Abstract
The high mutation rate of the human immunodeficiency virus type 1 (HIV-1) plays a major role in treatment resistance, from the development of vaccines to therapeutic drugs. In addressing the crux of the issue, various attempts to estimate the mutation rate of HIV-1 resulted in a large range of 10−5–10−3 errors/bp/cycle due to the use of different types of investigation methods. In this review, we discuss the different assay methods, their findings on the mutation rates of HIV-1 and how the locations of mutations can be further analyzed for their allosteric effects to allow for new inhibitor designs. Given that HIV is one of the fastest mutating viruses, it serves as a good model for the comprehensive study of viral mutations that can give rise to a more horizontal understanding towards overall viral drug resistance as well as emerging viral diseases.
1. Introduction
Among the retroviruses, HIV-1 is the most genetically diverse [1,2,3,4], due to its higher mutation rate (reviewed by [5,6]), genetic recombination, and fast viral replication [7,8,9], where multiple unique sequences can be isolated from within single patients [10,11,12,13,14]. This diversity allows for the emergence of resistance strains, and evasion from the host immune system and drug therapies. Under selection pressures (such as antiretroviral drugs), these drug-resistant mutations accumulate and become dominant strains [7,15], rendering treatment regimens ineffective and hindering the development of effective vaccines [16]. Thus, to combat viral diseases, HIV is an ideal model for studying drug resistance and emerging infections.
The genetic recombination of two single drug resistant strains can give rise to HIV-1 virions with multiple drug resistance [17,18,19,20]. Coupled with high mutation rates, the estimated production of 109 virions per day within an infected individual results in numerous mutant variants in a few viral generations [21], with drug-resistant strains completely replaced by wild type strains within 2–4 weeks after antiretroviral treatment [7].
The error rates and mutation frequencies of HIV-1 reverse transcriptase (RT) are reported to be approximately 10−5–10−3 errors/bp/cycle and 10−4–10−2 mutants/clones, respectively. However, the lack of consensus in mutation rates observed in HIV-1 suggests that multiple factors and contributors are involved (see Table 1 and Table 3).

Table 1.
Error rates of HIV-1 reverse transcriptase (RT) measured in cell-free fidelity assays.
In this review, we discuss the various experimental fidelity assays, their reported mutation rates, and the potential of non-active site allosteric mutations.
2. HIV-1 Reverse Transcriptase (RT)
HIV-1 RT is an asymmetric heterodimer consisting of the p66 (66 kDa) and the p51 (51 kDa) subunits [22] illustrated in Figure 1. It lacks the 3′ exonuclease proofreading activity [23], contributing up to 68% mutations in cell-based assays during early stage replication (minus-strand synthesis and RNA transcription) and 32% during late stage replication (plus-strand synthesis and DNA repair) [24]. HIV-1 RT has been found to contribute to HIV-1 mutagenesis [25], accounting for 59.7% of the mutations in the viral RNA and 2.0% in the viral DNA [26].
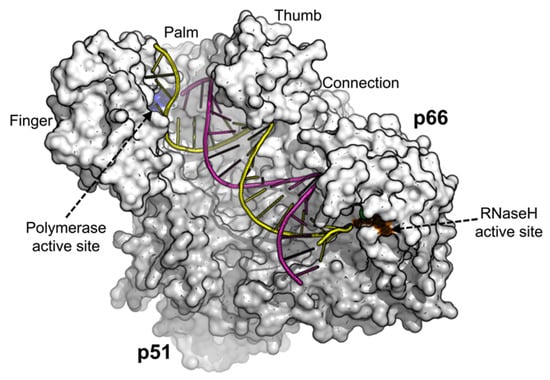
Figure 1.
Human immunodeficiency virus type 1 (HIV-1) Reverse Transcriptase structure complexed with DNA (pdb 1T05) [27]. The image was generated using PyMOL [28].
3. Comparison of Cell-Free and Cell-Based Fidelity Assays
The fidelity of HIV-1 RT is best assessed directly on patient samples, but ethical and biosafety requirements are insurmountable obstacles for many small labs/institutes. Even without such obstacles, the virus would have undergone multiple generations of replication within a patient between sampling, making it nearly impossible to determine the fidelity of a single round of replication. Thus, various cell-free and cell-based approaches were developed.
