Abstract
Across sub-Saharan Africa, key populations with elevated HIV-1 incidence and/or prevalence have been identified, but their contribution to disease spread remains unclear. We performed viral deep-sequence phylogenetic analyses to quantify transmission dynamics between the general population (GP), fisherfolk communities (FF), and women at high risk of infection and their clients (WHR) in central and southwestern Uganda. Between August 2014 and August 2017, 6185 HIV-1 positive individuals were enrolled in 3 GP and 10 FF communities, 3 WHR enrollment sites. A total of 2531 antiretroviral therapy (ART) naïve participants with plasma viral load >1000 copies/mL were deep-sequenced. One hundred and twenty-three transmission networks were reconstructed, including 105 phylogenetically highly supported source–recipient pairs. Only one pair involved a WHR and male participant, suggesting that improved population sampling is needed to assess empirically the role of WHR to the transmission dynamics. More transmissions were observed from the GP communities to FF communities than vice versa, with an estimated flow ratio of 1.56 (95% CrI 0.68–3.72), indicating that fishing communities on Lake Victoria are not a net source of transmission flow to neighboring communities further inland. Men contributed disproportionally to HIV-1 transmission flow regardless of age, suggesting that prevention efforts need to better aid men to engage with and stay in care.
1. Introduction
In 2019, the Joint United Nations program on HIV/AIDS (UNAIDS) reported an estimated 1.4 million people that were living with HIV-1 in Uganda [1]. Despite significant decreases in HIV-1 prevalence and incidence since the early 1990s, the epidemic in Uganda remains self-sustaining through heterosexual transmission in the general population (GP) [2]. Certain key populations in Uganda are particularly affected by HIV-1. These include fisherfolk (FF), women at high risk of HIV-1 infection (WHR), men who have sex with men, injection drug users, as well as men and women of the uniformed forces [3]. Average national HIV-1 incidence is estimated at less than 1 per 100 person years at risk (1/100 PYAR) [4], while incidence rates among FF and WHR are respectively estimated at 6/100 PYAR [5] and 3/100 PYAR [6]. Individuals in these groups also have a higher tendency to engage in high-risk sexual behavior, which led to the hypothesis that they may drive HIV-1 transmission in general, and sustain transmission in the GP [2,7].
UNAIDS and the WHO have recommended that countries develop combination HIV-1 intervention packages that are tailored to their high-risk/key populations in order to curb the HIV-1 epidemic in a cost-effective manner [8,9]. To achieve this, several strategies have been suggested. These include spatial mapping to identify geographic areas of higher-than-average HIV-1 prevalence [10] and demographic surveillance to characterize populations that are at higher risk of HIV-1 infection [11,12]. Recent studies in Uganda have employed spatial mapping and demographic surveillance prior to the implementation of combination interventions in high-risk populations like FF [13,14] and WHR [15]. In addition, phylogenetic analyses have been increasingly used to characterize aspects of HIV-1 transmission dynamics from nucleotide sequences of the virus. For instance, phylogenetic analysis helped to reconstruct the historical spread of HIV-1 [16,17,18], characterize HIV-1 transmission networks [19,20], identify traits associated with onward viral transmission in high-risk groups [21,22], and evaluate HIV-1 migration patterns [23,24]. In combination with sociodemographic or epidemiological data, these approaches have provided useful insights in assessing risk factors associated with HIV-1 spread [25] and identifying micro-epidemics [26], thus informing prevention strategies. Furthermore, they have been applied in source-attribution studies to assess the role of viral introductions from other communities [27,28], and to identify populations that serve as sinks or sources for HIV-1 transmission [29,30]. In particular, we previously estimated that high-prevalence FF communities located in central and southwestern Uganda are net recipients (sinks) of local HIV-1 transmission flow, based on phylogeographic analysis of 208 subtype A and 177 subtype D consensus sequences [29]. A second analysis estimated that there were 2.50 times (95% credibility interval (95%CrI) 1.02–7.30) more transmissions from inland to fishing areas than vice versa, based on a sample of 2652 viral deep-sequences and after accounting for human migration dynamics [30]. The two studies thus showed that the relatively small FF communities around Lake Victoria are not driving the surrounding HIV-1 epidemics regardless of their high HIV-1 prevalence.
Here, we test the finding that FF communities are net sinks and not sources of local transmission flow on an independent data set that includes a larger set of fishing villages. In addition, we aim to assess the extent to which key populations (FF and WHR) and socio-demographic factors contribute to the HIV-1 epidemic in the GP. To this end, the Medical Research Council/Uganda Virus Research Institute and London School of Hygiene & Tropical Medicine (MRC/UVRI and LSHTM) Uganda Research Unit team enrolled HIV-1 infected individuals in fishing villages along the shores of Lake Victoria, neighboring inland areas (approximately 10–40 km) and hotspots of women engaged in high-risk sexual behavior (see Figure 1 under Materials and Methods). The PANGEA (Phylogenetics And Networks for Generalized Epidemics in Africa) consortium (Appendix A and Appendix B) [31] generated HIV-1 deep-sequences from this convenience sample of infected individuals. The deep-sequence data was then used to reconstruct viral phylogenetic transmission networks that contained information on the direction of transmission. Pairs with phylogenetically highly supported evidence for the direction of transmission were identified in the networks, and these pairs were used to quantify HIV-1 transmission flows between the GP, FF, and WHR populations. In doing so, this study also demonstrates that HIV deep-sequence data can now be routinely used to reconstruct HIV-1 transmission networks, and provide important information on the direction of transmission within them.
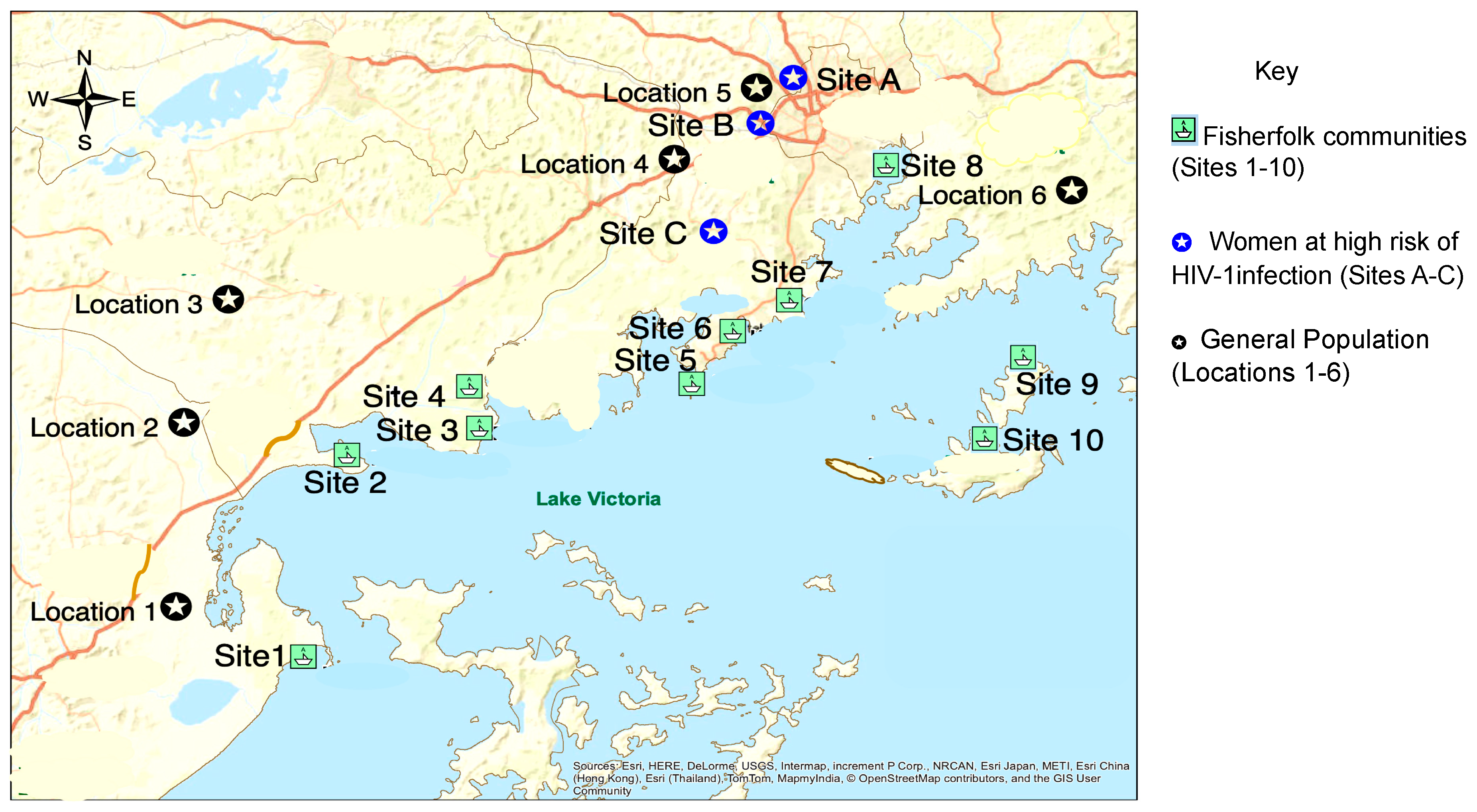
Figure 1.
