RNA-Binding Domains of Heterologous Viral Proteins Substituted for Basic Residues in the RSV Gag NC Domain Restore Specific Packaging of Genomic RNA
Abstract
:1. Introduction
2. Materials and Methods
2.1. Expression Vectors, Plasmids, and Cells
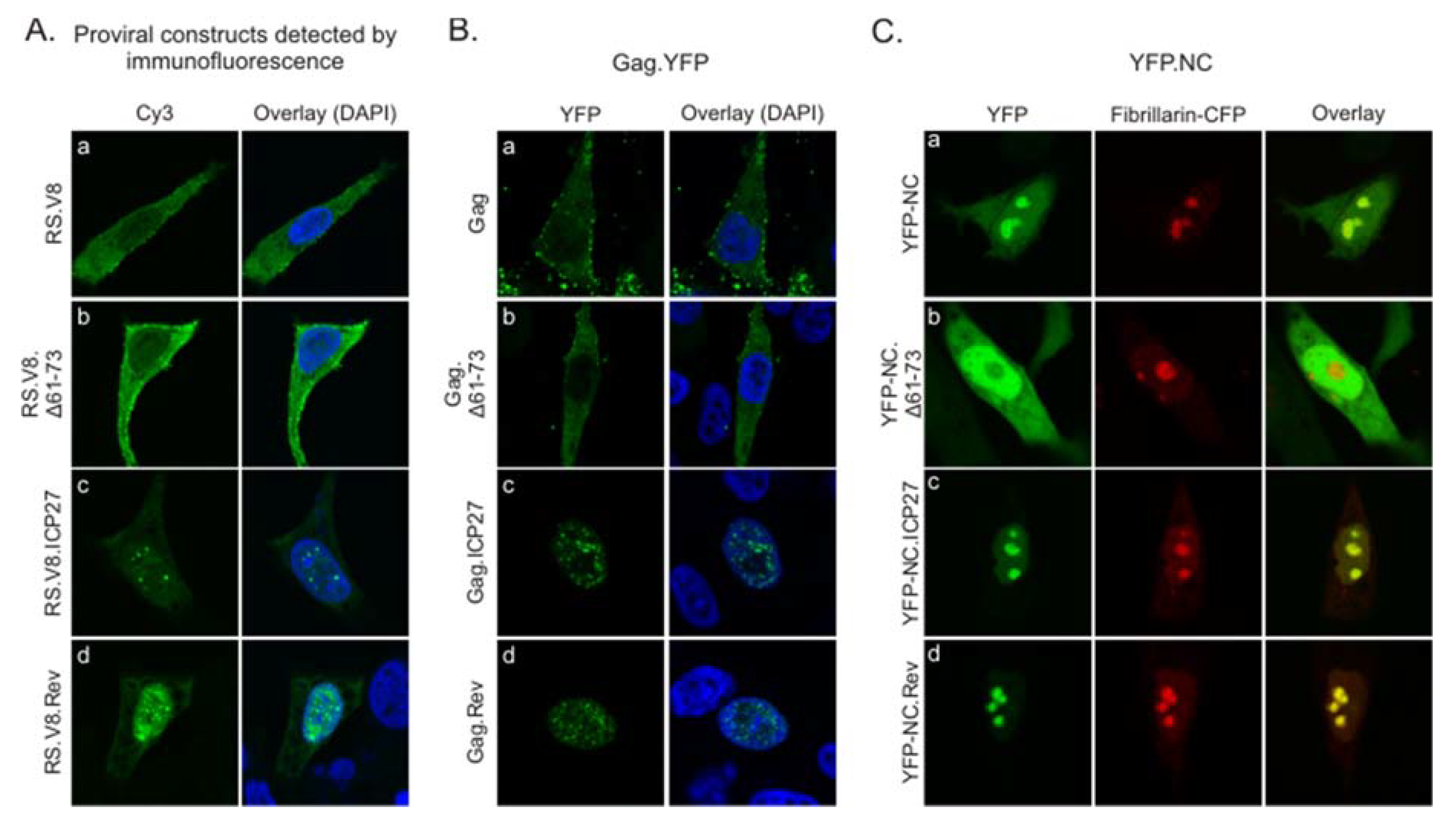
2.2. Immunofluorescence
2.3. Confocal Imaging
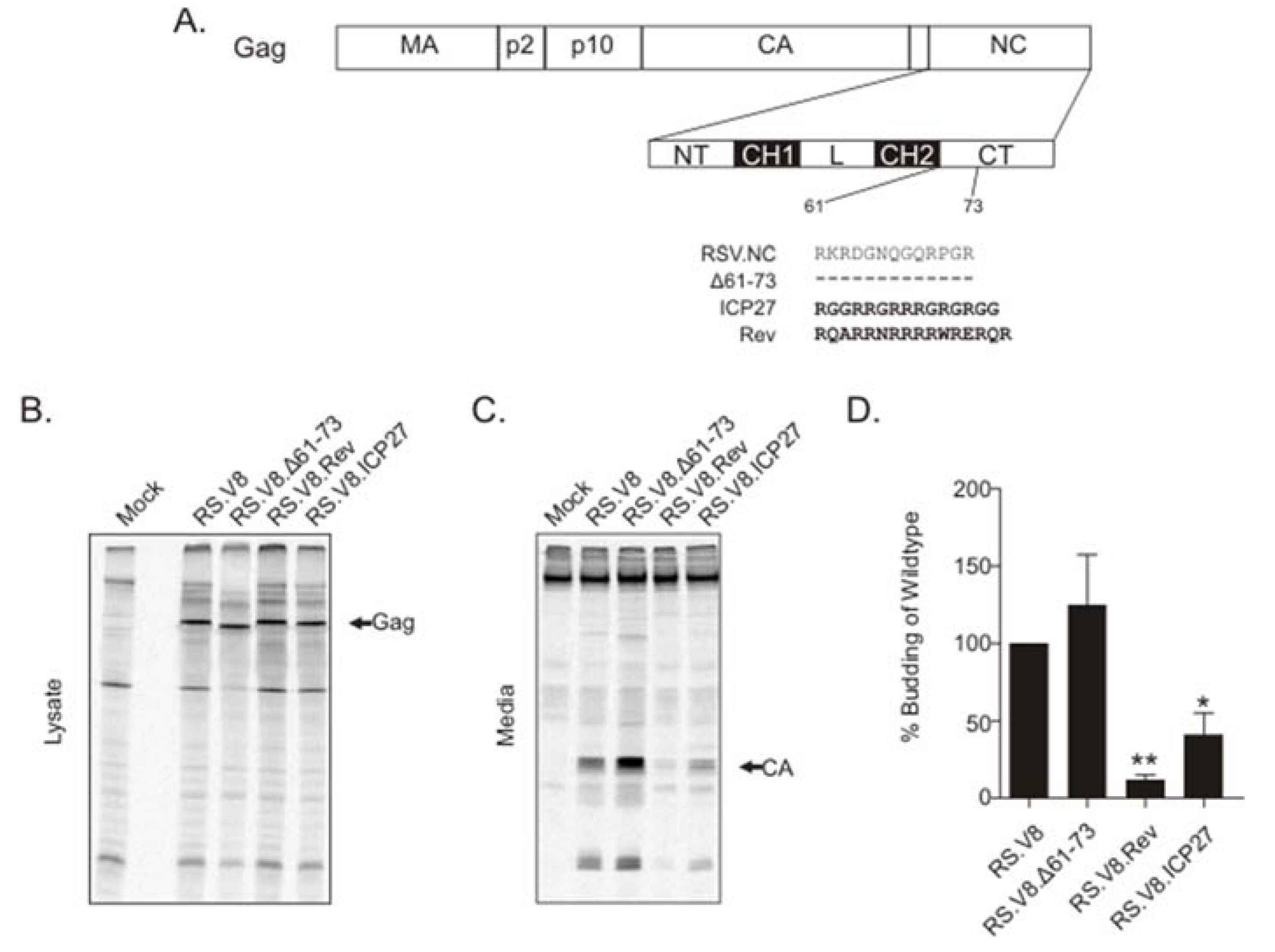
2.4. Budding Analysis
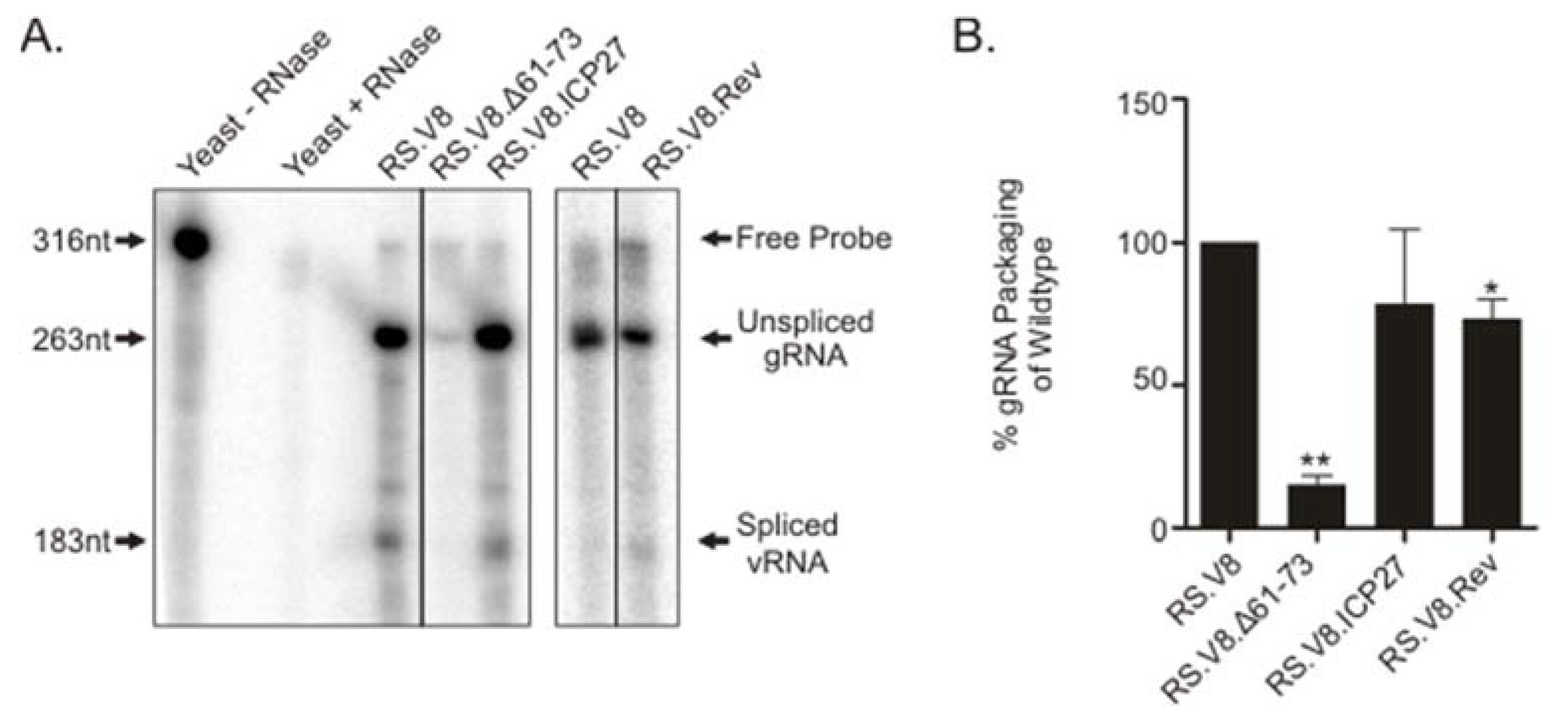
2.5. Ribonuclease Protection Assays
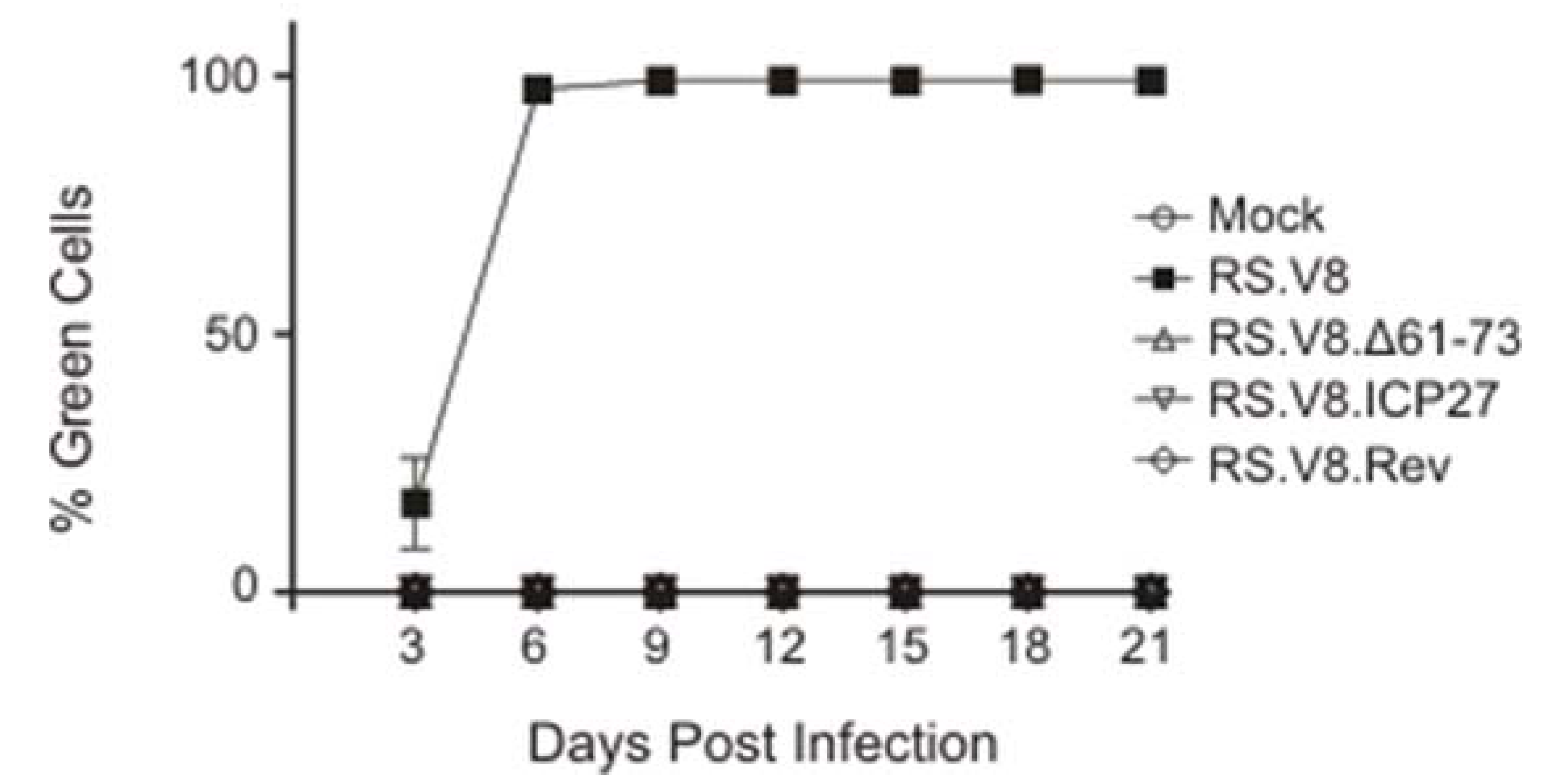
2.6. Viral Infectivity Assay
3. Results
3.1. Substitution of the ICP27 or Rev Basic Residue Motifs(BR) for Basic Residues in NC Altered the Budding of Virus Particles
3.2. Subcellular Localization of the ICP27 BR and Rev BR Gag Mutants
3.3. Heterologous BRs Substituted in the Gag NC Domain Restore gRNA Packaging
3.4. Viruses with Deletions of Basic Residues or Heterologous BR Substitutions in the NC Domain of Gag Are Noninfectious
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Kaddis Maldonado, R.J.; Parent, L.J. Orchestrating the Selection and Packaging of Genomic RNA by Retroviruses: An Ensemble of Viral and Host Factors. Viruses 2016, 8, 257. [Google Scholar] [CrossRef] [PubMed]
- Muriaux, D.; Darlix, J.L. Properties and functions of the nucleocapsid protein in virus assembly. RNA Biol. 2010, 7, 744–753. [Google Scholar] [CrossRef]
- Darlix, J.L.; de Rocquigny, H.; Mauffret, O.; Mely, Y. Retrospective on the all-in-one retroviral nucleocapsid protein. Virus Res. 2014, 193, 2–15. [Google Scholar] [CrossRef] [Green Version]
- Mirambeau, G.; Lyonnais, S.; Gorelick, R.J. Features, processing states, and heterologous protein interactions in the modulation of the retroviral nucleocapsid protein function. RNA Biol. 2010, 7, 724–734. [Google Scholar] [CrossRef] [Green Version]
- Olson, E.D.; Musier-Forsyth, K. Retroviral Gag protein-RNA interactions: Implications for specific genomic RNA packaging and virion assembly. Semin. Cell Dev. Biol. 2019, 86, 129–139. [Google Scholar] [CrossRef]
- Zhou, J.; McAllen, J.K.; Tailor, Y.; Summers, M.F. High affinity nucleocapsid protein binding to the muPsi RNA packaging signal of Rous sarcoma virus. J. Mol. Biol. 2005, 349, 976–988. [Google Scholar] [CrossRef]
- Scheifele, L.Z.; Rhoads, J.D.; Parent, L.J. Specificity of plasma membrane targeting by the rous sarcoma virus gag protein. J. Virol. 2003, 77, 470–480. [Google Scholar] [CrossRef] [Green Version]