3.1. Cell-Free Fidelity Assays
In vitro assays, termed “cell-free” assays to emphasize the absence of HIV replication in cells, reduce confounding factors (e.g., a balanced dNTP pool) to give higher reproducibility. However, such reductionist methods also remove factors that influence the fidelity of HIV RT, while employing high temperatures in PCR that could disrupt secondary structures of the template nucleic acid.
The first cell-free fidelity assays utilized synthetic polynucleotide templates to determine errors using a mismatched radio-labelled nucleotide [29]. However, these homopolymer templates do not accurately represent the natural heteropolymer template (such as a gene), resulting in the overestimation of mutation rates due to their repeating nature [30]. Confounding factors such as the slippage of primers and stacking interactions between nucleotides further influence the fidelity of base substitution [29].
For the visual identification of mutants and non-mutants, reporter genes such as the α-complementing region of the lacZ gene (lacZα) and the DNA of bacteriophage ΦX174 are commonly employed in base reversion assays that measure the error rate of RT on a single base [30]. However, RNA secondary structure influences are not considered, overlooking the spectrum of mutations and any potential mutational hotspots. To address this limitation, the cell-free forward mutation assay [31] involving gap filling by RT, allows the determination of mutation spectra and hotspots. The error rates and mutant frequencies of HIV-1 RT from previously reported cell-free fidelity assays are shown in Table 1 and Table 2, respectively.

Table 2.
Mutant frequencies of HIV-1 RT measured in cell-free fidelity assays.
3.2. Cell-Based Fidelity Assays
In vivo assays, termed “cell-based” assays here, attempt to mimic the conditions to include host and viral proteins absent in cell-free assays. However, this trades off reproducibility when different cells are utilized. The assays leverage on transfecting shuttle vectors containing HIV genes into mammalian cells, followed by the selection of mutants in suitable hosts (e.g., bacteria). However, silent mutations are not detected, possibly leading to an overestimated fidelity of HIV RT in both cell-based and cell-free assays. The error rates and mutant frequencies of HIV-1 RT from previously reported cell-based fidelity assays are shown in Table 3 and Table 4, respectively.

Table 3.
Error rates of HIV-1 RT measured in cell-based fidelity assays.

Table 4.
Mutant frequencies of HIV-1 RT measured in cell-based fidelity assays.
4. Studies on HIV-1 Genes
For a better study of HIV-1 RT, the mutation positions are important to provide mechanistic insights to the development of drug resistance [76]. The well-established mutation rate of HIV-1 RT is performed on lacZα [39,40,41,42,43,44,45,48,58,77], resulting in a gap of knowledge regarding how HIV-1 genes are mutated by RT.
We found only two studies that utilized HIV templates [36,74]. One involved the HIV-1 env gene (see Table 5) in cell-free assays, finding an error rate in the DNA of 1.90 × 10−4 errors/bp/cycle, in RNA at 2.00 × 10−4 errors/bp/cycle and RNA/DNA at 3.80 × 10−4 errors/bp/cycle. This is comparable to previous M13mp2 forward assays using the lacZα template in DNA at 1.69 × 10−4 and RNA at 1.45 × 10−4 error/bp/cycle [36]. Similarly, Geller and colleagues’ work on HIV env and int-vir-vpr RNA, found error rates of 0.36 × 10−4 and 0.75 × 10−4 error/bp/cycle, respectively [74].

Table 5.
Percentages of nucleotide mutations of HIV-1 RT on the HIV-1 gene and LacZα template.
Mutations reported in the first study were found to partially correlate with those found in AIDS patients [36], while the latter showed that sequence and secondary structure affected the activity of cytidine deamination and fidelity of HIV RT [74]. Together, they demonstrate the need to perform studies directly on HIV-1 genes.
5. Analysis of Allosteric Communication
Current anti-HIV drugs target the key viral enzymes such as reverse transcriptase, integrase, and protease [78], and, as such, occurrences of mutations hotspots within these targets are essential for understanding drug resistance and the design of new drugs.
Clinical mutations were found not only at the direct drug-binding sites but also non-functional regions [79], the latter of which can have indirect effects on drug resistance such as allosteric communication to the catalytic sites. The inhibition of such communication can be explored for novel classes of drugs, such as in the case of reverse transcriptase inhibitors (RTIs) [80], integrase inhibitors [81], and protease inhibitors (PIs) [82]. The non-nucleoside RTIs bind non-competitively to an allosteric site on p66 subunit to cause structural changes in the RT polymerase active site, hindering DNA polymerization. Our ongoing study has found further potential interferences targeting the p51 to affect overall RT activity [83]. On integrase, allosteric inhibitors impair the binding of integrase and the cellular cofactor LEDGF/p75 during HIV-1 replication to induce aberrant integrase multimerization [81]. Similarly, a potential allosteric protease inhibitor was found to bind to a site at the protease flap to equipotently inhibit both wild-type and certain drug-resistant variants [82].
