathogenic Burkholderia
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Genomic Analysis and Phylogenomics
2.2. Prophage Identification and Comparison
2.3. Bacterial Strains and Culture Media
2.4. Isolation of Spontaneously Induced Bacteriophages from Lysogenic Burkholderia vietnamiensis G4
2.5. Bacteriophage Host Range Determination of Phage G4P1 Isolated from B. vietnamiensis G4
2.6. Transmission Electron Microscopy (TEM)
2.7. Bacteriophage Genomic Analysis
2.8. Data Summary
3. Results
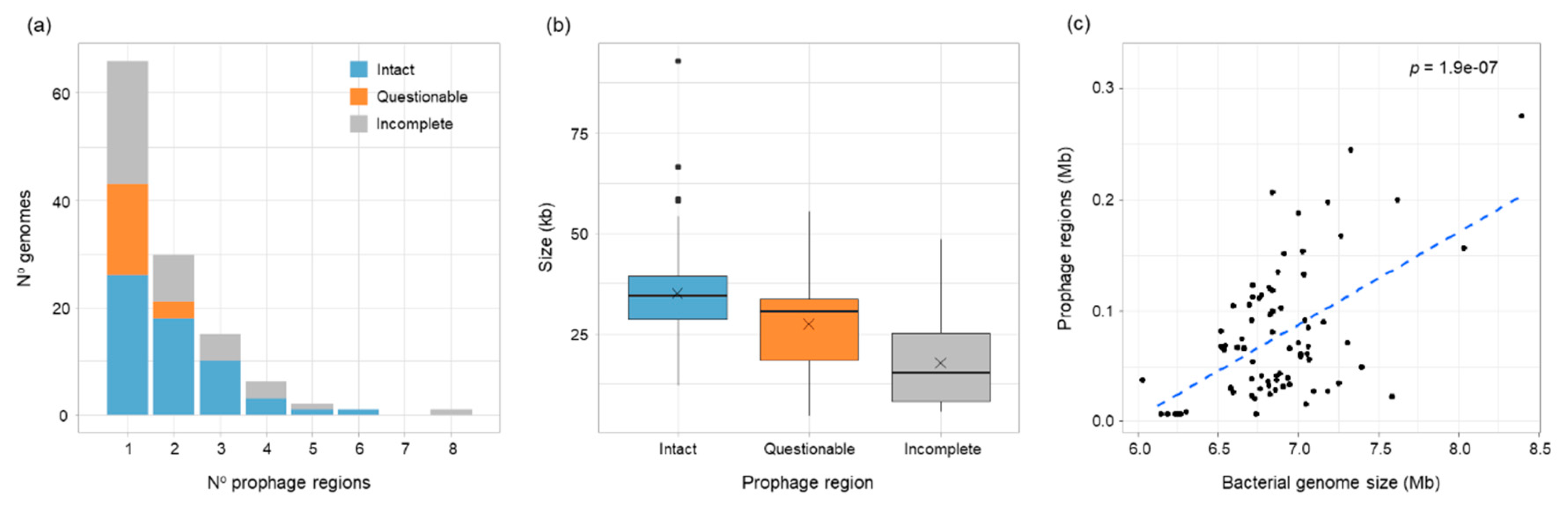
3.1. Prophage Carriage Was Common in B. vietnamiensis
3.2. Prophages Were Spontaneously Induced from B. vietnamiensis Strain G4
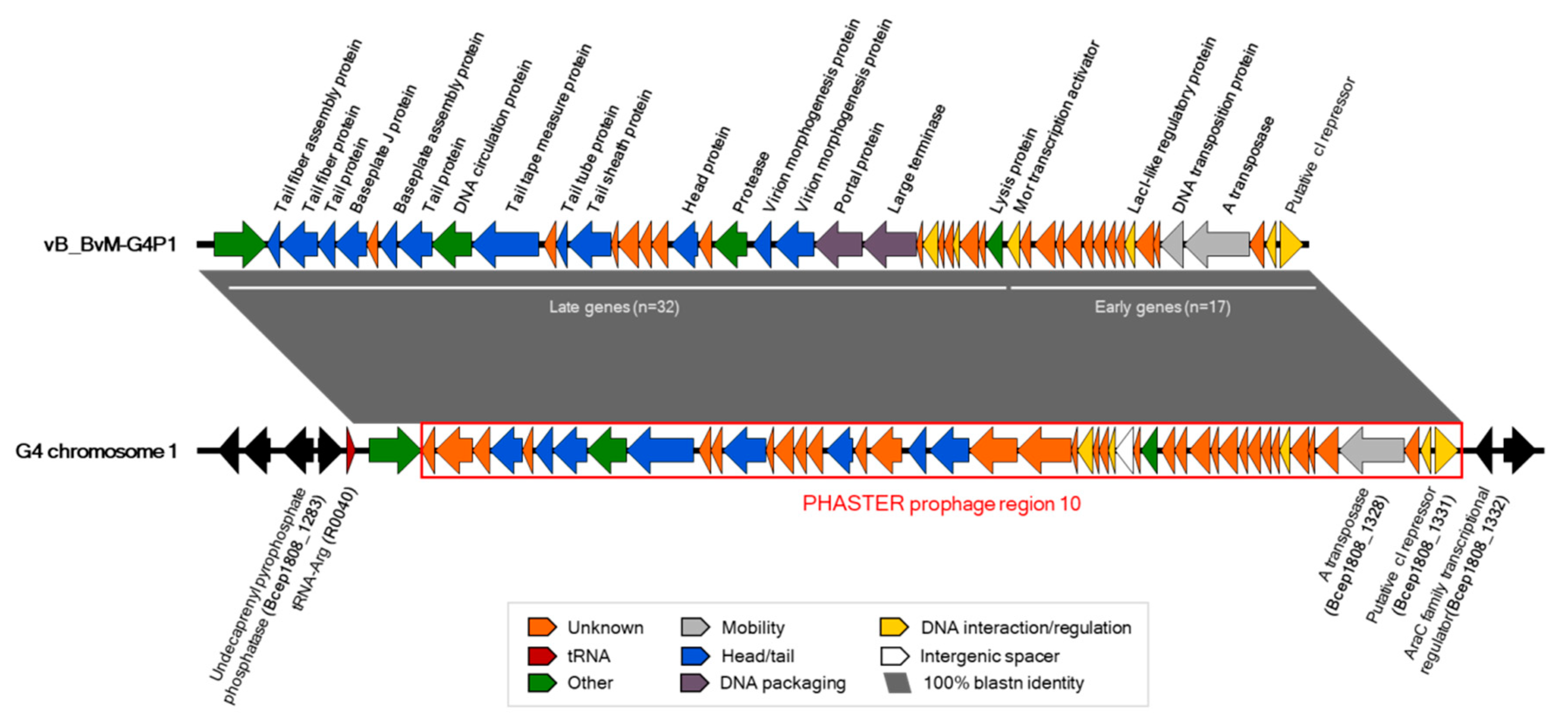
3.3. G4P1 was Localised to Chromosome 1 of B. vietnamiensis G4
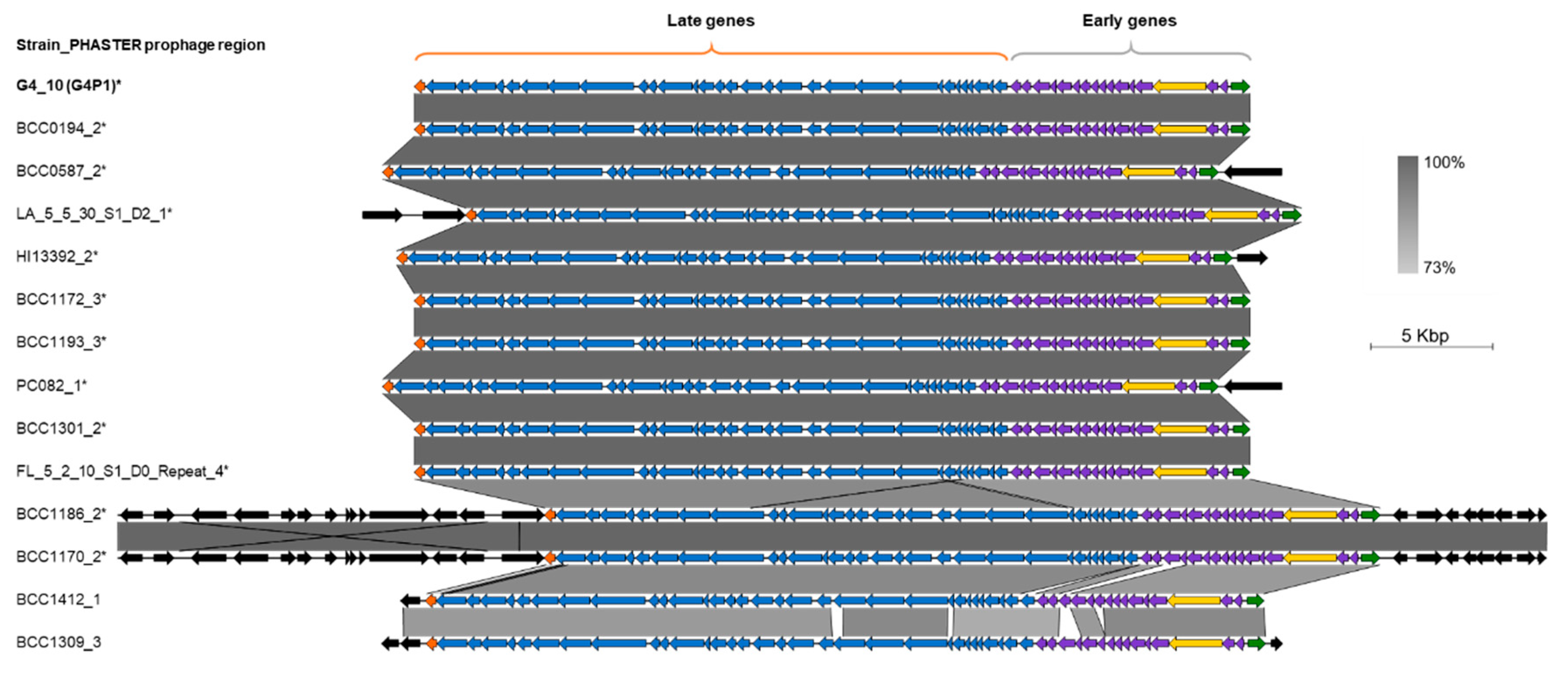
3.4. vB_BvM-G4P1 Was Found in Other B. vietnamiensis Strains
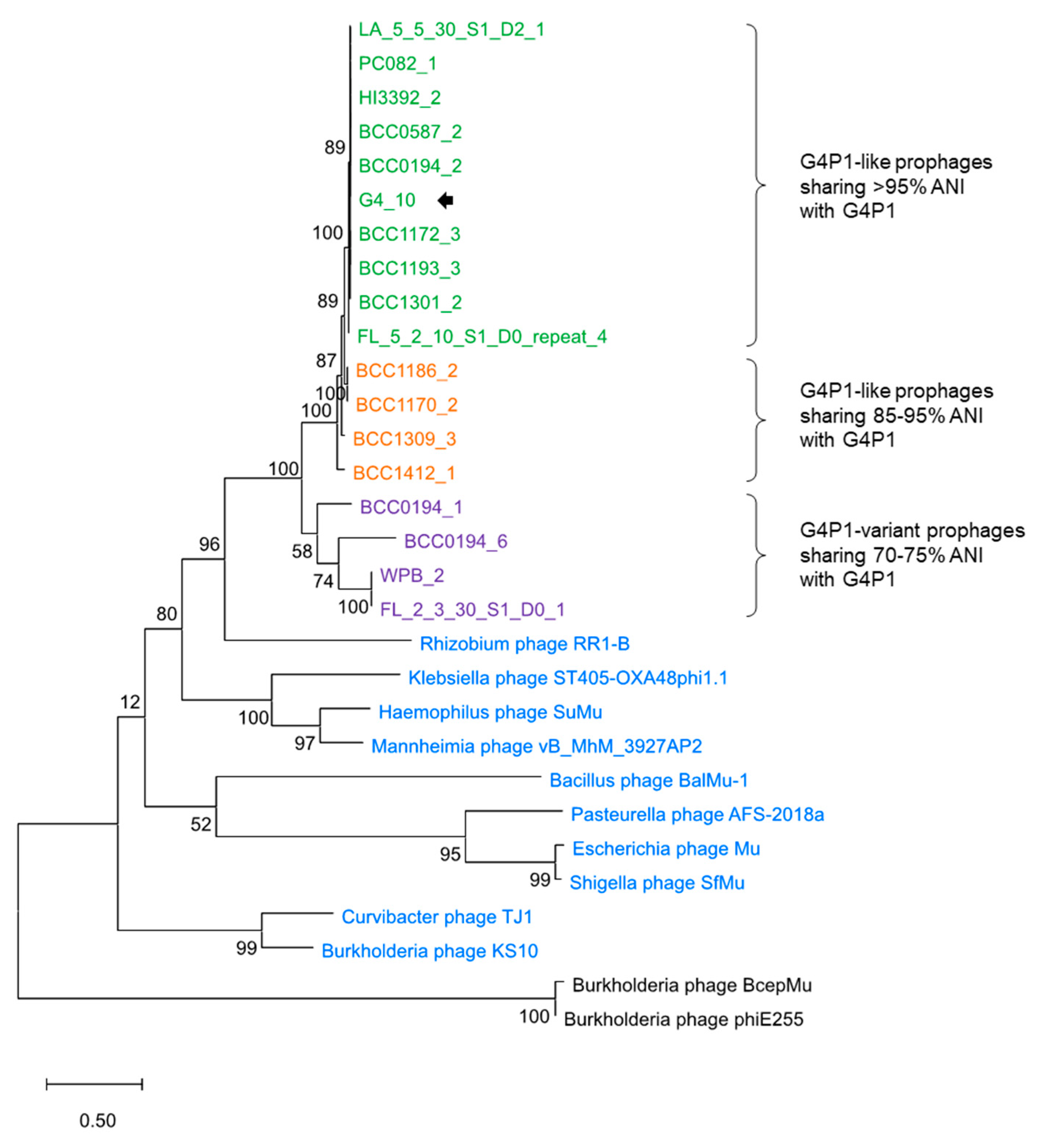
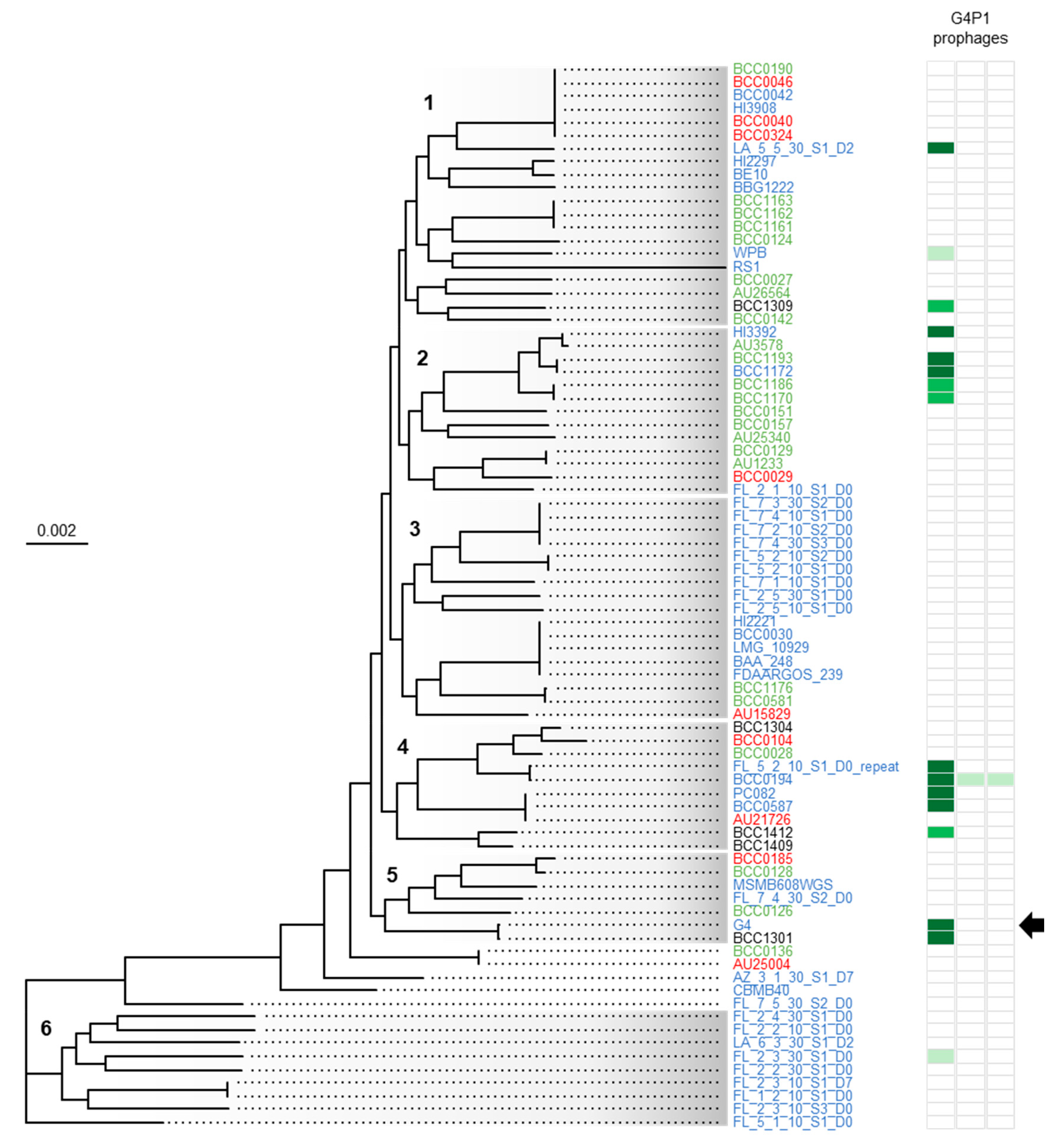
3.5. vB_BvM-G4P1 Was Widely Distributed Across the Population Structure of B. vietnamiensis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Clokie, M.R.; Millard, A.D.; Letarov, A.V.; Heaphy, S. Phages in nature. Bacteriophage 2011, 1, 31–45. [Google Scholar] [CrossRef] [PubMed]
- Harrison, E.; Brockhurst, M.A. Ecological and evolutionary benefits of temperate phage: What does or doesn’t kill you makes you stronger. Bioessays 2017, 39. [Google Scholar] [CrossRef] [PubMed]
- Casjens, S. Prophages and bacterial genomics: What have we learned so far? Mol. Microbiol. 2003, 49, 277–300. [Google Scholar] [CrossRef] [PubMed]
- Touchon, M.; Bernheim, A.; Rocha, E.P.C. Genetic and life-history traits associated with the distribution of prophages in bacteria. ISME J. 2016, 10, 2744–2754. [Google Scholar] [CrossRef] [PubMed]