Map showing the sampling sites. Sites 1 to 10 show the fisherfolk (FF) communities and sites A to C show areas where women at high risk of HIV-1 infection (WHR) and some of their clients were enrolled. Locations 1 to 6 show areas in the general population (GP) where study participants were enrolled. Almost half (40.9%) of HIV-1 positive participants enrolled in clinics and study sites (see Table 1) shown on the map in south central Uganda had their samples deep sequenced and subjected to phylogenetic analyses.
2. Materials and Methods
2.1. Study Design and Population
The MRC/UVRI and LSHTM Uganda Research Unit (MRC) conducted several population-based cross-sectional surveys in HIV-1 high-risk and general population groups in central and southwestern Uganda [29,32,33,34,35,36]. In this analysis, we combined data collected from several populations surveyed between August 2014 and August 2017. Figure 1 below shows a map of the study sites.
The FF study population included individuals from 10 FF communities [13,19,37,38,39] as shown in Figure 1. Individuals were approached in community health facilities (CHF) or during voluntary counselling and testing (VCT) campaigns, and were eligible to participate if they had an HIV-1 positive status and were at least 18 years old (FF communities 1–8) or 14 years (FF communities 9–10).
The WHR study population included participants of the MRC Good Health for Women’s Project (GHWP), and an extension of this study population to other commercial sex work areas (non-GHWP). The GHWP cohort was initiated in 2008, when initially 1027 women were recruited with the objective of studying the size, determinants, and dynamics of the HIV-1 epidemic, and remains an open cohort with approximately 3000 women enrolled to-date [6,40]. Women above 18 years of age who reported being involved in commercial sex work and/or high-risk sexual behavior, and their clients who consented to participate were enrolled in the study. Data were collected on socio-demographic characteristics and risk behavior, and consenting participants were tested for HIV-1.
The GP study population included individuals from rural communities surveyed through the MRC General Population Cohort (GPC) since 1989 [33,41]. We additionally sampled areas that neighbor the FF populations, as well as areas that are geographically close to the WHR recruitment area. In these communities, individuals were recruited in CHF or during VCT campaigns, and eligible if they were HIV-1 positive and were at least 16 years old. Further, individuals with HIV-1 infection who received care or were diagnosed through VCT were invited to participate. Participants completed structured questionnaires that captured general demographic, socioeconomic, marital, partnership histories and behavioral data. A biometric fingerprint-scanning device was used on all study participants to avoid duplicate enrolments [42].
2.2. Ethics Statement
This study was part of the MRC HIV-1 Molecular Epidemiology study which was approved by the Uganda Virus Research Institute Research and Ethics Committee (UVRI-REC) on the 24th/06/2013 (Federal Wide Assurance (FWA) No. 00001354, Project identification code: GC/127/13/06/27) and the Uganda National Council for Science and Technology (UNCST) on the 12th/09/2013 (FWA No. 00001293, Project identification code: HS 1432). All participants were recruited voluntarily and provided written informed consent.
2.3. Deep Sequencing and Assembly of HIV-1 Reads
First, 10 mL of blood was collected from the study participants by venipuncture, including those on antiretroviral therapy (ART). Next generation sequencing was performed on samples from infected participants who did not report ART use and who had sufficient plasma viraemia (viral load >1000 copies/mL). HIV-1 deep sequencing was performed using the Illumina Miseq platform at the Africa Health Research Institute (Durban, South Africa), and at the Wellcome Trust Sanger Institute (Hinxton, UK) [43,44]. Briefly, viral RNA was extracted from plasma samples using the QIAamp Viral RNA Minikit (Qiagen Inc., Valencia, CA, USA). Reverse transcription and amplification of the extracted viral RNA was done in a one-step RT-PCR reaction using a set of 4 HIV-1 primers that were designed for the amplification of HIV-1 genomes in four overlapping amplicons for all groups and subtypes [43]. Amplicons were pooled in equimolar quantities according to the manufacturer’s instructions for the library preparation step using the Nextera XT kit. Library validation was performed using the Agilent Bioanalyzer before sequencing was done using the Miseq 250-bp paired end technology as previously described [43]. Deep-sequencing reads were assembled using the Shiver pipeline [45].
2.4. Deep-Sequence Phylogenetic Analysis
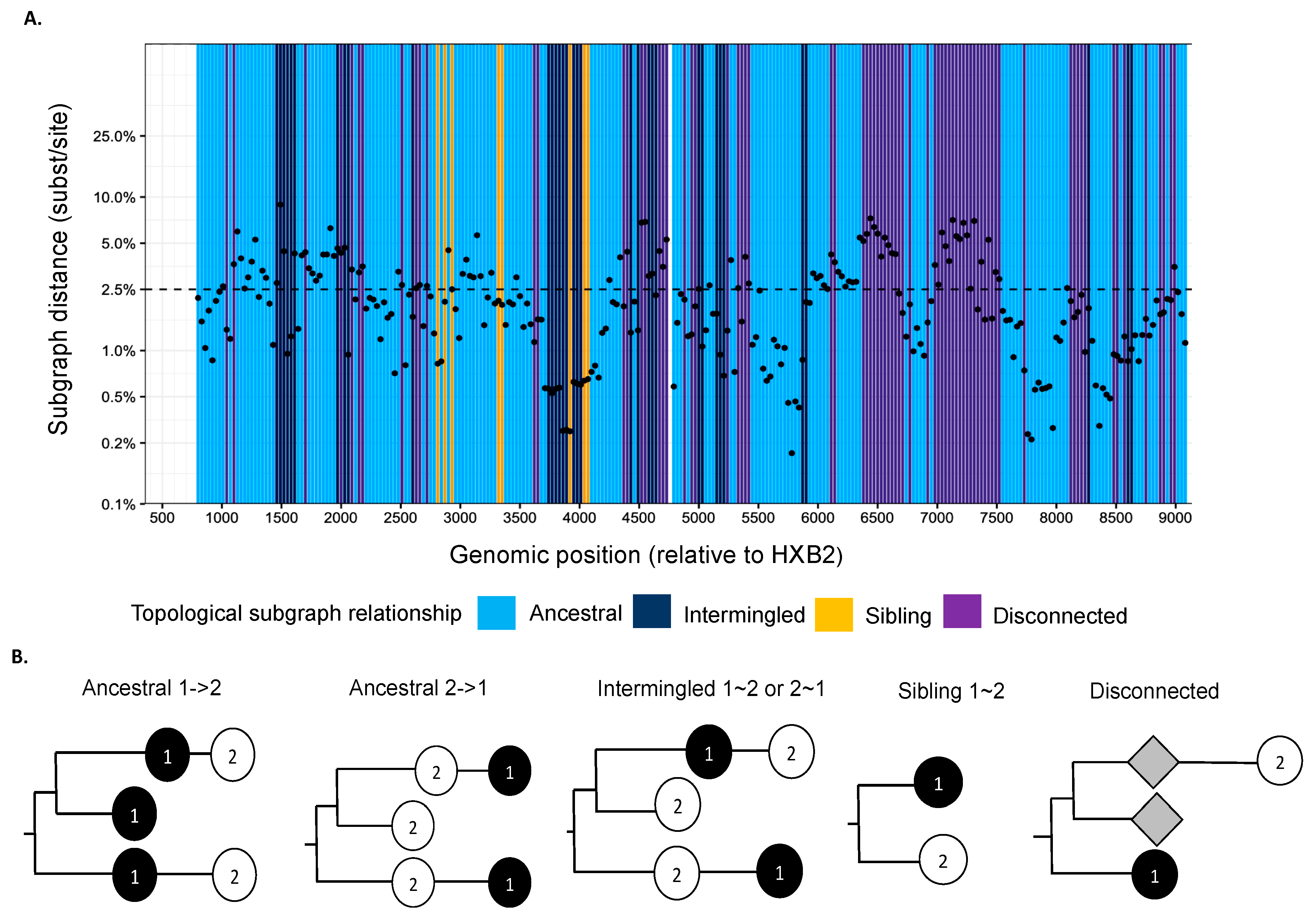
We used phyloscanner version 1.8.0 to reconstruct directed HIV-1 transmission networks from deep sequences [46]. Briefly, phyloscanner analysis starts with HIV deep-sequence reads from a set of infected individuals that are mapped against reference sequences of the entire genome. Across the genome, sliding windows are defined, and for each window, paired-end reads from all individuals that overlap the window are extracted and aligned, and deep-sequence phylogenies reconstructed. In our analysis, windows were of length 250 bp and incremented by 25 bp across the entire genome. Next, viral lineages in the deep-sequence phylogenies were attributed to each individual using maximum parsimony ancestral state reconstruction, which defined larger parts (called “subgraphs”) of the deep-sequence phylogenies that are attributed to viral evolution within each individual. Using Sanger sequencing, typically one sequence is available per person, and in this case, each subgraph consists of the tip sequence only that corresponds to the sampled individual. Using deep-sequence data, subgraphs of each individual typically comprise several tips and inner nodes in the phylogeny, and this information can be used to characterize the relationship of individuals in terms of distance between subgraphs and topological relationship between subgraphs. Figure 2 shows the subgraph distances (y-axis) and subgraph topologies (color) between a pair of individuals in the phylogenies that were reconstructed from the overlapping 250 bp read alignments across the genome. The subgraph distances are used to infer phylogenetic linkage, and the subgraph topologies are used to infer the likely direction of transmission between individuals (Figure 2b). A phyloscanner analysis returns for each pair of individuals phylogenetic scores for linkage and transmission direction that are calculated across all genomic windows [46,47,48]. The scores indicate the proportion of genomic windows with phylogenetic support for linkage and transmission direction, after accounting for overlap between the read alignments.