Given the resource constraints for experimentally testing every residue for allosteric communication, in silico screening can be performed on potential mutation hotspots. Following experimental validation via recombinant methods to establish the level and type of effect (detrimental or augmented catalytic activity), the site can then be targeted for intervention, especially if they are able to elicit drastic protein structural changes to affect functionality. One such example is in IgA1 and IgA2, which had varying allosteric communications due to different intermediate protein regions [84].
Recent advances have shown the p51 subunit of HIV-1 RT to induce flexibility on the DNA polymerase active site on p66, inhibiting RT function [85]. Similarly, Gag non-cleavage site mutations that are known to compensate for viral fitness in drug resistance were found to have allosteric communications [86] with the first protease cleavage site on Gag [87]. While further research is on-going, the example of non-cleavage Gag mutations involved in protease inhibitor resistance (despite an absence of mutations in the cleavage sites) demonstrates the importance of non-active site mutations that exhibit allosteric communications [75,88].
For this review, the locations in the various HIV target proteins were screened for their allosteric communication to the known active sites. To quantify the strength of allosteric effects caused by each residual mutation, allosteric-free energy (ΔΔgresidue) was calculated (using AlloSigMA, more details in [89,90,91]) for the other responding residues. Individual perturbations for the whole protein are shown in the allosteric signaling map [92,93] of Figure 2.
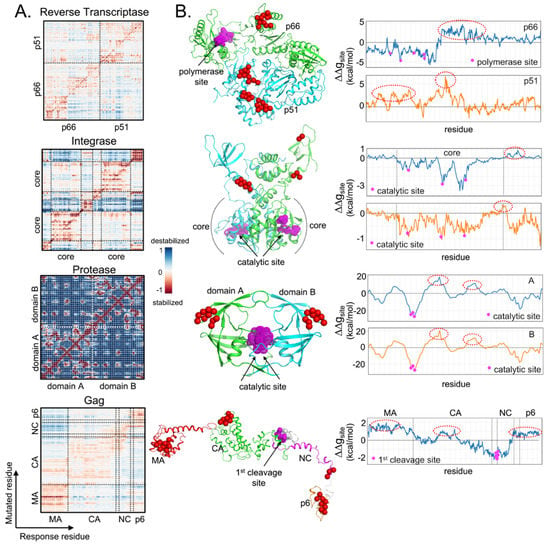
Figure 2.
Underlying allosteric communications were found within HIV-1 proteins. (A) Allosteric signaling maps (single-point mutation screening) of HIV-1 Reverse Transcriptase, Integrase, Protease, and Gag. Structural presentation using RT (pdb 3T19), IN (reconstructed from pdb 1K6Y and 1EX4), PR (pdb 2PC0), and Gag model from Su et al. [87]; (B) Allosteric-free energies (ΔΔgsite) on specific catalytic or cleavage sites (magenta spheres) were estimated based on individual perturbations at single residues (x-axis) to demonstrate the resulting stabilizing (ΔΔgsite < 0) or destabilizing (ΔΔgsite > 0) effects. The possible mutations which may potentially destabilize the sites of interest, are highlighted in red spheres and red dash ovals.
Asymmetrical effects in the HIV-1 RT structure further affirmed that mutations in p51 could stabilize the active site on p66, but not vice versa (Figure 2A). Since rigidity reduces HIV-1 RT activity [94], this opens up p51 as a potential new drug target. Given the estimated allosteric-free energies at the DNA polymerase active site (ΔΔgsite by averaging all ΔΔgresidue of the residues involving in the active site to demonstrate stabilizing (ΔΔgsite < 0) or destabilizing (ΔΔgsite > 0) effects), it showed that the active site is destabilized by mutations on the thumb domain of p66 (residues 260-321) and on p51 (residues 33–42, 68–78, and 96–114), highlighted in red spheres and red dash ovals in Figure 2B. These sites can thus be targeted for intervention.