- Arndt, D.; Grant, J.R.; Marcu, A.; Sajed, T.; Pon, A.; Liang, Y.; Wishart, D.S. Phaster: A better, faster version of the phast phage search tool. Nucleic Acids Res. 2016, 44, W16–W21. [Google Scholar] [CrossRef] [PubMed]
- Fouts, D.E. Phage_finder: Automated identification and classification of prophage regions in complete bacterial genome sequences. Nucleic Acids Res. 2006, 34, 5839–5851. [Google Scholar] [CrossRef] [PubMed]
- Kang, H.S.; McNair, K.; Cuevas, D.A.; Bailey, B.A.; Segall, A.M.; Edwards, R.A. Prophage genomics reveals patterns in phage genome organization and replication. BioRxiv 2017, 114819. [Google Scholar] [CrossRef]
- Depoorter, E.; Bull, M.J.; Peeters, C.; Coenye, T.; Vandamme, P.; Mahenthiralingam, E. Burkholderia: An update on taxonomy and biotechnological potential as antibiotic producers. Appl. Microbiol. Biotechnol. 2016, 100, 5215–5229. [Google Scholar] [CrossRef]
- Yabuuchi, E.; Kosako, Y.; Oyaizu, H.; Yano, I.; Hotta, H.; Hashimoto, Y.; Ezaki, T.; Arakawa, M. Proposal of burkholderia gen. Nov. And transfer of seven species of the genus pseudomonas homology group II to the new genus, with the type species burkholderia cepacia (palleroni and holmes 1981) comb. Nov. Microbiol. Immunol. 1992, 36, 1251–1275. [Google Scholar] [CrossRef]
- Wallner, A.; King, E.; Ngonkeu, E.L.M.; Moulin, L.; Béna, G. Genomic analyses of burkholderia cenocepacia reveal multiple species with differential host-adaptation to plants and humans. BMC Genom. 2019, 20, 803. [Google Scholar] [CrossRef]
- Zlosnik, J.E.; Zhou, G.; Brant, R.; Henry, D.A.; Hird, T.J.; Mahenthiralingam, E.; Chilvers, M.A.; Wilcox, P.; Speert, D.P. Burkholderia species infections in patients with cystic fibrosis in British Columbia, Canada. 30 years’ experience. Ann. Am. Thorac. Soc. 2015, 12, 70–78. [Google Scholar] [CrossRef] [PubMed]