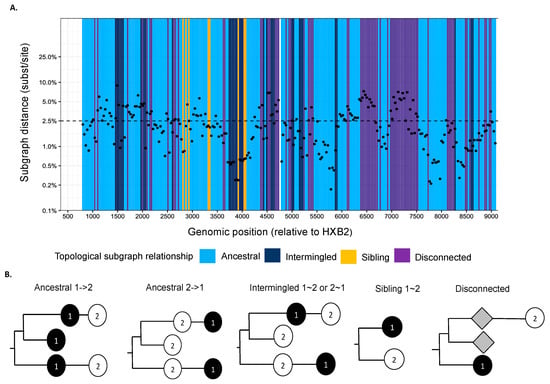
Figure 2.
(A) Phyloscan plot showing deep-sequence phylogenies reconstructed at 250 bp genomic window intervals across the genome. Ancestral state reconstruction was used to attribute phylogenetic lineages to individuals, and phylogenetic subgraphs were defined as groups of lineages that were attributed to the same individual. For a pair of individuals, the scan plots show the shortest patristic distance between subgraphs of both individuals (y-axis) and the topological relationship between subgraphs of both individuals (colors) across the genome (ancestral to one another (light blue), siblings (yellow), intermingled (dark blue), or disconnected (purple)). (B) Description of topological relationship between subgraphs in deep-sequence phylogenies. The phylogenetic topology or ordering of subgraphs is used to infer the likely direction of transmission between individuals [48]. Ancestral 1->2 means that at least one subgraph from individual 1 is phylogenetically adjacent to a subgraph from individual 2, and that all adjacent subgraphs from individual 1 are ancestral to those of individual 2. This phylogenetic pattern is consistent with transmission from individual 1 to individual 2, possibly via unsampled intermediates. Similarly, ancestral 2->1 means at least one subgraph from individual 1 is phylogenetically adjacent to a subgraph from individual 2, and that all adjacent subgraphs from individual 1 are descendent from those of individual 2. This phylogenetic pattern is consistent with transmission from individual 2 to individual 1, possibly via unsampled intermediates. Intermingled 1~2 means that at least one subgraph from individual 1 is phylogenetically adjacent to one subgraph from individual 2, and that there are adjacent subgraphs from individual 1 ancestral to those of individual 2, and vice versa. Sibling 1~2 means that at least one subgraph from individual 1 is phylogenetically adjacent to one subgraph from individual 2, and that the adjacent subgraphs are phylogenetically next to each other. These two patterns indicate that the two individuals are closely related, but there is no evidence for the direction of transmission. Disconnected refers to individuals who have subgraphs that are not adjacent to each other. This pattern suggests that the two individuals are phylogenetically unlinked.
2.5. Reconstruction of Transmission Networks
To reconstruct phylogenetic transmission networks, we first determined the deep-sequence phylogenetic relationships between all possible pairs of individuals in the sample with phyloscanner. Analysis proceeded in batches of 75 individuals, which considerably reduced the number of deep-sequence phylogenies that had to be estimated, and computational burden [47]. Second, from this output we identified pairs of individuals between whom phylogenetic linkage could not be excluded based on subgraph distance in deep-sequence phylogenies. Third, potentially phylogenetically linked pairs of individuals were grouped into larger phylogenetic transmission networks so that each individual was linked to at least one other individual. Fourth, each potential transmission network was separately analyzed with phyloscanner to substantiate membership of individuals in networks, and to estimate the phylogenetically likely direction of viral spread within networks. Fifth, we considered all possible transmission chains in these networks, which corresponded to all possible minimum (directed) spanning trees, similar as in [49,50]. We then identified the most likely transmission chain as the minimum spanning tree that maximized the product of phyloscanner transmission scores in the directions of the edges in the spanning tree [47]. Sixth, we extracted pairs of individuals with strong support for phylogenetic linkage and strong support for the direction of transmission in the phylogenetically most likely transmission chain (source–recipient pairs), using a threshold of 0.6 on the phyloscanner scores (see [47] for a justification of the threshold). The analysis protocol and phyloscanner runtime arguments were exactly as described in [47]. This analysis in general cannot rule out the presence of unsampled intermediates in the phylogenetically most likely transmission chains, and among source–recipient pairs.
2.6. Quantifying HIV-1 Transmission Flows
We used the phyloflows package, version 1.1.0, implemented in the R software [51] to quantify HIV-1 transmission flows within and between the three populations, while adjusting for sampling heterogeneity across our study populations. Briefly, a Bayesian model was used to estimate transmission flows in population groups under the assumption that each group was sampled at random, but with potentially different sampling rates [30]. A Markov Chain Monte Carlo (MCMC) algorithm was used to obtain samples from the posterior distribution of transmission flows, was run for 50,000 iterations, and the initial 5% were discarded as burn-in. Convergence was assessed with the Gelman–Rubin statistic [52], and all effective sample sizes of the marginal posterior transmission flows were well above 1000. Examplary MCMC outputs are shown in Supplementary Figure S1.
3. Results
3.1. Population-Based Sample of HIV-1 Deep Sequences
Between August 2014 and August 2017, a total of 6185 HIV-1 positive individuals were enrolled in CHF and VCT campaigns, of whom 2531 were deep-sequenced (at a sequencing depth of >30× over at least 5000 nt of the HIV-1 genome). A total of 3200 participants were from the GP of whom 1578 were male and 1622 female. Then, 1309 (40.9%) samples were deep-sequenced, comprising 636 men (40.3% of 1578) and 673 women (41.5% of 1622). Further, 2185 participants were from fishing sites of whom 1103 were male and 1082 were female. Then, 895 (41.0%) samples were deep-sequenced, comprising 468 men (42.4% of 1103) and 427 (39.5% of 1082) women. There were 720 women at high risk and 80 male clients recruited, of whom 301 (41.8%) women at high risk and 26 (32.5%) clients were deep-sequenced. Table 1 provides further demographic information on the recruited and sampled individuals. The proportion of men and women who had their samples deep sequenced in each study population were similar, with the exception of male clients of women at high risk.

Table 1.
Study population, and proportion of sampled and sequenced individuals.
3.2. Reconstructed HIV-1 Transmission Networks
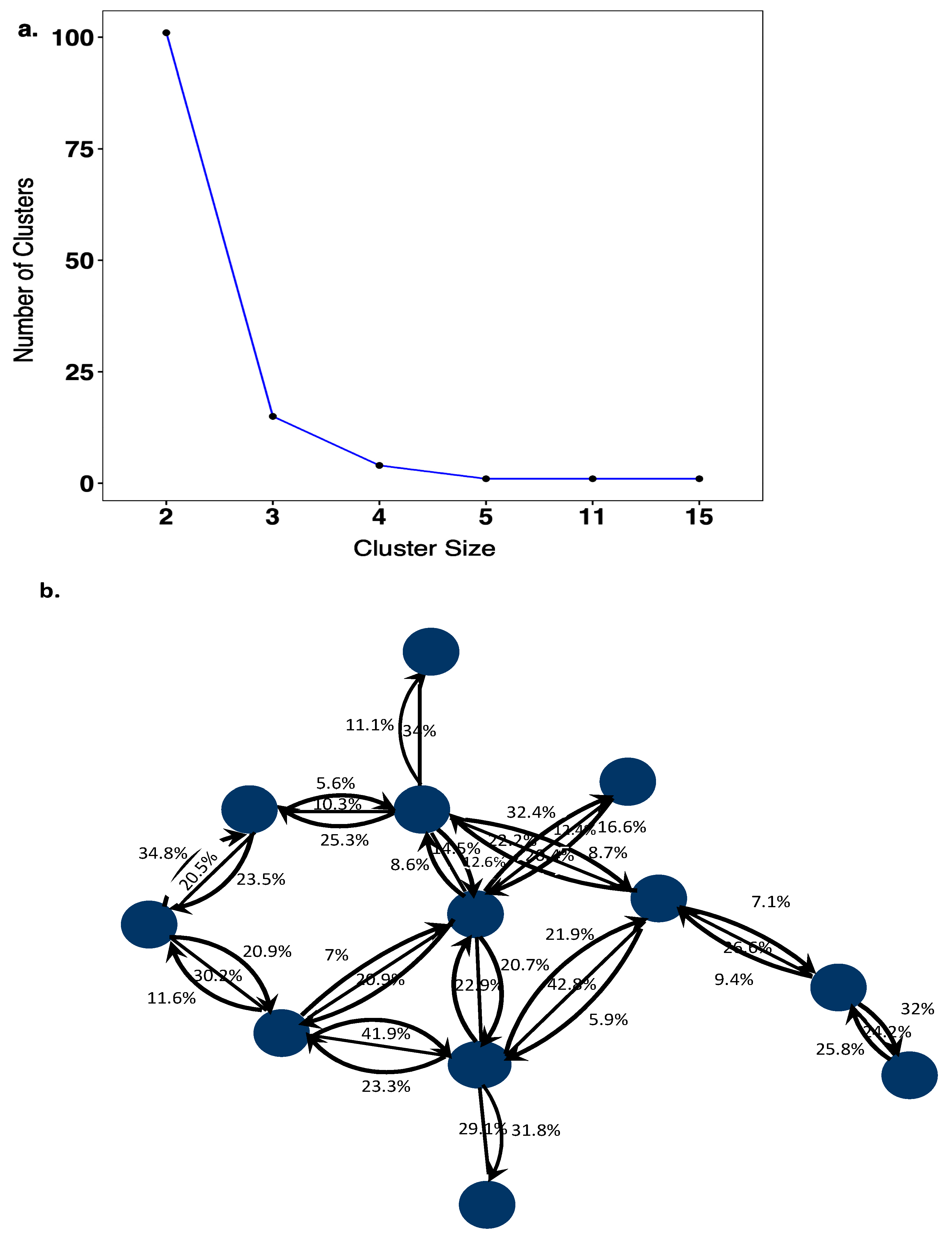
A total of 123 HIV-1 transmission networks were reconstructed with phyloscanner, of which 101 (82.1%) comprised of 2 individuals, 20 (16.3%) comprised of 3–5 individuals, 0 comprised 6–10 individuals, and 2 (1.6%) comprised of >10 individuals (Figure 3a). In larger networks, pairs of individuals between whom linkage was not excluded had larger subgraph distances (median distances were 0.5% in networks of size 2, 0.6% in networks of size 3–5, and 2.1% in networks of size >10; see Table S1), suggesting that in larger networks, slightly more phylogenetic distant individuals were connected. In the reconstructed transmission networks, a total of 105 source–recipient pairs with strong support for phylogenetic linkage and direction of transmission were identified based on the criteria described under methods. Further analysis was restricted to these highly supported source–recipient pairs. Figure 3b illustrates one of the largest identified networks. In the figure, source–recipient pairs are highlighted with arrows in black. No arrows were drawn when the corresponding two individuals were classified as phylogenetically unlinked in at least 60% of deep-sequence phylogenies across the genome.