Differing from RT, symmetrical allosteric communications between domains in HIV-1 integrase and protease are found [95,96], probably resulting from the homo-multimerization (Figure 2A). Mutations on the C-terminal domain (CTD, residues 220–221, 230–232, and 211–217) of integrase and at the “ears” regions of protease (residues 33–45 and 57–63, of which residues L33, E34, and M36 were reported to be resistant to several protease inhibitors [79]) have been found to affect the active sites of the two enzymes (Figure 2B). For the Gag protein, allosteric communications between several non-cleavage sites and the cleavage sites were found, affecting proteolysis [87], e.g., on Gag matrix (MA: residues E12, V35, E40, and L75), capsid (CA: residue H219), and p6 (L449 and P453) domains [88,97,98]. By analyzing mutational hotspots, additional sites with allosteric communications can be identified for pre-emptive drug design.
6. Implications on Sagacious Drug Design
The control of viral mutation rates has been proposed as an antiretroviral strategy [6]. In this review, we further propose identifying mutational hotspots and mutation rates of specific viral drug targets towards an application of rational drug design, that can include the development and pre-emptive design of novel drugs. While generic HIV-1 RT mutation rates are well-established from multiple studies reviewed in this article, there are still gaps when looking at specific HIV genes, especially given that sequence context and secondary structure are known to influence the fidelity of HIV-1 RT [74]. Within these targets, an in-depth understanding of the mutation rate and types of mutation (e.g., substitution, deletion and insertion) and predisposition to specific nucleotide changes (e.g., A to G) can shed light on viral drug resistance. Such knowledge allows for the targeting of regions with lower mutation rates while balancing possible allosteric effects or compensatory effects of viral fitness in a sagacious drug design strategy. For example, the M184V resistance mutation in HIV-1 RT is known to increase fidelity, impair viral fitness, and increase hyper-sensitization to NRTIs (such as amprenavir and efavirenz) [99]. Thus, it is possible to leverage on such features alongside structural understanding [76] to utilize combinatorial therapies to target the active site, using existing inhibitors to select for the mutation alongside other inhibitors to limit the escape mutations.
7. Conclusions and Future Perspective
Comparing cell-free and cell-based essays that contribute to the mutation rate of HIV-1, there is a lack of study of specific HIV-1 drug targets that would provide insights for rational drug designs, especially in the light of in silico allosteric analysis. Addressing such gaps has great promise for gaining an upper hand to develop novel intervention strategies against HIV.
Author Contributions
Writing—original draft preparation, J.Y.Y. and G.-R.G.; writing—review and editing, J.Y.Y., G.-R.G., C.T.-T.S., and S.K.-E.G.; overall supervision, S.K.-E.G. All authors have read and agreed to the published version of the manuscript.
Funding
The research is supported by core fund of A*STAR.
Acknowledgments
We thank other APD Lab members for their useful discussion and help in proof-reading.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Alizon, M.; Wain-Hobson, S.; Montagnier, L.; Sonigo, P. Genetic variability of the AIDS virus: Nucleotide sequence analysis of two isolates from African patients. Cell 1986, 46, 63–74. [Google Scholar] [CrossRef]