- Rushton, L.; Sass, A.; Baldwin, A.; Dowson, C.G.; Donoghue, D.; Mahenthiralingam, E. Key role for efflux in the preservative susceptibility and adaptive resistance of burkholderia cepacia complex bacteria. Antimicrob. Agents Chemother. 2013, 57, 2972–2980. [Google Scholar] [CrossRef] [PubMed]
- Mahenthiralingam, E.; Urban, T.A.; Goldberg, J.B. The multifarious, multireplicon burkholderia cepacia complex. Nat. Rev. Microbiol. 2005, 3, 144–156. [Google Scholar] [CrossRef] [PubMed]
- Coenye, T.; LiPuma, J.J.; Henry, D.; Hoste, B.; Vandemeulebroecke, K.; Gillis, M.; Speert, D.P.; Vandamme, P. Burkholderia cepacia genomovar vi, a new member of the burkholderia cepacia complex isolated from cystic fibrosis patients. Int. J. Syst. Evol. Microbiol. 2001, 51, 271–279. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Ren, H.; Hu, M.; Zhou, J.; Li, B.; Kong, N.; Zhang, Q.; Jin, Y.; Liang, L.; Yue, J. Characterization of burkholderia cepacia complex core genome and the underlying recombination and positive selection. Front. Genet. 2020, 11. [Google Scholar] [CrossRef]
- Mahenthiralingam, E.; Drevinek, P. Comparative genomics of burkholderia species. In Burkholderia: Molecular Biology and Genomics; Coenye, T., Vandamme, P., Eds.; Horizon Bioscience: Norwich, UK, 2007; pp. 53–79. [Google Scholar]
- Semler, D.; Lynch, K.; Dennis, J. The promise of bacteriophage therapy for burkholderia cepacia complex respiratory infections. Front. Cell. Infect. Microbiol. 2012, 1. [Google Scholar] [CrossRef]
- Kenna, D.T.D.; Lilley, D.; Coward, A.; Martin, K.; Perry, C.; Pike, R.; Hill, R.; Turton, J.F. Prevalence of burkholderia species, including members of burkholderia cepacia complex, among UK cystic and non-cystic fibrosis patients. J. Med. Microbiol. 2017, 66, 490–501. [Google Scholar] [CrossRef]