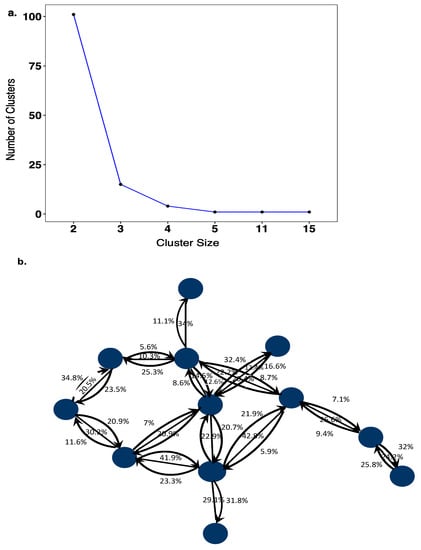
Figure 3.
(a) Cluster size distribution of HIV-1 transmission networks. The cluster size distribution of transmission networks consisted of 101 sequences that were linked as pairs, 15 clusters of 3 linked sequences, 4 clusters of 4 linked sequences, 1 cluster of 5 linked sequences, 1 cluster of 11 linked sequences and 1 cluster of 15 linked sequences. (b) Transmission pairs with phylogenetic evidence for the direction of HIV-1 transmission. The blue colored circles represent sequences that are linked to one another with phyloscanner scores (%) showing support for transmission in either direction between individual sequences. Source–recipient pairs that had phyloscanner scores >60% for linkage and >60% for one direction were considered highly supported.
3.3. HIV-1 Transmission within and between Study Participants of the General Population, Fisherfolk, and the Women at High Risk Cohort and their Clients
The phylogenetically reconstructed networks included 105 source–recipient pairs (Table 2, column three). The majority of reconstructed source–recipient pairs were within the population groupings. Thirty-six (34.3%) were from GP to GP; 33 (31.4%) were from FF to FF, and 10 (9.5%) were from WHR to WHR. Between population groups, we found 14 (13.3%) source–recipient pairs from GP to FF, 9 (8.6%) pairs from the FF to the GP, 2 (1.9%) pairs from FF to WHR, and 1 pair (1.0%) from WHR to the GP.

Table 2.
Phylogenetically observed HIV-1 transmission flows.
Considering gender, 50 (47.6%) source–recipient pairs were male to female, 12 (11.4%) were male to male, 24 (22.9%) were female to male, 10 (9.5%) were female to female among WHR, and 9 (8.6%) were female to female and involved at most one WHR (Supplementary Table S2). Observing male–male pairs was not significantly more likely than observing female–female pairs outside WHR. Thus, the most likely explanation for the presence of phylogenetically linked same-sex pairs is that intermediate individuals or common source cases of the opposite gender remained unsampled. In particular, there was only 1 male–female pair among the 13 highly supported source–recipient pairs that involved WHR, suggesting that a substantial number of WHR male clients were not captured in our populations. Due to this under-sampling of male clients, pairs from the WHR population were excluded from the subsequent HIV-1 transmission flow analysis. Column four in Table 2 lists the phylogenetically reconstructed source–recipient pairs after excluding same-sex pairs.
We estimated HIV-1 transmission flows within and between the FF and GP study participants, while accounting for slightly different sequence sampling fractions of HIV-1 positive participants. Overall, transmission was assortative within the FF and GP participants, with an estimated assortativity coefficient of 0.32 (95% CrI 0.10–0.53). More specifically, the estimated transmission flows when same-sex pairs were included are listed in Table 2 column five, and the estimated transmission flows when same-sex pairs were excluded are listed in Table 2 column six. Within and between groups of study participants, the estimated HIV-1 transmission flows were 45.5% (95% credibility interval (CrI) 34.1–57.0%) from FF to FF, 12.7% (95% CrI 6.1–21.3%) from FF to GP, 19.5% (95% CrI 11.4–29.3%) from GP to FF, and 22.3% (95% CrI 13.4–32.7%) from GP to GP in the analysis excluding same-sex pairs. The estimated flows were significantly different when same-sex pairs were excluded, because 20 (95.2%) of 21 same-sex source–recipient pairs were within the GP. All phylogenetically reconstructed transmissions between GP and FF locations involved a male and a female, and so there were more transmissions from GP to FF locations compared to the opposite direction regardless of excluding same-sex source–recipient pairs. The estimated flow ratio of transmission flows from GP to FF divided by flows from FF to GP was 1.56 (95% CrI 0.68–3.72).
We next estimated the sources of HIV-1 acquisition among the FF and GP study participants after adjusting for sequence sampling differences (Table 3). For any HIV-1 infection found in the FF sample, the probability that it originated from other FF was 70.4% (95% CrI 56.2–81.9%), and the probability that it originated from the GP was 29.6% (95% CrI 18.1–43.8%) when same-sex pairs were excluded. For any HIV-1 infection found in the GP sample, the probability that it originated from the GP was 80.2% (95% CrI 66.8–90.0%), and the probability that it originated from the FF was 19.8% (95% CrI 10.0–33.2%) when same-sex pairs were excluded.

Table 3.
Phylogenetically estimated sources of HIV-1 acquisition among FF and GP study participants.
3.4. HIV Transmission by Gender
After excluding same-sex pairs and accounting for differences in sequence sampling fractions of men and women, an estimated 68.0% (56.6–78.1%) of transmissions in the sample originated from men and 32.0% (21.9–43.4%) from women.
3.5. HIV-1 Transmission by Age Groups
We further characterized the age-specific HIV-1 transmission flows among the phylogenetically highly supported source–recipient pairs (Table 4). We sought to use the same age band as in de Oliveira et al. [25] to enable a comparison between the population sampled in KwaZulu-Natal, South Africa, and our settings in central and southwestern Uganda. However, with 74 reconstructed source–recipient pairs between men and women, we reduced these further to age bands 18–24 years and 25–59 years at time of diagnosis. In the sample, transmission was not assortative within the two age groups in both the male-to-female and female-to-male direction, with estimated assortativity coefficients of 0.07 (95% CrI −0.18–0.33) and −0.08 (−0.40–0.28) respectively. More detailed analysis of the transmission flows showed that most (n = 18, 25.0%) phylogenetically reconstructed transmissions occurred from men of 25–59 years to women of 25–59 years, followed by transmissions (n = 13, 18.1%) from men of 25–59 years to women of 18–24 years. The estimated transmission flows among participants after adjusting for sequence sampling differences are reported in Table 4 column 4. Considering transmission flows from men to women compared to vice versa, the estimated flow ratios were 3.16 (95% CrI 0.92–14.44) from men aged 18–24 years to women aged 18–24 years compared to vice versa, 1.29 (95% CrI 0.47–3.69) from men aged 18–24 years compared to vice versa, 2.63 (95% CrI 0.96–8.28) from men aged 25–59 compared to vice versa, and 2.29 (95% CrI 1.02–5.52) from men aged 25–59 to women aged 25–59 compared to vice versa.

Table 4.
Phylogenetically estimated transmission flows by age.
We also estimated the sources of infection among women aged 18–24 years and women aged 25–59 years in our sample, and similarly for men (Table 5). Considering women aged 18–24 years in the sample, an estimated 34.7% (17.1–55.9%) of infections originated from men aged 18–24 years, and 65.3% (95% CrI 44.1–82.9%) of infections originated from men aged 25–59 years. Considering women aged 25–59 years in the sample, an estimated 27.7% (14.1–45.7%) of infections originated from men aged 18–24 years, and 72.3% (54.3–85.9%) of infections originated from men aged 25–59 years. Considering men aged 18–24 years in the sample, an estimated 24.2% (6.3–54.0%) of infections originated from women aged 18–24 years, and 75.8% (46.0–93.7%) of infections originated from women aged 25–59 years. Considering men aged 25–59 years in the sample, an estimated 32.6% (12.7–59.9%) of infections originated from women aged 18–24 years, and 67.4% (40.1–87.3%) of infections originated from women aged 25–59 years.

Table 5.
Phylogenetically estimated sources of HIV-1 acquisition among male and female study participants by age.