- Garbitt-Hirst, R.; Kenney, S.P.; Parent, L.J. Genetic evidence for a connection between Rous sarcoma virus gag nuclear trafficking and genomic RNA packaging. J. Virol. 2009, 83, 6790–6797. [Google Scholar] [CrossRef] [Green Version]
- Butterfield-Gerson, K.L.; Scheifele, L.Z.; Ryan, E.P.; Hopper, A.K.; Parent, L.J. Importin-beta family members mediate alpharetrovirus gag nuclear entry via interactions with matrix and nucleocapsid. J. Virol. 2006, 80, 1798–1806. [Google Scholar] [CrossRef] [Green Version]
- Gudleski, N.; Flanagan, J.M.; Ryan, E.P.; Bewley, M.C.; Parent, L.J. Directionality of nucleocytoplasmic transport of the retroviral gag protein depends on sequential binding of karyopherins and viral RNA. Proc. Natl. Acad. Sci. USA 2010, 107, 9358–9363. [Google Scholar] [CrossRef] [Green Version]
- Lochmann, T.L.; Bann, D.V.; Ryan, E.P.; Beyer, A.R.; Mao, A.; Cochrane, A.; Parent, L.J. NC-mediated nucleolar localization of retroviral gag proteins. Virus Res. 2013, 171, 304–318. [Google Scholar] [CrossRef] [Green Version]
- Scheifele, L.Z.; Garbitt, R.A.; Rhoads, J.D.; Parent, L.J. Nuclear entry and CRM1-dependent nuclear export of the Rous sarcoma virus Gag polyprotein. Proc. Natl. Acad. Sci. USA 2002, 99, 3944–3949. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scheifele, L.Z.; Ryan, E.P.; Parent, L.J. Detailed mapping of the nuclear export signal in the Rous sarcoma virus Gag protein. J. Virol. 2005, 79, 8732–8741. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kenney, S.P.; Lochmann, T.L.; Schmid, C.L.; Parent, L.J. Intermolecular interactions between retroviral Gag proteins in the nucleus. J. Virol. 2008, 82, 683–691. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rice, B.L.; Kaddis, R.J.; Stake, M.S.; Lochmann, T.L.; Parent, L.J. Interplay between the alpharetroviral Gag protein and SR proteins SF2 and SC35 in the nucleus. Front. Microbiol. 2015, 6, 925. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bowles, N.E.; Damay, P.; Spahr, P.F. Effect of rearrangements and duplications of the Cys-His motifs of Rous sarcoma virus nucleocapsid protein. J. Virol. 1993, 67, 623–631. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meric, C.; Gouilloud, E.; Spahr, P.F. Mutations in Rous sarcoma virus nucleocapsid protein p12 (NC): Deletions of Cys-His boxes. J. Virol. 1988, 62, 3328–3333. [Google Scholar] [CrossRef] [Green Version]