- Hahn, B.H.; Shaw, G.M.; Taylor, M.E.; Redfield, R.R.; Markham, P.D.; Salahuddin, S.Z.; Wong-Staal, F.; Gallo, R.C.; Parks, E.S.; Parks, W.P. Genetic variation in HTLV-III/LAV over time in patients with AIDS or at risk for AIDS. Science 1986, 232, 1548–1553. [Google Scholar] [CrossRef] [PubMed]
- Saag, M.S.; Hahn, B.H.; Gibbons, J.; Li, Y.; Parks, E.S.; Parks, W.P.; Shaw, G.M. Extensive variation of human immunodeficiency virus type-1 in vivo. Nature 1988, 334, 440–444. [Google Scholar] [CrossRef] [PubMed]
- Balfe, P.; Simmonds, P.; Ludlam, C.A.; Bishop, J.O.; Brown, A.J. Concurrent evolution of human immunodeficiency virus type 1 in patients infected from the same source: Rate of sequence change and low frequency of inactivating mutations. J. Virol. 1990, 64, 6221–6233. [Google Scholar] [CrossRef] [PubMed]
- Mansky, L.M. Retrovirus mutation rates and their role in genetic variation. J. Gen. Virol. 1998, 79, 1337–1345. [Google Scholar] [CrossRef] [PubMed]
- Menéndez-Arias, L. Mutation Rates and Intrinsic Fidelity of Retroviral Reverse Transcriptases. Viruses 2009, 1, 1137–1165. [Google Scholar] [CrossRef]
- Wei, X.; Ghosh, S.K.; Taylor, M.E.; Johnson, V.A.; Emini, E.A.; Deutsch, P.; Lifson, J.D.; Bonhoeffer, S.; Nowak, M.A.; Hahn, B.H. Viral dynamics in human immunodeficiency virus type 1 infection. Nature 1995, 373, 117–122. [Google Scholar] [CrossRef]
- Hu, W.S.; Temin, H.M. Retroviral recombination and reverse transcription. Science 1990, 250, 1227–1233. [Google Scholar] [CrossRef]
- Coffin, J.M. HIV population dynamics in vivo: Implications for genetic variation, pathogenesis, and therapy. Science 1995, 267, 483–489. [Google Scholar] [CrossRef]
- Fisher, A.G.; Ensoli, B.; Looney, D.; Rose, A.; Gallo, R.C.; Saag, M.S.; Shaw, G.M.; Hahn, B.H.; Wong-Staal, F. Biologically diverse molecular variants within a single HIV-1 isolate. Nature 1988, 334, 444–447. [Google Scholar] [CrossRef]
- Meyerhans, A.; Cheynier, R.; Albert, J.; Seth, M.; Kwok, S.; Sninsky, J.; Morfeldt-Månson, L.; Asjö, B.; Wain-Hobson, S. Temporal fluctuations in HIV quasispecies in vivo are not reflected by sequential HIV isolations. Cell 1989, 58, 901–910. [Google Scholar] [CrossRef]
- Hedskog, C.; Mild, M.; Jernberg, J.; Sherwood, E.; Bratt, G.; Leitner, T.; Lundeberg, J.; Andersson, B.; Albert, J. Dynamics of HIV-1 Quasispecies during Antiviral Treatment Dissected Using Ultra-Deep Pyrosequencing. PLoS ONE 2010, 5, e11345. [Google Scholar] [CrossRef] [PubMed]
- Ode, H.; Matsuda, M.; Matsuoka, K.; Hachiya, A.; Hattori, J.; Kito, Y.; Yokomaku, Y.; Iwatani, Y.; Sugiura, W. Quasispecies Analyses of the HIV-1 Near-full-length Genome With Illumina MiSeq. Front. Microbiol. 2015, 6, 1258. [Google Scholar] [CrossRef] [PubMed]
- Dampier, W.; Nonnemacher, M.R.; Mell, J.; Earl, J.; Ehrlich, G.D.; Pirrone, V.; Aiamkitsumrit, B.; Zhong, W.; Kercher, K.; Passic, S.; et al. HIV-1 Genetic Variation Resulting in the Development of New Quasispecies Continues to Be Encountered in the Peripheral Blood of Well-Suppressed Patients. PLoS ONE 2016, 11, e0155382. [Google Scholar] [CrossRef] [PubMed]
- Domingo, E.; Holland, J.J. Rna Virus Mutations and Fitness for Survival. Annu. Rev. Microbiol. 1997, 51, 151–178. [Google Scholar] [CrossRef] [PubMed]
- Gaschen, B.; Taylor, J.; Yusim, K.; Foley, B.; Gao, F.; Lang, D.; Novitsky, V.; Haynes, B.; Hahn, B.H.; Bhattacharya, T.; et al. Diversity considerations in HIV-1 vaccine selection. Science 2002, 296, 2354–2360. [Google Scholar] [CrossRef]