4. Discussion
In this study, we reconstructed phylogenetic transmission networks from HIV-1 deep sequences in a large convenience sample of HIV-1 infected individuals from high-risk and general population groups in central and southwestern Uganda. Based on the reconstructed transmission networks, we estimated HIV-1 transmission flows within and between distinct populations (GP, FF, and WHR) in our sample, and by gender and age. We also estimated the sources of HIV-1 infection in these population groups. Several molecular epidemiology studies have used nucleotide sequence data generated by Sanger sequencing to characterize HIV-1 transmission dynamics in FF communities [19,53] and other high-risk groups [23]. Here, we were able to infer directed HIV-1 transmission networks through viral phylogenetic analysis, because deep-sequencing output generated multiple sequence reads from each sampled individual, from which the direction of transmission could be inferred at an accuracy sufficient for population-level analyses [46,48]. We estimated a higher proportion of HIV-1 transmissions among participants from the FF (45.5% (34.1–57.0%) compared to participants from the GP (22.3% (13.4–32.7%) after adjusting for heterogeneity in the proportion of participants that were sequenced. These estimates do not necessarily imply that a larger proportion of transmissions occur in FF populations compared to GP populations, because the fraction of the FF populations surveyed was larger than the fraction of the GP populations surveyed. However, sampling fractions cancel out in the flow ratio statistics. Specifically, if denotes the true transmission flows from population group to population group and the transmission flows among recruited individuals of groups and then:
where denotes the probability of recruiting an infected individual from population group into the convenience sample. This equation assumes that individuals were equally likely to be recruited whether they were a source of further infections or not. Thus, the flow ratios that we inferred in our convenience sample can be interpreted as estimates of the corresponding population flow ratios. We found that the flow ratio of transmissions from GP to FF compared to vice versa was 1.56 (95% CrI (0.68–3.72). Although not statistically significant, our inferences provide further evidence that high-prevalence fishing communities along Lake Victoria are a sink and not a source of the local epidemic [29,30].
We found a substantial number of viral introductions into FF communities. Considering that participants were disproportionally recruited in FF populations, we expect that the estimated proportion of GP infections in the sample attributable to GP sources (80.2%) is a lower bound of the proportion of infections in the GP population that is attributable to GP sources. Similarly, we expect that the estimated proportion of FF infections in the sample attributable to FF sources (70.4%) is an upper bound of the proportion of infections in the FF population that is attributable to FF sources. In a study done in Rakai [30], differences in survey sampling were adjusted for, and the estimated proportion of FF infections attributable to FF sources was 54.8% (95% CrI 42.2–69.0%). Thus, taken together, the two studies indicate very high levels of viral introductions to the HIV-1 epidemic in FF populations for the duration of both studies, from August 2011 to August 2017.
HIV-1 transmission in the GP and across populations into the FF communities was mostly driven by individuals above 25 years of age relative to those younger than 25 years. We found that an estimated 65.3% (44.1–82.9%) of women aged 18–24 years in the sample were infected by men above 25 years, consistent with findings from a recent study conducted in South Africa [25]. However, we also found that an estimated 75.8% (46.0–93.7%) of men aged 18–24 years in the sample were infected by women above 25 years. There was no combination of age groups in which transmissions from women to men outweighed those from men to women, suggesting that women in the sample are “sinks” of HIV-1 transmission regardless of age. These observations challenge the concept of a transmission cycle wherein young women are infected by older men, and then transmit to men of similar age [25].
This study has limitations. First, individuals were recruited in health care facilities and during VCT campaigns, and so this study did not include infected individuals who had not yet linked to care. Second, our convenience sample preferentially included infected individuals from FF populations, rendering it challenging to extrapolate transmission flow estimates from study participants to the population the participants were recruited from. However, the flow ratio statistics in Table 2 and Table 4 are invariant to differential sampling of population groups, and can thus be used to characterize population-level HIV-1 spread. Third, few WHR male clients were recruited, and we were not able to substantiate the transmission probabilities from this population as a source of infections. Recruitment of male clients and partners of WHR in the parent study was challenging due to stigma or other unknown socio-behavioral factors that the men in this group associated with study participation. We observed from our socio-demographic data that WHR were a mobile group of individuals who interacted with high-risk male individuals in the GP and FF particularly during peak seasons of high fish stock, suggesting that a non-negligible proportion of infections may originate from WHR. Fourth, virus from less than 50% of enrolled participants could be deep-sequenced, excluding those that did not have sufficient viraemia for HIV-1 nucleic acid amplification and deep sequencing. Our sample comprised study participants who were ART naïve or on therapy with unsuppressed virus, and it is possible that transmissions from individuals who reached viral suppression soon after infection are under-represented in our data. Generation of additional HIV-1 deep sequence data based on sequencing protocols that amplify proviral HIV-1 DNA or sequence virus from low viraemic specimens [54,55] is a consideration for our future analyses. Fifth, transmissions including individuals with co-infections may have been excluded by the definition of source–recipient pairs. It is important to note that while the FF communities investigated in this study were net recipients for viral transmission (sinks) from GP population groups, the possibility of fishing villages being sources of HIV-1 transmission to other populations cannot be ruled out.
Phylogenetic reconstruction of HIV transmission networks is increasingly common to characterize HIV dynamics, typically based on tools such as HIV-TRACE [56] or Cluster Picker [57]. These tools can be applied to Sanger sequence data, and the reconstructed networks represent sets of phylogenetically linked individuals whose viral genetic relatedness suggest a direct or indirect epidemiologic link. In contrast, the deep-sequence approach adopted here provides further information into the direction of transmission between individuals in such networks using additional signal in the topology of deep-sequence phylogenetic trees. However, while the deep-sequence approach is advantageous in several aspects, there are important potential limitations to consider in future applications. Most obviously, deep-sequence phylogenetic analyses require deep-sequence data, which is only beginning to become more broadly available [31,58,59]. It is also important to consider the substantially larger requirements on storage (on the order of hundreds of gigabytes for our population-based study), as well as analysis time (~300 phylogenies reconstructed for each potential transmission network across sliding windows of the entire HIV genome is computationally intensive). Finally, due to the complexity involved in handling deep-sequence data, available analysis software does not easily install on Windows systems [46,60]. However, these challenges are primarily of logistic nature and we see no fundamental reason that prevents use of HIV deep-sequence technology to support the End the HIV initiative in the US as well as similar efforts [61,62].
In conclusion, the reconstructed patterns of HIV-1 transmission found across cohorts of HIV-1 infected individuals in central and southwestern Uganda support previous findings [29,30] that high-prevalence fishing communities on Lake Victoria are net recipients of HIV-1 transmission flow in the local epidemic, and not a net source of transmission flow to the neighboring low-prevalence communities located further inland. We suggest that geographically targeted interventions that focus on high-prevalence FF populations should not restrict the roll-out of preventative measures in neighboring areas that could be sources of HIV-1 infection. Furthermore, this study supports calls to re-design prevention interventions to more effectively aid men of all ages to engage with and stay in care, and thereby reduce the disproportionate infection burden among women. Two-pronged intervention approaches that target both viral transmission sources to reduce transmission and sink populations to reduce risk of infection appear optimal in reducing the future HIV-1 incidence in key and general population groups.
Supplementary Materials
The following are available online at https://www.mdpi.com/1999-4915/12/3/331/s1, Figure S1: Markov Chain Monte Carlo (MCMC) simulation trajectories for estimated transmission flows within/between populations after adjusting for sampling heterogeneity, Table S1: Mean pairwise distances in phylogenetically reconstructed transmission networks, Table S2: Phylogenetically reconstructed source–recipient pairs by location and gender.
Author Contributions
Conceptualization, D.S. and O.R.; methodology, N.B., Y.M. and U.B.; software, O.R., X.X. and A.S.; validation, A.S. and N.B.; formal analysis, N.B.; investigation, N.B.; resources, D.S.; Sequence preparation and assembly, T.G.; writing—original draft preparation, N.B.; writing—review and editing, O.R., J.S., P.K., L.A.-D., D.P.; visualization, N.B and A.S.; supervision, D.S. and P.K.; PANGEA project support, L.A.-D.; funding acquisition, D.S., C.F. and P.K. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the UK Medical Research Council (MRC) and the UK Department for International Development (DFID) under the MRC/DFID Concordat agreement. PANGEA is funded by the Bill & Melinda Gates Foundation (consecutive grants OPP1084362 and OPP1175094).
Acknowledgments
We thank two anonymous reviewers, the PANGEA steering committee, Maria J. Wawer, Janet Seeley and Pontiano Kaleebu for helpful comments on this work. Computations were done at the Imperial College Research Computing Service (https://www.imperial.ac.uk/admin-services/ict/self-service/research-support/rcs/). N.B. was supported by the PANGEA training fellowship to facilitate computations and data analysis. We are grateful to all the study participants and the healthcare professionals who were involved in the study.
Conflicts of Interest
OR reports grant funding from the Bill & Melinda Gates Foundation.
Appendix A

Table A1.
PANGEA 2 Steering Committee.
Table A1.