- Zhou, J.; Bean, R.L.; Vogt, V.M.; Summers, M. Solution structure of the Rous sarcoma virus nucleocapsid protein: muPsi RNA packaging signal complex. J. Mol. Biol. 2007, 365, 453–467. [Google Scholar] [CrossRef] [Green Version]
- Dupraz, P.; Oertle, S.; Meric, C.; Damay, P.; Spahr, P.F. Point mutations in the proximal Cys-His box of Rous sarcoma virus nucleocapsid protein. J. Virol. 1990, 64, 4978–4987. [Google Scholar] [CrossRef] [Green Version]
- René, B.; Mauffret, O.; Fossé, P. Retroviral nucleocapsid proteins and DNA strand transfers. Biochim. Open 2018, 7, 10–25. [Google Scholar] [CrossRef]
- Lee, E.G.; Linial, M.L. Basic residues of the retroviral nucleocapsid play different roles in gag-gag and Gag-Psi RNA interactions. J. Virol. 2004, 78, 8486–8495. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, E.G.; Alidina, A.; May, C.; Linial, M.L. Importance of basic residues in binding of rous sarcoma virus nucleocapsid to the RNA packaging signal. J. Virol. 2003, 77, 2010–2020. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bowzard, J.B.; Bennett, R.P.; Krishna, N.K.; Ernst, S.M.; Rein, A.; Wills, J.W. Importance of basic residues in the nucleocapsid sequence for retrovirus Gag assembly and complementation rescue. J. Virol. 1998, 72, 9034–9044. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lengyel, J.; Guy, C.; Leong, V.; Borge, S.; Rice, S.A. Mapping of functional regions in the amino-terminal portion of the herpes simplex virus ICP27 regulatory protein: Importance of the leucine-rich nuclear export signal and RGG Box RNA-binding domain. J. Virol. 2002, 76, 11866–11879. [Google Scholar] [CrossRef] [Green Version]
- Mears, W.E.; Lam, V.; Rice, S.A. Identification of nuclear and nucleolar localization signals in the herpes simplex virus regulatory protein ICP27. J. Virol. 1995, 69, 935–947. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sandri-Goldin, R.M. ICP27 mediates HSV RNA export by shuttling through a leucine-rich nuclear export signal and binding viral intronless RNAs through an RGG motif. Genes Dev. 1998, 12, 868–879. [Google Scholar] [CrossRef]
- Mears, W.E.; Rice, S.A. The RGG box motif of the herpes simplex virus ICP27 protein mediates an RNA-binding activity and determines in vivo methylation. J. Virol. 1996, 70, 7445–7453. [Google Scholar] [CrossRef] [Green Version]