- Kellam, P.; Larder, B.A. Retroviral recombination can lead to linkage of reverse transcriptase mutations that confer increased zidovudine resistance. J. Virol. 1995, 69, 669–674. [Google Scholar] [CrossRef]
- Gu, Z.; Gao, Q.; Faust, E.A.; Wainberg, M.A. Possible involvement of cell fusion and viral recombination in generation of human immunodeficiency virus variants that display dual resistance to AZT and 3TC. J. Gen. Virol. 1995, 76, 2601–2605. [Google Scholar] [CrossRef]
- Moutouh, L.; Corbeil, J.; Richman, D.D. Recombination leads to the rapid emergence of HIV-1 dually resistant mutants under selective drug pressure. Proc. Natl. Acad. Sci. USA 1996, 93, 6106–6111. [Google Scholar] [CrossRef]
- Burke, D.S. Recombination in HIV: An important viral evolutionary strategy. Emerg. Infect. Dis. 1997, 3, 253–259. [Google Scholar] [CrossRef]
- Santoro, M.M.; Perno, C.F. HIV-1 Genetic Variability and Clinical Implications. ISRN Microbiol. 2013, 2013, 481314. [Google Scholar] [CrossRef] [PubMed]
- Lightfoote, M.M.; Coligan, J.E.; Folks, T.M.; Fauci, A.S.; Martin, M.A.; Venkatesan, S. Structural characterization of reverse transcriptase and endonuclease polypeptides of the acquired immunodeficiency syndrome retrovirus. J. Virol. 1986, 60, 771–775. [Google Scholar] [CrossRef] [PubMed]
- Roberts, J.D.; Bebenek, K.; Kunkel, T.A. The accuracy of reverse transcriptase from HIV-1. Science 1988, 242, 1171–1173. [Google Scholar] [CrossRef]
- Kim, T.; Mudry, R.A.; Rexrode, C.A.; Pathak, V.K. Retroviral mutation rates and A-to-G hypermutations during different stages of retroviral replication. J. Virol. 1996, 70, 7594–7602. [Google Scholar] [CrossRef] [PubMed]
- O’Neil, P.K.; Sun, G.; Yu, H.; Ron, Y.; Dougherty, J.P.; Preston, B.D. Mutational Analysis of HIV-1 Long Terminal Repeats to Explore the Relative Contribution of Reverse Transcriptase and RNA Polymerase II to Viral Mutagenesis. J. Biol. Chem. 2002, 277, 38053–38061. [Google Scholar] [CrossRef] [PubMed]
- Cuevas, J.M.; Geller, R.; Garijo, R.; López-Aldeguer, J.; Sanjuán, R. Extremely High Mutation Rate of HIV-1 In Vivo. PLoS Biol. 2015, 13, e1002251. [Google Scholar] [CrossRef]
- Tuske, S.; Sarafianos, S.G.; Clark, A.D.; Ding, J.; Naeger, L.K.; White, K.L.; Miller, M.D.; Gibbs, C.S.; Boyer, P.L.; Clark, P.; et al. Structures of HIV-1 RT–DNA complexes before and after incorporation of the anti-AIDS drug tenofovir. Nature Structural & Molecular Biology 2004, 11, 469–474. [Google Scholar]
- Schrodinger, L. The PyMOL Molecular Graphics System. 2019. Available online: http://www.pymol.org (accessed on 5 February 2020).
- Loeb, L.A.; Kunkel, T.A. Fidelity of DNA synthesis. Annu. Rev. Biochem. 1982, 51, 429–457. [Google Scholar] [CrossRef]
- Weymouth, L.A.; Loeb, L.A. Mutagenesis during in vitro DNA synthesis. Proc. Natl. Acad. Sci. USA 1978, 75, 1924–1928. [Google Scholar] [CrossRef]
- Bebenek, K.; Kunkel, T.A. Analyzing fidelity of DNA polymerases. In Methods in Enzymology; DNA Replication; Elsevier: Amsterdam, The Netherlands, 1995; Volume 262, pp. 217–232. [Google Scholar]
- Preston, B.D.; Poiesz, B.J.; Loeb, L.A. Fidelity of HIV-1 reverse transcriptase. Science 1988, 242, 1168–1171. [Google Scholar] [CrossRef]
- Takeuchi, Y.; Nagumo, T.; Hoshino, H. Low fidelity of cell-free DNA synthesis by reverse transcriptase of human immunodeficiency virus. J. Virol. 1988, 62, 3900–3902. [Google Scholar] [CrossRef] [PubMed]
- Weber, J.; Grosse, F. Fidelity of human immunodeficiency virus type I reverse transcriptase in copying natural DNA. Nucleic Acids Res. 1989, 17, 1379–1393. [Google Scholar] [CrossRef] [PubMed]