PANGEA 2 Steering Committee.
| Name | Institution | Representatives & Roles | Email Address |
|---|---|---|---|
| Lucie Abeler-Dörner | University of Oxford | Project manager | lucie.abeler-dorner@bdi.ox.ac.uk |
| Helen Ayles | PopART/Zambart | PopART | helen@zambart.org.zm |
| David Bonsall | University of Oxford | Sequencing lab | david.bonsall@bdi.ox.ac.uk |
| Rory Bowden | University of Oxford | Sequencing lab | rbowden@well.ox.ac.uk |
| Vincent Calvez | Institut Pasteur | TasP trial | vincent.calvez@me.com |
| Max Essex | Harvard Botswana | Botswana studies | messex@hsph.harvard.edu |
| Sarah Fidler | PopART/Imperial College London | PopART | s.fidler@imperial.ac.uk |
| Christophe Fraser | University of Oxford | Principal Investigator PANGEA 2, Executive Committee and PopART Phylogenetics | christophe.fraser@bdi.ox.ac.uk |
| Kate Grabowski | Johns Hopkins University | Executive Committee and Rakai | mgrabows@jhu.edu |
| Tanya Golubchik | University of Oxford | Data manager | tanya.golubchik@bdi.ox.ac.uk |
| Richard Hayes | PopART/LSHTM | PopART | Richard.Hayes@lshtm.ac.uk |
| Joshua Herbeck | University of Washington | Partners PrEP; Partners in Prevention | herbeck@uw.edu |
| Joseph Kagaayi | Rakai Health Sciences Program | Rakai Health Sciences Program | jkagayi@rhsp.org |
| Pontiano Kaleebu | MRC/UVRI Uganda | MRC studies | pontiano.kaleebu@mrcuganda.org |
| Jairam Lingappa | University of Washington | Partners PrEP; Partners in Prevention | lingappa@uw.edu |
| Sikhulile Moyo | Botswana Harvard AIDS Institute Partnership | Botswana studies | sikhulilemoyo@gmail.com |
| Vladimir Novitsky | Harvard University | Botswana studies | vnovi@hsph.harvard.edu |
| Deenan Pillay | Africa Health Research Institute (AHRI)/University College London | Principal Investigator PANGEA-1/Executive Committee, AHRI studies | dpillay@ahri.org |
| Thomas Quinn | Johns Hopkins University | Rakai Health Sciences Program | tquinn2@jhmi.edu |
| Andrew Rambaut | University of Edinburgh | Executive Committee | a.rambaut@ed.ac.uk |
| Oliver Ratmann | Imperial College London | Analysis | oliver.ratmann@imperial.ac.uk |
| Janet Seeley | MRC/UVRI Uganda/LSHTM | MRC Uganda | Janet.Seeley@LSHTM.ac.uk |
| Deogratius Ssemwanga | MRC/UVRI Uganda | MRC studies | Deogratius.Ssemwanga@mrcuganda.org |
| Frank Tanser | Africa Health Research Institute | AHRI studies | ftanser@lincoln.ac.uk |
| Maria Wawer | Johns Hopkins University | Rakai Health Sciences Program | mwawer1@jhu.edu |
Appendix B

Table A2.
PANGEA 1 Steering Committee.
Table A2.
PANGEA 1 Steering Committee.
| Name | Institution | Study Team Representatives | Email Address |
|---|---|---|---|
| Myron Cohen | University of North Carolina | -------- | myron_cohen@med.unc.edu |
| Tulio D’Oliveira | University of KwaZulu-Natal | -------- | tuliodna@gmail.com |
| Ann Dennis | University of North Carolina | -------- | ann_dennis@med.unc.edu |
| Max Essex | Harvard Botswana | Botswana studies | messex@hsph.harvard.edu |
| Sarah Fidler | PopART/Imperial College London | PopART Phylogenetics | s.fidler@imperial.ac.uk |
| Dan Frampton | University College London | -------- | d.frampton@ucl.ac.uk |
| Christophe Fraser | University of Oxford | PopART Phylogenetics | christophe.fraser@well.ox.ac.uk |
| Tanya Golubchik | University of Oxford | tanya.golubchik@bdi.ox.ac.uk | |
| Richard Hayes | PopART/LSHTM | PopART Phylogenetics | Richard.Hayes@lshtm.ac.uk |
| Josh Herbeck | University of Washington | Partners PrEP; Partners in Prevention | herbeck@uw.edu |
| Anne Hoppe | University College London | Project Manager PANGEA 1/EARNEST | a.hoppe@ucl.ac.uk; hoppe.anne@gmail.com |
| Pontiano Kaleebu | MRC/UVRI Uganda | MRC studies | pontiano.kaleebu@mrcuganda.org |
| Paul Kellam | Cambridge University | -------- | paul.kellam@kymab.com |
| Cissy Kityo | EARNEST/JCRC Uganda | EARNEST | ckityo@jcrc.org.ug |
| Andrew Leigh-Brown | University of Edinburgh | -------- | A.Leigh-Brown@ed.ac.uk |
| Jairam Lingappa | University of Washington | Partners PrEP; Partners in Prevention | lingappa@uw.edu |
| Vladimir Novitsky | Harvard University | Botswana studies | vnovi@hsph.harvard.edu |
| Nick Paton | EARNEST/University of Singapore | EARNEST | nick.paton@ucl.ac.uk |
| Deenan Pillay | Africa Health Research Institute/University College London | Principal Investigator PANGEA 1/Africa Health Research Institute studies | dpillay@ahri.org |
| Tom Quinn | Johns Hopkins University | Rakai Health Sciences Program | tquinn2@jhmi.edu |
| Oliver Ratmann | Imperial College London | -------- | oliver.ratmann@imperial.ac.uk |
| Deogratius Ssemwanga | MRC/UVRI Uganda | MRC studies | Deogratius.Ssemwanga@mrcuganda.org |
| Frank Tanser | Africa Health Research Institute | -------- | ftanser@gmail.com |
| Maria Wawer | Johns Hopkins University | Rakai Health Sciences Program | mwawer1@jhu.edu |
References
- UNAIDS DATA 2019. Available online: https://www.unaids.org/sites/default/files/media_asset/2019-UNAIDS-data_en.pdf (accessed on 19 December 2019).
- Uganda AIDS Commision. Uganda HIV/AIDS Country Progress Report July 2017–June 2018; Uganda AIDS Commission: Kampala, Uganda, 2018; p. 91.
- Ministry of Health and Uganda AIDS Commission. Multi-Sectoral HIV Programming for MARPS in Uganda: Review of Profiles, Sizes and Programme Coverage; Review Report; Ministry of Health and Uganda AIDS Commission: Kampala, Uganda, 2014.
- Ministry of Health Uganda. Population-Based HIV Impact Assessement (UPHIA) 2016–2017. Summary Sheet: Preliminary Findings; Ministry of Health Uganda: Kampala, Uganda, 2017.
- Kamali, A.; Nsubuga, R.N.; Ruzagira, E.; Bahemuka, U.; Asiki, G.; Price, M.A.; Newton, R.; Kaleebu, P.; Fast, P. Heterogeneity of HIV incidence: A comparative analysis between fishing communities and in a neighbouring rural general population, Uganda, and implications for HIV control. Sex. Transm. Infect. 2016, 92, 447–454. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kasamba, I.; Nash, S.; Seeley, J.; Weiss, H.A. HIV incidence among women at high risk of HIV infection attending a dedicated clinic in Kampala, Uganda: 2008–2017. Sex. Transm. Dis. 2019, 46, 407–415. [Google Scholar] [CrossRef] [PubMed]
- Wabwire-Mangen, F.; Odiit, M.; Kirungi, W.; Kisitu, D.K.; Wanyama, J.O. HIV Modes of Transmission and Prevention Response Analysis; Uganda National AIDS Commission: Kampala, Uganda, 2009.
- UNAIDS Fast-Track—Ending the AIDS Epidemic by 2030. Available online: http://www.unaids.org/en/resources/documents/2014/JC2686_WAD2014report (accessed on 16 February 2018).