- Sciabica, K.S.; Dai, Q.J.; Sandri-Goldin, R.M. ICP27 interacts with SRPK1 to mediate HSV splicing inhibition by altering SR protein phosphorylation. EMBO J. 2003, 22, 1608–1619. [Google Scholar] [CrossRef] [Green Version]
- Fankhauser, C.; Izaurralde, E.; Adachi, Y.; Wingfield, P.; Laemmli, U.K. Specific complex of human immunodeficiency virus type 1 rev and nucleolar B23 proteins: Dissociation by the Rev response element. Mol. Cell. Biol. 1991, 11, 2567–2575. [Google Scholar] [CrossRef] [Green Version]
- Daelemans, D.; Costes, S.V.; Cho, E.H.; Erwin-Cohen, R.A.; Lockett, S.; Pavlakis, G.N. In vivo HIV-1 Rev multimerization in the nucleolus and cytoplasm identified by fluorescence resonance energy transfer. J. Biol. Chem. 2004, 279, 50167–50175. [Google Scholar] [CrossRef] [Green Version]
- Berger, J.; Aepinus, C.; Dobrovnik, M.; Fleckenstein, B.; Hauber, J.; Bohnlein, E. Mutational analysis of functional domains in the HIV-1 Rev trans-regulatory protein. Virology 1991, 183, 630–635. [Google Scholar] [CrossRef]
- Himly, M.; Foster, D.N.; Bottoli, I.; Iacovoni, J.S.; Vogt, P.K. The DF-1 chicken fibroblast cell line: Transformation induced by diverse oncogenes and cell death resulting from infection by avian leukosis viruses. Virology 1998, 248, 295–304. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parent, L.J.; Cairns, T.M.; Albert, J.A.; Wilson, C.B.; Wills, J.W.; Craven, R.C. RNA dimerization defect in a Rous sarcoma virus matrix mutant. J. Virol. 2000, 74, 164–172. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fujiwara, T.; Oda, K.; Yokota, S.; Takatsuki, A.; Ikehara, Y. Brefeldin A causes disassembly of the Golgi complex and accumulation of secretory proteins in the endoplasmic reticulum. J. Biol. Chem. 1988, 263, 18545–18552. [Google Scholar] [PubMed]
- Matic, I.; van Hagen, M.; Schimmel, J.; Macek, B.; Ogg, S.C.; Tatham, M.H.; Hay, R.T.; Lamond, A.I.; Mann, M.; Vertegaal, A.C.O. In vivo identification of human small ubiquitin-like modifier polymerization sites by high accuracy mass spectrometry and an in vitro to in vivo strategy. Mol. Cell. Proteom. 2008, 7, 132–144. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weldon, R.A.; Erdie, C.R.; Oliver, M.G.; Wills, J.W. Incorporation of chimeric gag protein into retroviral particles. J. Virol. 1990, 64, 4169. [Google Scholar] [CrossRef] [Green Version]
- Callahan, E.M.; Wills, J.W. Repositioning basic residues in the M domain of the Rous sarcoma virus gag protein. J. Virol. 2000, 74, 11222–11229. [Google Scholar] [CrossRef] [Green Version]