- Pope, C.E.; Short, P.; Carter, P.E. Species distribution of burkholderia cepacia complex isolates in cystic fibrosis and non-cystic fibrosis patients in New Zealand. J. Cyst. Fibros. 2010, 9, 442–446. [Google Scholar] [CrossRef]
- Lupo, A.; Isis, E.; Tinguely, R.; Endimiani, A. Clonality and antimicrobial susceptibility of burkholderia cepacia complex isolates collected from cystic fibrosis patients during 1998–2013 in Bern, Switzerland. New Microbiol. 2015, 38, 281–288. [Google Scholar]
- Medina-Pascual, M.J.; Valdezate, S.; Carrasco, G.; Villalon, P.; Garrido, N.; Saez-Nieto, J.A. Increase in isolation of burkholderia contaminans from spanish patients with cystic fibrosis. Clin. Microbiol. Infect. 2015, 21, 150–156. [Google Scholar] [CrossRef]
- Lipuma, J.J. The changing microbial epidemiology in cystic fibrosis. Clin. Microbiol. Rev. 2010, 23, 299–323. [Google Scholar] [CrossRef] [PubMed]
- Shinjo, R.; Uesaka, K.; Ihara, K.; Sakazaki, S.; Yano, K.; Kondo, M.; Tanaka, A. Draft genome sequence of burkholderia vietnamiensis strain rs1, a nitrogen-fixing endophyte isolated from sweet potato. Microbiol. Res. Announc. 2018, 7, e00820–00818. [Google Scholar] [CrossRef] [PubMed]
- Gillis, M.; Van Van, T.; Bardin, R.; Goor, M.; Hebbar, P.; Willems, A.; Segers, P.; Kersters, K.; Heulin, T.; Fernandez, M.P. Polyphasic taxonomy in the genus burkholderia leading to an emended description of the genus and proposition of burkholderia vietnamiensis sp. Nov. For n2-fixing isolates from rice in Vietnam. Int. J. Syst. Evol. Microbiol. 1995, 45, 274–289. [Google Scholar] [CrossRef]
- O’Sullivan, L.A.; Mahenthiralingam, E. Biotechnological potential within the genus burkholderia. Lett. Appl. Microbiol. 2005, 41, 8–11. [Google Scholar] [CrossRef] [PubMed]