- UNAIDS/WHO Working Group on Global HIV/AIDS and STI Surveillance; Joint United Nations Programme on HIV/AIDS; World Health Organization. Guidelines for Second Generation HIV Surveillance: An Update: Know Your Epidemic; WHO Press: Geneva, Switzerland, 2013; ISBN 978-92-4-150582-6. [Google Scholar]
- Cuadros, D.F.; Li, J.; Branscum, A.J.; Akullian, A.; Jia, P.; Mziray, E.N.; Tanser, F. Mapping the spatial variability of HIV infection in Sub-Saharan Africa: Effective information for localized HIV prevention and control. Sci. Rep. 2017, 7, 9093. [Google Scholar] [CrossRef] [PubMed]
- Barankanira, E.; Molinari, N.; Niyongabo, T.; Laurent, C. Spatial analysis of HIV infection and associated individual characteristics in Burundi: Indications for effective prevention. BMC Public Health 2016, 16, 118. [Google Scholar] [CrossRef] [PubMed]
- Anderson, S.-J.; Cherutich, P.; Kilonzo, N.; Cremin, I.; Fecht, D.; Kimanga, D.; Harper, M.; Masha, R.L.; Ngongo, P.B.; Maina, W.; et al. Maximising the effect of combination HIV prevention through prioritisation of the people and places in greatest need: A modelling study. Lancet 2014, 384, 249–256. [Google Scholar] [CrossRef]
- Kuteesa, M.O.; Weiss, H.A.; Abaasa, A.; Nash, S.; Nsubuga, R.N.; Newton, R.; Seeley, J.; Kamali, A. Feasibility of conducting HIV combination prevention interventions in fishing communities in Uganda: A pilot cluster randomised trial. PLoS ONE 2019, 14, e0210719. [Google Scholar] [CrossRef]
- Kagaayi, J.; Chang, L.W.; Ssempijja, V.; Grabowski, M.K.; Ssekubugu, R.; Nakigozi, G.; Kigozi, G.; Serwadda, D.M.; Gray, R.H.; Nalugoda, F.; et al. Impact of combination HIV interventions on HIV incidence in hyperendemic fishing communities in Uganda: A prospective cohort study. Lancet HIV 2019, 6, e680–e687. [Google Scholar] [CrossRef]
- Ssewamala, F.M.; Sensoy Bahar, O.; Tozan, Y.; Nabunya, P.; Mayo-Wilson, L.J.; Kiyingi, J.; Kagaayi, J.; Bellamy, S.; McKay, M.M.; Witte, S.S. A combination intervention addressing sexual risk-taking behaviors among vulnerable women in Uganda: Study protocol for a cluster randomized clinical trial. BMC Women’s Health 2019, 19, 111. [Google Scholar] [CrossRef]
- Rambaut, A.; Robertson, D.L.; Pybus, O.G.; Peeters, M.; Holmes, E.C. Human immunodeficiency virus. Phylogeny and the origin of HIV-1. Nature 2001, 410, 1047–1048. [Google Scholar] [CrossRef]
- Sharp, P.M.; Hahn, B.H. Origins of HIV and the AIDS pandemic. Cold Spring Harb. Perspect. Med. 2011, 1, a006841. [Google Scholar] [CrossRef]
- Faria, N.R.; Rambaut, A.; Suchard, M.A.; Baele, G.; Bedford, T.; Ward, M.J.; Tatem, A.J.; Sousa, J.D.; Arinaminpathy, N.; Pépin, J.; et al. The early spread and epidemic ignition of HIV-1 in human populations. Science 2014, 346, 56–61. [Google Scholar] [CrossRef] [PubMed]
- Kiwuwa-Muyingo, S.; Nazziwa, J.; Ssemwanga, D.; Ilmonen, P.; Njai, H.; Ndembi, N.; Parry, C.; Kitandwe, P.K.; Gershim, A.; Mpendo, J.; et al. HIV-1 transmission networks in high risk fishing communities on the shores of Lake Victoria in Uganda: A phylogenetic and epidemiological approach. PLoS ONE 2017, 12, e0185818. [Google Scholar] [CrossRef] [PubMed]
- Aldous, J.L.; Pond, S.K.; Poon, A.; Jain, S.; Qin, H.; Kahn, J.S.; Kitahata, M.; Rodriguez, B.; Dennis, A.M.; Boswell, S.L.; et al. Characterizing HIV Transmission Networks Across the United States. Clin. Infect. Dis. 2012, 55, 1135–1143. [Google Scholar] [CrossRef] [PubMed]
- Leigh Brown, A.J.; Lycett, S.J.; Weinert, L.; Hughes, G.J.; Fearnhill, E.; Dunn, D.T. UK HIV drug resistance collaboration transmission network parameters estimated from HIV sequences for a nationwide epidemic. J. Infect. Dis. 2011, 204, 1463–1469. [Google Scholar] [CrossRef]
- Lewis, F.; Hughes, G.J.; Rambaut, A.; Pozniak, A.; Leigh Brown, A.J. Episodic Sexual Transmission of HIV revealed by molecular phylodynamics. PLoS Med. 2008, 5, e50. [Google Scholar] [CrossRef]
- Yebra, G.; Ragonnet-Cronin, M.; Ssemwanga, D.; Parry, C.M.; Logue, C.H.; Cane, P.A.; Kaleebu, P.; Brown, A.J.L. Analysis of the history and spread of HIV-1 in Uganda using phylodynamics. J. Gen. Virol. 2015, 96, 1890–1898. [Google Scholar] [CrossRef]
- Gray, R.R.; Tatem, A.J.; Lamers, S.; Hou, W.; Laeyendecker, O.; Serwadda, D.; Sewankambo, N.; Gray, R.H.; Wawer, M.; Quinn, T.C.; et al. Spatial phylodynamics of HIV-1 epidemic emergence in east Africa. AIDS 2009, 23, F9–F17. [Google Scholar] [CrossRef]
- de Oliveira, T.; Kharsany, A.B.; Gräf, T.; Cawood, C.; Khanyile, D.; Grobler, A.; Puren, A.; Madurai, S.; Baxter, C.; Karim, Q.A.; et al. Transmission networks and risk of HIV infection in KwaZulu-Natal, South Africa: A community-wide phylogenetic study. Lancet HIV 2017, 4, e41–e50. [Google Scholar] [CrossRef]
- Coltart, C.; Tanser, F.; Gareta, D.; Ntuli, S.; de Oliveira, T.; Pillay, D.; Johnson, A.; Hue, S. The identification of a micro-epidemic in a hyper-endemic HIV setting using molecular epidemiology. J. Int. AIDS Soc. Mexico 2018, 21, e25148. [Google Scholar]
- Rasmussen, D.A.; Wilkinson, E.; Vandormael, A.; Tanser, F.; Pillay, D.; Stadler, T.; de Oliveira, T. Tracking external introductions of HIV using phylodynamics reveals a major source of infections in rural KwaZulu-Natal, South Africa. Virus Evol. 2018, 4, vey037. [Google Scholar] [CrossRef]
- Grabowski, M.K.; Lessler, J.; Redd, A.D.; Kagaayi, J.; Laeyendecker, O.; Ndyanabo, A.; Nelson, M.I.; Cummings, D.A.T.; Bwanika, J.B.; Mueller, A.C.; et al. The role of viral introductions in sustaining community-based HIV epidemics in rural Uganda: Evidence from spatial clustering, phylogenetics, and egocentric transmission models. PLoS Med. 2014, 11, e1001610. [Google Scholar] [CrossRef] [PubMed]
- Bbosa, N.; Ssemwanga, D.; Nsubuga, R.N.; Salazar-Gonzalez, J.F.; Salazar, M.G.; Nanyonjo, M.; Kuteesa, M.; Seeley, J.; Kiwanuka, N.; Bagaya, B.S.; et al. Phylogeography of HIV-1 suggests that Ugandan fishing communities are a sink for, not a source of, virus from general populations. Sci. Rep. 2019, 9, 1051. [Google Scholar] [CrossRef] [PubMed]
- Ratmann, O.; Kagaayi, J.; Hall, M.; Golubchick, T.; Kigozi, G.; Xi, X.; Wymant, C.; Nakigozi, G.; Abeler-Dörner, L.; Bonsall, D.; et al. Quantifying HIV transmission flow between high-prevalence hotspots and surrounding communities: A population-based study in Rakai, Uganda. Lancet HIV 2020, 7, e173–e183. [Google Scholar] [CrossRef]
- Abeler-Dörner, L.; Grabowski, M.K.; Rambaut, A.; Pillay, D.; Fraser, C. PANGEA consortium PANGEA-HIV 2: Phylogenetics and networks for generalised epidemics in Africa. Curr. Opin. HIV AIDS 2019, 14, 173–180. [Google Scholar] [CrossRef] [PubMed]
- Asiki, G.; Mpendo, J.; Abaasa, A.; Agaba, C.; Nanvubya, A.; Nielsen, L.; Seeley, J.; Kaleebu, P.; Grosskurth, H.; Kamali, A. HIV and syphilis prevalence and associated risk factors among fishing communities of Lake Victoria, Uganda. Sex. Transm. Infect. 2011, 87, 511–515. [Google Scholar] [CrossRef] [PubMed]
- Asiki, G.; Murphy, G.; Nakiyingi-Miiro, J.; Seeley, J.; Nsubuga, R.N.; Karabarinde, A.; Waswa, L.; Biraro, S.; Kasamba, I.; Pomilla, C.; et al. The general population cohort in rural south-western Uganda: A platform for communicable and non-communicable disease studies. Int. J. Epidemiol. 2013, 42, 129–141. [Google Scholar] [CrossRef] [PubMed]
- Omooja, J.; Nannyonjo, M.; Sanyu, G.; Nabirye, S.E.; Nassolo, F.; Lunkuse, S.; Kapaata, A.; Segujja, F.; Kateete, D.P.; Ssebaggala, E.; et al. Rates of HIV-1 virological suppression and patterns of acquired drug resistance among fisherfolk on first-line antiretroviral therapy in Uganda. J. Antimicrob. Chemother. 2019, 74, 3021–3029. [Google Scholar] [CrossRef]
- Salome, T.; Kasamba, I.; Mayanja, B.N.; Kazooba, P.; Were, J.; Kaleebu, P.; Munderi, P. The effect of Tenofovir on renal function among Ugandan adults on long-term antiretroviral therapy: A cross-sectional enrolment analysis. AIDS Res. Ther. 2016, 13, 28. [Google Scholar] [CrossRef]