- Ji, J.; Loeb, L.A. Fidelity of HIV-1 reverse transcriptase copying RNA in vitro. Biochemistry 1992, 31, 954–958. [Google Scholar] [CrossRef]
- Ji, J.; Loeb, L.A. Fidelity of HIV-1 Reverse Transcriptase Copying a Hypervariable Region of the HIV-1 env Gene. Virology 1994, 199, 323–330. [Google Scholar] [CrossRef]
- Stuke, A.W.; Ahmad-Omar, O.; Hoefer, K.; Hunsmann, G.; Jentsch, K.D. Mutations in the SIV env and the M13 lacZa gene generated in vitro by reverse transcriptases and DNA polymerases. Arch. Virol. 1997, 142, 1139–1154. [Google Scholar] [CrossRef]
- Rezende, L.F.; Curr, K.; Ueno, T.; Mitsuya, H.; Prasad, V.R. The Impact of Multidideoxynucleoside Resistance-Conferring Mutations in Human Immunodeficiency Virus Type 1 Reverse Transcriptase on Polymerase Fidelity and Error Specificity. J. Virol. 1998, 72, 2890–2895. [Google Scholar] [CrossRef]
- Drosopoulos, W.C.; Prasad, V.R. Increased Misincorporation Fidelity Observed for Nucleoside Analog Resistance Mutations M184V and E89G in Human Immunodeficiency Virus Type 1 Reverse Transcriptase Does Not Correlate with the Overall Error Rate Measured In Vitro. J. Virol. 1998, 72, 4224–4230. [Google Scholar] [CrossRef]
- Rezende, L.F.; Drosopoulos, W.C.; Prasad, V.R. The influence of 3TC resistance mutation M184I on the fidelity and error specificity of human immunodeficiency virus type 1 reverse transcriptase. Nucleic Acids Res. 1998, 26, 3066–3072. [Google Scholar] [CrossRef]
- Boyer, P.L.; Hughes, S.H. Effects of Amino Acid Substitutions at Position 115 on the Fidelity of Human Immunodeficiency Virus Type 1 Reverse Transcriptase. J. Virol. 2000, 74, 6494–6500. [Google Scholar] [CrossRef][Green Version]
- Shah, F.S.; Curr, K.A.; Hamburgh, M.E.; Parniak, M.A.; Mitsuya, H.; Arnez, J.G.; Prasad, V.R. Differential influence of nucleoside analog-resistance mutations K65R and L74V on the overall mutation rate and error specificity of human immunodeficiency virus type 1 reverse transcriptase. J. Biol. Chem. 2000, 35, 27037–27044. [Google Scholar] [CrossRef]
- Rezende, L.F.; Kew, Y.; Prasad, V.R. The effect of increased processivity on overall fidelity of human immunodeficiency virus type 1 reverse transcriptase. J. Biomed. Sci. 2001, 8, 197–205. [Google Scholar] [CrossRef] [PubMed]
- Boyer, P.L.; Stenbak, C.R.; Hoberman, D.; Linial, M.L.; Hughes, S.H. In Vitro Fidelity of the prototype primate foamy virus (PFV) RT compared to HIV-1 RT. Virology 2007, 367, 253–264. [Google Scholar] [CrossRef] [PubMed]
- Alvarez, M.; Matamoros, T.; Menéndez-Arias, L. Increased thermostability and fidelity of DNA synthesis of wild-type and mutant HIV-1 group O reverse transcriptases. J. Mol. Biol. 2009, 392, 872–884. [Google Scholar] [CrossRef][Green Version]
- Álvarez, M.; Barrioluengo, V.; Afonso-Lehmann, R.N.; Menéndez-Arias, L. Altered error specificity of RNase H-deficient HIV-1 reverse transcriptases during DNA-dependent DNA synthesis. Nucleic Acids Res. 2013, 41, 4601–4612. [Google Scholar] [CrossRef][Green Version]
- Álvarez, M.; Sebastián-Martín, A.; García-Marquina, G.; Menéndez-Arias, L. Fidelity of classwide-resistant HIV-2 reverse transcriptase and differential contribution of K65R to the accuracy of HIV-1 and HIV-2 reverse transcriptases. Sci. Rep. 2017, 7, 44834. [Google Scholar] [CrossRef]
- Sebastián-Martín, A.; Barrioluengo, V.; Menéndez-Arias, L. Transcriptional inaccuracy threshold attenuates differences in RNA-dependent DNA synthesis fidelity between retroviral reverse transcriptases. Sci. Rep. 2018, 8, 627. [Google Scholar] [CrossRef]