- Craven, R.C.; Leure-duPree, A.E.; Weldon, R.A., Jr.; Wills, J.W. Genetic analysis of the major homology region of the Rous sarcoma virus Gag protein. J. Virol. 1995, 69, 4213–4227. [Google Scholar] [CrossRef] [Green Version]
- Garbitt, R.A.; Bone, K.R.; Parent, L.J. Insertion of a classical nuclear import signal into the matrix domain of the Rous sarcoma virus Gag protein interferes with virus replication. J. Virol. 2004, 78, 13534–13542. [Google Scholar] [CrossRef] [Green Version]
- Mears, W.E.; Rice, S.A. The herpes simplex virus immediate-early protein ICP27 shuttles between nucleus and cytoplasm. Virology 1998, 242, 128–137. [Google Scholar] [CrossRef] [Green Version]
- Bohnlein, E.; Berger, J.; Hauber, J. Functional mapping of the human immunodeficiency virus type 1 Rev RNA binding domain: New insights into the domain structure of Rev and Rex. J. Virol. 1991, 65, 7051–7055. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, E.G.; Linial, M.L. Deletion of a Cys-His motif from the Alpharetrovirus nucleocapsid domain reveals late domain mutant-like budding defects. Virology 2006, 347, 226–233. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gallis, B.; Linial, M.; Eisenman, R. An avian oncovirus mutant deficient in genomic RNA: Characterization of the packaged RNA as cellular messenger RNA. Virology 1979, 94, 146–161. [Google Scholar] [CrossRef]
- Anderson, D.J.; Stone, J.; Lum, R.; Linial, M.L. The packaging phenotype of the SE21Q1b provirus is related to high proviral expression and not trans-acting factors. J. Virol. 1995, 69, 7319. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shank, P.R.; Linial, M. Avian oncovirus mutant (SE21Q1b) deficient in genomic RNA: Characterization of a deletion in the provirus. J. Virol. 1980, 36, 450–456. [Google Scholar] [CrossRef] [Green Version]
- Perkins, A.; Cochrane, A.W.; Ruben, S.M.; Rosen, C.A. Structural and functional characterization of the human immunodeficiency virus rev protein. J. Acquir. Immune Defic. Syndr. 1989, 2, 256–263. [Google Scholar]
- Malik, P.; Tabarraei, A.; Kehlenbach, R.H.; Korfali, N.; Iwasawa, R.; Graham, S.V.; Schirmer, E.C. Herpes simplex virus ICP27 protein directly interacts with the nuclear pore complex through Nup62, inhibiting host nucleocytoplasmic transport pathways. J. Biol. Chem. 2012, 287, 12277–12292. [Google Scholar] [CrossRef] [Green Version]