- Namale, G.; Kamacooko, O.; Bagiire, D.; Mayanja, Y.; Abaasa, A.; Kilembe, W.; Price, M.; Ssemwanga, D.; Lunkuse, S.; Nanyonjo, M.; et al. Sustained virological response and drug resistance among female sex workers living with HIV on antiretroviral therapy in Kampala, Uganda: A cross-sectional study. Sex. Transm. Infect. 2019, 95, 405–411. [Google Scholar] [CrossRef]
- Nampijja, M.; Webb, E.L.; Kaweesa, J.; Kizindo, R.; Namutebi, M.; Nakazibwe, E.; Oduru, G.; Kabuubi, P.; Kabagenyi, J.; Kizito, D.; et al. The lake victoria island intervention study on worms and allergy-related diseases (LaVIISWA): Study protocol for a randomised controlled trial. Trials 2015, 16, 187. [Google Scholar] [CrossRef]
- Sanya, R.E.; Nkurunungi, G.; Hoek Spaans, R.; Nampijja, M.; O’Hara, G.; Kizindo, R.; Oduru, G.; Kabuubi Nakawungu, P.; Niwagaba, E.; Abayo, E.; et al. The impact of intensive versus standard anthelminthic treatment on allergy-related outcomes, helminth infection intensity and helminth-related morbidity in Lake Victoria fishing communities, Uganda: Results from the LaVIISWA cluster randomised trial. Clin. Infect. Dis. 2018, 68, 1665–1674. [Google Scholar] [CrossRef] [PubMed]
- Abaasa, A.; Asiki, G.; Obuku Ekii, A.; Wanyenze, J.; Pala, P.; van Dam, G.J.; Corstjens, P.L.A.M.; Hughes, P.; Ding, S.; Pantaleo, G.; et al. Effect of high-intensity versus low-intensity praziquantel treatment on HIV disease progression in HIV and Schistosoma mansoni co-infected patients: A randomised controlled trial. Wellcome Open Res. 2018, 3, 81. [Google Scholar] [CrossRef] [PubMed]
- Vandepitte, J.; Bukenya, J.; Weiss, H.A.; Nakubulwa, S.; Francis, S.C.; Hughes, P.; Hayes, R.; Grosskurth, H. HIV and other sexually transmitted infections in a cohort of women involved in high-risk sexual behavior in Kampala, Uganda. Sex. Transm. Dis. 2011, 38, 316–323. [Google Scholar] [CrossRef] [PubMed]
- Kapaata, A.; Lyagoba, F.; Ssemwanga, D.; Magambo, B.; Nanyonjo, M.; Levin, J.; Mayanja, B.N.; Mugasa, C.; Parry, C.M.; Kaleebu, P. HIV-1 subtype distribution trends and evidence of transmission clusters among incident cases in a rural clinical cohort in southwest Uganda, 2004–2010. AIDS Res. Hum. Retrovir. 2013, 29, 520–527. [Google Scholar] [CrossRef] [PubMed]
- Abaasa, A.; Mbonye, M.; Asiki, G.; Ruzagira, E.; Price, M.; Fast, P.; Priddy, F.; Kaleebu, P.; Kamaali, A. Use of Fingerprinting Technology in HIV Prevention Studies. Experience from Fishing Communities in South-Western Uganda; HIV Research for Prevention (HIVR4P) conference; Poster Session: Chicago, IL, USA, 2016. [Google Scholar]
- Gall, A.; Morris, C.; Kellam, P.; Berry, N. Complete genome sequence of the WHO international standard for HIV-1 RNA determined by deep sequencing. Genome Announc. 2014, 2, e01254-13. [Google Scholar] [CrossRef] [PubMed]
- Yebra, G.; Frampton, D.; Gallo Cassarino, T.; Raffle, J.; Hubb, J.; Ferns, R.B.; Waters, L.; Tong, C.Y.W.; Kozlakidis, Z.; Hayward, A.; et al. A high HIV-1 strain variability in London, UK, revealed by full-genome analysis: Results from the ICONIC project. PLoS ONE 2018, 13, e0192081. [Google Scholar] [CrossRef]
- Wymant, C.; Blanquart, F.; Golubchik, T.; Gall, A.; Bakker, M.; Bezemer, D.; Croucher, N.J.; Hall, M.; Hillebregt, M.; Ong, S.H.; et al. Easy and accurate reconstruction of whole HIV genomes from short-read sequence data with shiver. Virus Evol. 2018, 4, vey007. [Google Scholar] [CrossRef]
- Wymant, C.; Hall, M.; Ratmann, O.; Bonsall, D.; Golubchik, T.; de Cesare, M.; Gall, A.; Cornelissen, M.; Fraser, C. STOP-HCV Consortium, The maela pneumococcal collaboration, and the BEEHIVE collaboration PHYLOSCANNER: Inferring transmission from within- and between-host pathogen genetic diversity. Mol. Biol. Evol. 2018, 35, 719–733. [Google Scholar] [CrossRef]
- Ratmann, O.; Grabowski, M.K.; Hall, M.; Golubchik, T.; Wymant, C.; Abeler-Dörner, L.; Bonsall, D.; Hoppe, A.; Brown, A.L.; de Oliveira, T.; et al. Inferring HIV-1 transmission networks and sources of epidemic spread in Africa with deep-sequence phylogenetic analysis. Nat. Commun. 2019, 10, 1411. [Google Scholar] [CrossRef]
- Romero-Severson, E.O.; Bulla, I.; Leitner, T. Phylogenetically resolving epidemiologic linkage. Proc. Natl. Acad. Sci. USA 2016, 113, 2690–2695. [Google Scholar] [CrossRef]
- Campbell, E.M.; Jia, H.; Shankar, A.; Hanson, D.; Luo, W.; Masciotra, S.; Owen, S.M.; Oster, A.M.; Galang, R.R.; Spiller, M.W.; et al. Detailed transmission network analysis of a large opiate-driven outbreak of HIV infection in the United States. J. Infect. Dis. 2017, 216, 1053–1062. [Google Scholar] [CrossRef] [PubMed]
- Bartlett, S.R.; Wertheim, J.O.; Bull, R.A.; Matthews, G.V.; Lamoury, F.M.J.; Scheffler, K.; Hellard, M.; Maher, L.; Dore, G.J.; Lloyd, A.R.; et al. A molecular transmission network of recent hepatitis C infection in people with and without HIV: Implications for targeted treatment strategies. J. Viral Hepat. 2017, 24, 404–411. [Google Scholar] [CrossRef] [PubMed]
- Phyloscanner. Available online: https://github.com/BDI-pathogens/phyloscanner/tree/master/phyloflows (accessed on 21 December 2019).
- Gelman, A.; Rubin, D.B. Markov chain Monte Carlo methods in biostatistics. Stat. Methods Med. Res. 1996, 5, 339–355. [Google Scholar] [CrossRef] [PubMed]
- Nazziwa, J.; Njai, H.F.; Ndembi, N.; Birungi, J.; Lyagoba, F.; Gershim, A.; Nakiyingi-Miiro, J.; Nielsen, L.; Mpendo, J.; Nanvubya, A.; et al. Short communication: HIV type 1 transmitted drug resistance and evidence of transmission clusters among recently infected antiretroviral-naive individuals from Ugandan fishing communities of Lake Victoria. AIDS Res. Hum. Retrovir. 2013, 29, 788–795. [Google Scholar] [CrossRef] [PubMed]
- Alves, B.M.; Siqueira, J.D.; Prellwitz, I.M.; Botelho, O.M.; Da Hora, V.P.; Sanabani, S.; Recordon-Pinson, P.; Fleury, H.; Soares, E.A.; Soares, M.A. Estimating HIV-1 genetic diversity in Brazil through next-generation sequencing. Front. Microbiol. 2019, 10, 749. [Google Scholar] [CrossRef]
- Iwase, S.C.; Miyazato, P.; Katsuya, H.; Islam, S.; Yang, B.T.J.; Ito, J.; Matsuo, M.; Takeuchi, H.; Ishida, T.; Matsuda, K.; et al. HIV-1 DNA-capture-seq is a useful tool for the comprehensive characterization of HIV-1 provirus. Sci. Rep. 2019, 9, 12326. [Google Scholar] [CrossRef]
- Kosakovsky Pond, S.L.; Weaver, S.; Leigh Brown, A.J.; Wertheim, J.O. HIV-TRACE (TRAnsmission cluster engine): A tool for large scale molecular epidemiology of HIV-1 and other rapidly evolving pathogens. Mol. Biol. Evol. 2018, 35, 1812–1819. [Google Scholar] [CrossRef]
- Ragonnet-Cronin, M.; Hodcroft, E.; Hué, S.; Fearnhill, E.; Delpech, V.; Brown, A.J.L.; Lycett, S. UK HIV drug resistance database automated analysis of phylogenetic clusters. BMC Bioinform. 2013, 14, 317. [Google Scholar] [CrossRef]
- Gibson, K.M.; Jair, K.; Castel, A.D.; Bendall, M.L.; Wilbourn, B.; Jordan, J.A.; Crandall, K.A.; Pérez-Losada, M. DC Cohort Executive Committee A cross-sectional study to characterize local HIV-1 dynamics in Washington, DC using next-generation sequencing. Sci. Rep. 2020, 10, 1989. [Google Scholar] [CrossRef]
- Hauser, A.; Hofmann, A.; Meixenberger, K.; Altmann, B.; Hanke, K.; Bremer, V.; Bartmeyer, B.; Bannert, N. Increasing proportions of HIV-1 non-B subtypes and of NNRTI resistance between 2013 and 2016 in Germany: Results from the national molecular surveillance of new HIV-diagnoses. PLoS ONE 2018, 13, e0206234. [Google Scholar] [CrossRef]
- Skums, P.; Zelikovsky, A.; Singh, R.; Gussler, W.; Dimitrova, Z.; Knyazev, S.; Mandric, I.; Ramachandran, S.; Campo, D.; Jha, D.; et al. QUENTIN: Reconstruction of disease transmissions from viral quasispecies genomic data. Bioinformatics 2018, 34, 163–170. [Google Scholar] [CrossRef] [PubMed]
- Giroir, B.P. The time is now to end the hiv epidemic. Am. J. Public Health 2020, 110, 22–24. [Google Scholar] [CrossRef] [PubMed]
- Fauci, A.S.; Redfield, R.R.; Sigounas, G.; Weahkee, M.D.; Giroir, B.P. Ending the HIV epidemic: A plan for the United States. JAMA 2019, 321, 844. [Google Scholar] [CrossRef] [PubMed]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).