- Fries, M.R.; Forney, L.J.; Tiedje, J.M. Phenol-and toluene-degrading microbial populations from an aquifer in which successful trichloroethene cometabolism occurred. Appl. Environ. Microbiol. 1997, 63, 1523–1530. [Google Scholar] [CrossRef]
- Nelson, M.J.; Montgomery, S.O.; Mahaffey, W.R.; Pritchard, P.H. Biodegradation of trichloroethylene and involvement of an aromatic biodegradative pathway. Appl. Environ. Microbiol. 1987, 53, 949–954. [Google Scholar] [CrossRef]
- Nzula, S.; Vandamme, P.; Govan, J.R. Sensitivity of the burkholderia cepacia complex and pseudomonas aeruginosa to transducing bacteriophages. FEMS Immunol. Med. Microbiol. 2000, 28, 307–312. [Google Scholar] [CrossRef]
- Mullins, A.J.; Murray, J.A.H.; Bull, M.J.; Jenner, M.; Jones, C.; Webster, G.; Green, A.E.; Neill, D.R.; Connor, T.R.; Parkhill, J.; et al. Genome mining identifies cepacin as a plant-protective metabolite of the biopesticidal bacterium burkholderia ambifaria. Nat. Microbiol. 2019, 4, 996–1005. [Google Scholar] [CrossRef]
- Connor, T.R.; Loman, N.J.; Thompson, S.; Smith, A.; Southgate, J.; Poplawski, R.; Bull, M.J.; Richardson, E.; Ismail, M.; Thompson, S.E.; et al. Climb (the cloud infrastructure for microbial bioinformatics): An online resource for the medical microbiology community. Microbial. Genom. 2016, 2, e000086. [Google Scholar] [CrossRef]
- Krueger, F. Trim galore! A Wrapper Tool around Cutadapt and Fastqc to Consistently Apply Quality and Adapter Trimming to Fastq Files. Available online: https://www.Bioinformatics.Babraham.Ac.Uk/projects/trim_galore/ (accessed on 7 January 2019).
- Andrews, S. Fastqc: A Quality Control Tool for High Throughput Sequence Data. Available online: http://www.Bioinformatics.Babraham.Ac.Uk/projects/fastqc (accessed on 7 January 2019).