- Rojas, S.; Corbin-Lickfett, K.A.; Escudero-Paunetto, L.; Sandri-Goldin, R.M. ICP27 Phosphorylation Site Mutants Are Defective in Herpes Simplex Virus 1 Replication and Gene Expression. J. Virol. 2010, 84, 2200. [Google Scholar] [CrossRef] [Green Version]
- Wills, J.W.; Craven, R.C. Form, function, and use of retroviral gag proteins. AIDS 1991, 5, 639–654. [Google Scholar] [CrossRef]
- Ma, Y.M.; Vogt, V.M. Rous sarcoma virus Gag protein-oligonucleotide interaction suggests a critical role for protein dimer formation in assembly. J. Virol. 2002, 76, 5452–5462. [Google Scholar] [CrossRef] [Green Version]
- Sandri-Goldin, R.M. The many roles of the regulatory protein ICP27 during herpes simplex virus infection. Front. Biosci. 2008, 13, 5241–5256. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tunnicliffe, R.B.; Tian, X.; Storer, J.; Sandri-Goldin, R.M.; Golovanov, A.P. Overlapping motifs on the herpes viral proteins ICP27 and ORF57 mediate interactions with the mRNA export adaptors ALYREF and UIF. Sci. Rep. 2018, 8, 15005. [Google Scholar] [CrossRef]
- Naji, S.; Ambrus, G.; Cimermančič, P.; Reyes, J.R.; Johnson, J.R.; Filbrandt, R.; Huber, M.D.; Vesely, P.; Krogan, N.J.; Yates, J.R., 3rd; et al. Host cell interactome of HIV-1 Rev includes RNA helicases involved in multiple facets of virus production. Mol. Cell. Proteom. MCP 2012, 11, M111.015313. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Modem, S.; Thipparthi, R.R. Cellular Proteins and HIV-1 Rev Function. Curr. HIV Res. 2009, 7, 91–100. [Google Scholar] [CrossRef]
- Lunde, B.M.; Moore, C.; Varani, G. RNA-binding proteins: Modular design for efficient function. Nat. Rev. Mol. Cell Biol. 2007, 8, 479–490. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Balcerak, A.; Trebinska-Stryjewska, A.; Konopinski, R.; Wakula, M.; Grzybowska, E.A. RNA–protein interactions: Disorder, moonlighting and junk contribute to eukaryotic complexity. Open Biol. 2019. [Google Scholar] [CrossRef] [Green Version]
- Jeong, S. SR Proteins: Binders, Regulators, and Connectors of RNA. Mol. Cells 2017, 40, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Ghaemi, Z.; Guzman, I.; Gnutt, D.; Luthey-Schulten, Z.; Gruebele, M. Role of Electrostatics in Protein–RNA Binding: The Global vs the Local Energy Landscape. J. Phys. Chem. B 2017, 121, 8437–8446. [Google Scholar] [CrossRef]
- Baba, S.; Takahashi, K.; Koyanagi, Y.; Yamamoto, N.; Takaku, H.; Gorelick, R.J.; Kawai, G. Role of the zinc fingers of HIV-1 nucleocapsid protein in maturation of genomic RNA. J. Biochem. 2003, 134, 637–639. [Google Scholar] [CrossRef]
- Prats, A.C.; Housset, V.; de Billy, G.; Cornille, F.; Prats, H.; Roques, B.; Darlix, J.L. Viral RNA annealing activities of the nucleocapsid protein of Moloney murine leukemia virus are zinc independent. Nucleic Acids Res. 1991, 19, 3533–3541. [Google Scholar] [CrossRef]
- Allain, B.; Lapadat-Tapolsky, M.; Berlioz, C.; Darlix, J.L. Transactivation of the minus-strand DNA transfer by nucleocapsid protein during reverse transcription of the retroviral genome. EMBO J. 1994, 13, 973–981. [Google Scholar] [CrossRef] [PubMed]
- Barat, C.; Schatz, O.; Le Grice, S.; Darlix, J.L. Analysis of the interactions of HIV1 replication primer tRNA(Lys,3) with nucleocapsid protein and reverse transcriptase. J. Mol. Biol. 1993, 231, 185–190. [Google Scholar] [CrossRef] [PubMed]
- Darlix, J.L.; Vincent, A.; Gabus, C.; de Rocquigny, H.; Roques, B. Trans-activation of the 5’ to 3’ viral DNA strand transfer by nucleocapsid protein during reverse transcription of HIV1 RNA. C. R. Acad. Sci. III 1993, 316, 763–771. [Google Scholar] [PubMed]
- Drummond, J.E.; Mounts, P.; Gorelick, R.J.; Casas-Finet, J.R.; Bosche, W.J.; Henderson, L.E.; Waters, D.J.; Arthur, L.O. Wild-type and mutant HIV type 1 nucleocapsid proteins increase the proportion of long cDNA transcripts by viral reverse transcriptase. AIDS Res. Hum. Retroviruses 1997, 13, 533–543. [Google Scholar] [CrossRef]
- Guo, J.; Wu, T.; Anderson, J.; Kane, B.F.; Johnson, D.G.; Gorelick, R.J.; Henderson, L.E.; Levin, J.G. Zinc finger structures in the human immunodeficiency virus type 1 nucleocapsid protein facilitate efficient minus- and plus-strand transfer. J. Virol. 2000, 74, 8980–8988. [Google Scholar] [CrossRef] [Green Version]
- Thomas, J.A.; Gorelick, R.J. Nucleocapsid protein function in early infection processes. Virus Res. 2008, 134, 39–63. [Google Scholar] [CrossRef] [Green Version]
- Rye-McCurdy, T.D.; Nadaraia-Hoke, S.; Gudleski-O’Regan, N.; Flanagan, J.M.; Parent, L.J.; Musier-Forsyth, K. Mechanistic differences between nucleic acid chaperone activities of the Gag proteins of Rous sarcoma virus and human immunodeficiency virus type 1 are attributed to the MA domain. J. Virol. 2014, 88, 7852–7861. [Google Scholar] [CrossRef] [Green Version]