- Wick, R.R.; Judd, L.M.; Gorrie, C.L.; Holt, K.E. Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 2017, 13, e1005595. [Google Scholar] [CrossRef]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. Spades: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. J. Comput. Mol. Cell Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef] [PubMed]
- Wick, R.R.; Schultz, M.B.; Zobel, J.; Holt, K.E. Bandage: Interactive visualization of de novo genome assemblies. Bioinform. Oxford Engl. 2015, 31, 3350–3352. [Google Scholar] [CrossRef] [PubMed]
- Jain, C.; Rodriguez-R, L.M.; Phillippy, A.M.; Konstantinidis, K.T.; Aluru, S. High throughput ani analysis of 90k prokaryotic genomes reveals clear species boundaries. Nat. Commun. 2018, 9, 5114. [Google Scholar] [CrossRef] [PubMed]
- Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef] [PubMed]
- Page, A.J.; Cummins, C.A.; Hunt, M.; Wong, V.K.; Reuter, S.; Holden, M.T.G.; Fookes, M.; Falush, D.; Keane, J.A.; Parkhill, J. Roary: Rapid large-scale prokaryote pan genome analysis. Bioinformatics 2015, 31, 3691–3693. [Google Scholar] [CrossRef]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. Fasttree: Computing large minimum evolution trees with profiles instead of a distance matrix. Mol. Biol. Evol. 2009, 26, 1641–1650. [Google Scholar] [CrossRef]
- Weiser, R.; Green, A.E.; Bull, M.J.; Cunningham-Oakes, E.; Jolley, K.A.; Maiden, M.C.J.; Hall, A.J.; Winstanley, C.; Weightman, A.J.; Donoghue, D.; et al. Not all pseudomonas aeruginosa are equal: Strains from industrial sources possess uniquely large multireplicon genomes. Microb. Genom. 2019. [Google Scholar] [CrossRef]
- Rutherford, K.; Parkhill, J.; Crook, J.; Horsnell, T.; Rice, P.; Rajandream, M.-A.; Barrell, B. Artemis: Sequence visualization and annotation. Bioinformatics 2000, 16, 944–945. [Google Scholar] [CrossRef]
- R-Core-Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2013. [Google Scholar]
- Pritchard, L.; Glover, R.H.; Humphris, S.; Elphinstone, J.G.; Toth, I.K. Genomics and taxonomy in diagnostics for food security: Soft-rotting enterobacterial plant pathogens. Anal. Methods 2016, 8, 12–24. [Google Scholar] [CrossRef]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. Blast+: Architecture and applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef]
- Sullivan, M.J.; Petty, N.K.; Beatson, S.A. Easyfig: A genome comparison visualizer. Bioinform. Oxford Engl. 2011, 27, 1009–1010. [Google Scholar] [CrossRef] [PubMed]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal w and clustal x version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. Mega7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Jones, D.T.; Taylor, W.R.; Thornton, J.M. The rapid generation of mutation data matrices from protein sequences. Comput. Appl. Biosci. 1992, 8, 275–282. [Google Scholar] [CrossRef] [PubMed]
- Mahenthiralingam, E.; Baldwin, A.; Dowson, C.G. Burkholderia cepacia complex bacteria: Opportunistic pathogens with important natural biology. J. Appl. Microbiol. 2008, 104, 1539–1551. [Google Scholar] [CrossRef] [PubMed]
- Adams, M.H. Bacteriophages; Interscience Publishers Inc.: New York, NY, USA, 1959. [Google Scholar]
- Seed, K.D.; Dennis, J.J. Isolation and characterisation of bacteriophages of the burkholderia cepacia complex. FEMS Microbiol. Lett. 2005, 251, 273–280. [Google Scholar] [CrossRef]
- Nzakizwanayo, J.; Hanin, A.; Alves, D.R.; McCutcheon, B.; Dedi, C.; Salvage, J.; Knox, K.; Stewart, B.; Metcalfe, A.; Clark, J.; et al. Bacteriophage can prevent encrustation and blockage of urinary catheters by proteus mirabilis. Antimicrob. Agents Chemother. 2015, 60, 1530–1536. [Google Scholar] [CrossRef]
- Lane, D.J.; Pace, B.; Olsen, G.J.; Stahl, D.A.; Sogin, M.L.; Pace, N.R. Rapid determination of 16s ribosomal rna sequences for phylogenetic analyses. Proc. Natl. Acad. Sci. USA 1985, 82, 6955–6959. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. The sequence alignment/map format and samtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- Quinlan, A.R.; Hall, I.M. Bedtools: A flexible suite of utilities for comparing genomic features. Bioinformatics 2010, 26, 841–842. [Google Scholar] [CrossRef]
- Mitchell, A.L.; Attwood, T.K.; Babbitt, P.C.; Blum, M.; Bork, P.; Bridge, A.; Brown, S.D.; Chang, H.-Y.; El-Gebali, S.; Fraser, M.I.; et al. Interpro in 2019: Improving coverage, classification and access to protein sequence annotations. Nucl. Acids Res. 2019, 47, D351–D360. [Google Scholar] [CrossRef] [PubMed]
- Jones, P.; Binns, D.; Chang, H.-Y.; Fraser, M.; Li, W.; McAnulla, C.; McWilliam, H.; Maslen, J.; Mitchell, A.; Nuka, G.; et al. Interproscan 5: Genome-scale protein function classification. Bioinform. Oxford Engl. 2014, 30, 1236–1240. [Google Scholar] [CrossRef] [PubMed]
- Kropinski, A.M.; Prangishvili, D.; Lavigne, R. Position paper: The creation of a rational scheme for the nomenclature of viruses of bacteria and archaea. Environ. Microbiol. 2009, 11, 2775–2777. [Google Scholar] [CrossRef] [PubMed]
- Winsor, G.L.; Khaira, B.; Van Rossum, T.; Lo, R.; Whiteside, M.D.; Brinkman, F.S.L. The burkholderia genome database: Facilitating flexible queries and comparative analyses. Bioinformatics 2008, 24, 2803–2804. [Google Scholar] [CrossRef]
- Niu, Y.D.; Cook, S.R.; Wang, J.; Klima, C.L.; Hsu, Y.H.; Kropinski, A.M.; Turner, D.; McAllister, T.A. Comparative analysis of multiple inducible phages from mannheimia haemolytica. BMC Microbiol. 2015, 15, 175. [Google Scholar] [CrossRef]
- Ronning, C.M.; Losada, L.; Brinkac, L.; Inman, J.; Ulrich, R.L.; Schell, M.; Nierman, W.C.; DeShazer, D. Genetic and phenotypic diversity in burkholderia: Contributions by prophage and phage-like elements. BMC Microbiol. 2010, 10, 202. [Google Scholar] [CrossRef]
- Langley, R.; Kenna, D.T.; Vandamme, P.; Ure, R.; Govan, J.R. Lysogeny and bacteriophage host range within the burkholderia cepacia complex. J. Med. Microbiol. 2003, 52, 483–490. [Google Scholar] [CrossRef]
- Hens, D.K.; Chatterjee, N.C.; Kumar, R. New temperate DNA phage bcp15 acts as a drug resistance vector. Arch. Virol. 2006, 151, 1345–1353. [Google Scholar] [CrossRef]
- Summer, E.J.; Gonzalez, C.F.; Carlisle, T.; Mebane, L.M.; Cass, A.M.; Savva, C.G.; LiPuma, J.; Young, R. Burkholderia cenocepacia phage bcepmu and a family of mu-like phages encoding potential pathogenesis factors. J. Mol. Biol. 2004, 340, 49–65. [Google Scholar] [CrossRef]
- Lynch, K.H.; Stothard, P.; Dennis, J.J. Genomic analysis and relatedness of p2-like phages of the burkholderia cepacia complex. BMC Genom. 2010, 11, 599. [Google Scholar] [CrossRef]
- Woods, D.E.; Jeddeloh, J.A.; Fritz, D.L.; DeShazer, D. Burkholderia thailandensis e125 harbors a temperate bacteriophage specific for burkholderia mallei. J. Bacteriol. 2002, 184, 4003–4017. [Google Scholar] [CrossRef] [PubMed]
- Pratama, A.A.; van Elsas, J.D. A novel inducible prophage from the mycosphere inhabitant paraburkholderia terrae bs437. Sci. Rep. 2017, 7, 9156. [Google Scholar] [CrossRef] [PubMed]
- Dedrick, R.M.; Guerrero-Bustamante, C.A.; Garlena, R.A.; Russell, D.A.; Ford, K.; Harris, K.; Gilmour, K.C.; Soothill, J.; Jacobs-Sera, D.; Schooley, R.T.; et al. Engineered bacteriophages for treatment of a patient with a disseminated drug-resistant mycobacterium abscessus. Nat. Med. 2019, 25, 730–733. [Google Scholar] [CrossRef] [PubMed]
- Taylor, J.B.; Hogue, L.A.; LiPuma, J.J.; Walter, M.J.; Brody, S.L.; Cannon, C.L. Entry of burkholderia organisms into respiratory epithelium: Cftr, microfilament and microtubule dependence. J. Cyst. Fibros. 2010, 9, 36–43. [Google Scholar] [CrossRef] [PubMed]
- Kalish, L.A.; Waltz, D.A.; Dovey, M.; Potter-Bynoe, G.; McAdam, A.J.; Lipuma, J.J.; Gerard, C.; Goldmann, D. Impact of burkholderia dolosa on lung function and survival in cystic fibrosis. Am. J. Respir. Crit. Care Med. 2006, 173, 421–425. [Google Scholar] [CrossRef] [PubMed]
- Toussaint, A.; Rice, P.A. Transposable phages, DNA reorganization and transfer. Curr. Opin. Microbiol. 2017, 38, 88–94. [Google Scholar] [CrossRef]
- Langley, R.J.; Kenna, D.; Bartholdson, J.; Campopiano, D.J.; Govan, J.R.W. Temperate bacteriophages dk4 and bcepmu from burkholderia cenocepacia j2315 are identical. FEMS Immunol. Med. Microbiol. 2005, 45, 349–350. [Google Scholar] [CrossRef][Green Version]
- Goudie, A.D.; Lynch, K.H.; Seed, K.D.; Stothard, P.; Shrivastava, S.; Wishart, D.S.; Dennis, J.J. Genomic sequence and activity of ks10, a transposable phage of the burkholderia cepacia complex. BMC Genom. 2008, 9, 615. [Google Scholar] [CrossRef]
- Van Truong Thi, B.; Pham Khanh, N.H.; Namikawa, R.; Miki, K.; Kondo, A.; Dang Thi, P.T.; Kamei, K. Genomic characterization of ralstonia solanacearum phage varphirs138 of the family siphoviridae. Arch. Virol. 2016, 161, 483–486. [Google Scholar] [CrossRef]
- Summer, E.J.; Gill, J.J.; Upton, C.; Gonzalez, C.F.; Young, R. Role of phages in the pathogenesis of burkholderia, or ‘where are the toxin genes in burkholderia phages? ’ Curr. Opin. Microbiol. 2007, 10, 410–417. [Google Scholar] [CrossRef]
- Roszniowski, B.; McClean, S.; Drulis-Kawa, Z. Burkholderia cenocepacia prophages-prevalence, chromosome location and major genes involved. Viruses 2018, 10, 297. [Google Scholar] [CrossRef] [PubMed]
- Rezaei Javan, R.; Ramos-Sevillano, E.; Akter, A.; Brown, J.; Brueggemann, A.B. Prophages and satellite prophages are widespread in streptococcus and may play a role in pneumococcal pathogenesis. Nat. Commun. 2019, 10, 4852. [Google Scholar] [CrossRef] [PubMed]
- Crispim, J.S.; Dias, R.S.; Vidigal, P.M.P.; de Sousa, M.P.; da Silva, C.C.; Santana, M.F.; de Paula, S.O. Screening and characterization of prophages in desulfovibrio genomes. Sci. Rep. 2018, 8, 9273. [Google Scholar] [CrossRef] [PubMed]