- Stewart-Maynard, K.M.; Cruceanu, M.; Wang, F.; Vo, M.-N.; Gorelick, R.J.; Williams, M.C.; Rouzina, I.; Musier-Forsyth, K. Retroviral Nucleocapsid Proteins Display Nonequivalent Levels of Nucleic Acid Chaperone Activity. J. Virol. 2008, 82, 10129. [Google Scholar] [CrossRef] [Green Version]
- Chadwick, D.R.; Lever, A.M. Antisense RNA sequences targeting the 5’ leader packaging signal region of human immunodeficiency virus type-1 inhibits viral replication at post-transcriptional stages of the life cycle. Gene Ther. 2000, 7, 1362–1368. [Google Scholar] [CrossRef] [Green Version]
- Ingemarsdotter, C.K.; Zeng, J.; Long, Z.; Lever, A.M.L.; Kenyon, J.C. An RNA-binding compound that stabilizes the HIV-1 gRNA packaging signal structure and specifically blocks HIV-1 RNA encapsidation. Retrovirology 2018, 15, 25. [Google Scholar] [CrossRef]
- Andersson, A.-M.C.; Schwerdtfeger, M.; Holst, P.J. Virus-Like-Vaccines against HIV. Vaccines 2018, 6, 10. [Google Scholar] [CrossRef] [Green Version]
- Zhao, C.; Ao, Z.; Yao, X. Current Advances in Virus-Like Particles as a Vaccination Approach against HIV Infection. Vaccines 2016, 4, 2. [Google Scholar] [CrossRef]
- Doan, L.X.; Li, M.; Chen, C.; Yao, Q. Virus-like particles as HIV-1 vaccines. Rev. Med. Virol. 2005, 15, 75–88. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rice, B.L.; Lochmann, T.L.; Parent, L.J. RNA-Binding Domains of Heterologous Viral Proteins Substituted for Basic Residues in the RSV Gag NC Domain Restore Specific Packaging of Genomic RNA. Viruses 2020, 12, 370. https://doi.org/10.3390/v12040370
Rice BL, Lochmann TL, Parent LJ. RNA-Binding Domains of Heterologous Viral Proteins Substituted for Basic Residues in the RSV Gag NC Domain Restore Specific Packaging of Genomic RNA. Viruses. 2020; 12(4):370. https://doi.org/10.3390/v12040370
Chicago/Turabian StyleRice, Breanna L., Timothy L. Lochmann, and Leslie J. Parent. 2020. "RNA-Binding Domains of Heterologous Viral Proteins Substituted for Basic Residues in the RSV Gag NC Domain Restore Specific Packaging of Genomic RNA" Viruses 12, no. 4: 370. https://doi.org/10.3390/v12040370
APA StyleRice, B. L., Lochmann, T. L., & Parent, L. J. (2020). RNA-Binding Domains of Heterologous Viral Proteins Substituted for Basic Residues in the RSV Gag NC Domain Restore Specific Packaging of Genomic RNA. Viruses, 12(4), 370. https://doi.org/10.3390/v12040370