- Pratama, A.A.; Chaib De Mares, M.; van Elsas, J.D. Evolutionary history of bacteriophages in the genus paraburkholderia. Front. Microbiol. 2018, 9, 835. [Google Scholar] [CrossRef]
- Czajkowski, R.; Ozymko, Z.; Lojkowska, E. Isolation and characterization of novel soilborne lytic bacteriophages infecting dickeya spp. Biovar 3 (‘d. Solani’). Plant Pathol. 2014, 63, 758–772. [Google Scholar] [CrossRef]
- Fan, X.; Li, Y.; He, R.; Li, Q.; He, W. Comparative analysis of prophage-like elements in helicobacter sp. Genomes. PeerJ 2016, 4, e2012. [Google Scholar] [CrossRef]
- Kalatzis, P.G.; Rorbo, N.I.; Castillo, D.; Mauritzen, J.J.; Jorgensen, J.; Kokkari, C.; Zhang, F.; Katharios, P.; Middelboe, M. Stumbling across the same phage: Comparative genomics of widespread temperate phages infecting the fish pathogen vibrio anguillarum. Viruses 2017, 9, 122. [Google Scholar] [CrossRef]
- Bobay, L.M.; Rocha, E.P.; Touchon, M. The adaptation of temperate bacteriophages to their host genomes. Mol. Biol. Evol. 2013, 30, 737–751. [Google Scholar] [CrossRef]

| Host Species | BCC# | Alternative Strain Name | Origin (Code) | G4 Culture Supernatant | G4P1 |
|---|---|---|---|---|---|
| Burkholderia cepacia complex | |||||
| B. ambifaria | BCC0191 | HI 2345; J82 | Soil (ENV) | +/− | +/− |
| BCC0192 | Ral-3; R-8863 | Rhizosphere (ENV) | + | + | |
| BCC0197 | ATCC 51671 | Leaves (ENV) | + | + | |
| BCC0203 | BCF/HG1-A; LMG-P 24640 | Environmental (ENV) | + | + | |
| BCC0338 | ATCC 53266; LMG 17828 | Roots (ENV) | + | + | |
| BCC0410 | MVP/C1 64 | Maize (ENV) | + | + | |
| BCC1212 | MC40-6 | Rhizosphere (ENV) | + | + | |
| B. cenocepacia | BCC0019 | LMG 18829; PC184/NEH4 | Cystic fibrosis (CF) | + | + |
| BCC1202 | AU1054 | Cystic fibrosis (CF) | − | − | |
| BCC1210 | MC0-3 | Rhizosphere (ENV) | + | − | |
| B. cepacia | BCC0001 | ATCC 25416; LMG1222-T | Onion (ENV) | − | − |
| B. contaminans | BCC0362 | R-9929; CEP0964 | Cystic fibrosis (CF) | +/− | + |
| B. dolosa | BCC1343 | AU0794 | Cystic fibrosis (CF) | Not tested | + |
| BCC1356 | AU3556; clinical isolate of the SLC6 epidemic strain | Cystic fibrosis (CF) | Not tested | + | |
| BCC1357 | AU1568; clinical isolate of the SLC6 epidemic strain | Cystic fibrosis (CF) | Not tested | + | |
| BCC1359 | AU3960 | Cystic fibrosis (CF) | + | + | |
| BCC1360 | AU4298; clinical isolate of the SLC6 epidemic strain | Cystic fibrosis (CF) | Not tested | + | |
| BCC1361 | AU2130; clinical isolate of the SLC6 epidemic strain | Cystic fibrosis (CF) | Not tested | + | |
| B. lata | BCC0803 | ATCC 17660; LMG 22485T; R-18194; 383 | Soil (ENV) | − | − |
| B. multivorans | BCC0005 | MA; LMG 18822; C5393 | Cystic fibrosis (CF) | − | − |
| BCC0011 | C1576 | Cystic fibrosis (CF) | − | − | |
| BCC1421 | ATCC 17616; LMG17588 | Soil (ENV) | − | − | |
| B. pyrrocinia | BCC0180 | LMG 14191-T | Soil (ENV) | − | − |
| B. vietnamiensis | BCC0027 | LMG 18835; PC259; JCM-APRIL93; CEP0040 | Cystic fibrosis (CF) | + | + |
| BCC0030 | LMG 10929; FC0369 | Riceroot (ENV) | + | + | |
| BCC0324 | J1742 | Non-CF (CLIN) | − | − | |
| BCC1162 | CEP1224 | Cystic fibrosis (CF) | − | − | |
| BCC1304 | − | Industry (IND) | − | +/− | |
| BCC1309 | − | Industry (IND) | − | − | |
| Non-Burkholderia cepacia complex | |||||
| B. gladioli | BCC0238 | MA4 | Cystic fibrosis (CF) | − | − |
| B. thailandensis | BCC0779 | LMG 20219; ATCC 700388; E264 | Soil (ENV) | − | − |
| Paraburkholderia | |||||
| P. phymatum | BCC1607 | LMG 22487; PsJN | Environmental (ENV) | − | − |
| P. phytofirmans | BCC1604 | LMG 21445 | Environmental (ENV) | − | − |
| P. graminis | BCC0774 | ATCC 700544; LMG 18924 | Soil (ENV) | − | − |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Weiser, R.; Yap, Z.L.; Otter, A.; Jones, B.V.; Salvage, J.; Parkhill, J.; Mahenthiralingam, E. A Novel Inducible Prophage from Burkholderia vietnamiensis G4 Is Widely Distributed across the Species and Has Lytic Activity against Pathogenic Burkholderia. Viruses 2020, 12, 601. https://doi.org/10.3390/v12060601
Weiser R, Yap ZL, Otter A, Jones BV, Salvage J, Parkhill J, Mahenthiralingam E. A Novel Inducible Prophage from Burkholderia vietnamiensis G4 Is Widely Distributed across the Species and Has Lytic Activity against Pathogenic Burkholderia. Viruses. 2020; 12(6):601. https://doi.org/10.3390/v12060601
Chicago/Turabian StyleWeiser, Rebecca, Zhong Ling Yap, Ashley Otter, Brian V. Jones, Jonathan Salvage, Julian Parkhill, and Eshwar Mahenthiralingam. 2020. "A Novel Inducible Prophage from Burkholderia vietnamiensis G4 Is Widely Distributed across the Species and Has Lytic Activity against Pathogenic Burkholderia" Viruses 12, no. 6: 601. https://doi.org/10.3390/v12060601
APA StyleWeiser, R., Yap, Z. L., Otter, A., Jones, B. V., Salvage, J., Parkhill, J., & Mahenthiralingam, E. (2020). A Novel Inducible Prophage from Burkholderia vietnamiensis G4 Is Widely Distributed across the Species and Has Lytic Activity against Pathogenic Burkholderia. Viruses, 12(6), 601. https://doi.org/10.3390/v12060601