- Bebenek, K.; Abbotts, J.; Roberts, J.D.; Wilson, S.H.; Kunkel, T.A. Specificity and mechanism of error-prone replication by human immunodeficiency virus-1 reverse transcriptase. J. Biol. Chem. 1989, 264, 16948–16956. [Google Scholar]
- Hübner, A.; Kruhoffer, M.; Grosse, F.; Krauss, G. Fidelity of human immunodeficiency virus type I reverse transcriptase in copying natural RNA. J. Mol. Biol. 1992, 223, 595–600. [Google Scholar] [CrossRef]
- Boyer, J.C.; Bebenek, K.; Kunkel, T.A. Unequal human immunodeficiency virus type 1 reverse transcriptase error rates with RNA and DNA templates. Proc. Natl. Acad. Sci. USA 1992, 89, 6919–6923. [Google Scholar] [CrossRef]
- Bebenek, K.; Abbotts, J.; Wilson, S.H.; Kunkel, T.A. Error-prone polymerization by HIV-1 reverse transcriptase. Contribution of template-primer misalignment, miscoding, and termination probability to mutational hot spots. J. Biol. Chem. 1993, 268, 10324–10334. [Google Scholar] [PubMed]
- Beard, W.A.; Stahl, S.J.; Kim, H.R.; Bebenek, K.; Kumar, A.; Strub, M.P.; Becerra, S.P.; Kunkel, T.A.; Wilson, S.H. Structure/function studies of human immunodeficiency virus type 1 reverse transcriptase. Alanine scanning mutagenesis of an alpha-helix in the thumb subdomain. J. Biol. Chem. 1994, 269, 28091–28097. [Google Scholar] [PubMed]
- Bebenek, K.; Beard, W.A.; Casas-Finet, J.R.; Kim, H.-R.; Darden, T.A.; Wilson, S.H.; Kunkel, T.A. Reduced Frameshift Fidelity and Processivity of HIV-1 Reverse Transcriptase Mutants Containing Alanine Substitutions in Helix H of the Thumb Subdomain. J. Biol. Chem. 1995, 270, 19516–19523. [Google Scholar] [CrossRef] [PubMed]
- Beard, W.A.; Minnick, D.T.; Wade, C.L.; Prasad, R.; Won, R.L.; Kumar, A.; Kunkel, T.A.; Wilson, S.H. Role of the “Helix Clamp” in HIV-1 Reverse Transcriptase Catalytic Cycling as Revealed by Alanine-scanning Mutagenesis. J. Biol. Chem. 1996, 271, 12213–12220. [Google Scholar] [CrossRef] [PubMed]
- Kim, B.; Hathaway, T.R.; Loeb, L.A. Fidelity of Mutant HIV-1 Reverse Transcriptases: Interaction with the Single-Stranded Template Influences the Accuracy of DNA Synthesis. Biochemistry 1998, 37, 5831–5839. [Google Scholar] [CrossRef] [PubMed]
- Kim, B.; Ayran, J.C.; Sagar, S.G.; Adman, E.T.; Fuller, S.M.; Tran, N.H.; Horrigan, J. New Human Immunodeficiency Virus, Type 1 Reverse Transcriptase (HIV-1 RT) Mutants with Increased Fidelity of DNA Synthesis. Accuracy, template binding, and processivity. J. Biol. Chem. 1999, 274, 27666–27673. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Lewis, D.A.; Bebenek, K.; Beard, W.A.; Wilson, S.H.; Kunkel, T.A. Uniquely Altered DNA Replication Fidelity Conferred by an Amino Acid Change in the Nucleotide Binding Pocket of Human Immunodeficiency Virus Type 1 Reverse Transcriptase. J. Biol. Chem. 1999, 274, 32924–32930. [Google Scholar] [CrossRef] [PubMed]
- Jonckheere, H.; Clercq, E.D.; Anné, J. Fidelity analysis of HIV-1 reverse transcriptase mutants with an altered amino-acid sequence at residues Leu74, Glu89, Tyr115, Tyr183 and Met184. Eur. J. Biochem. 2000, 267, 2658–2665. [Google Scholar] [CrossRef]
- Weiss, K.K.; Isaacs, S.J.; Tran, N.H.; Adman, E.T.; Kim, B. Molecular Architecture of the Mutagenic Active Site of Human Immunodeficiency Virus Type 1 Reverse Transcriptase: Roles of the β8-αE Loop in Fidelity, Processivity, and Substrate Interactions. Biochemistry 2000, 39, 10684–10694. [Google Scholar] [CrossRef]
- Fisher, T.S.; Prasad, V.R. Substitutions of Phe61 Located in the Vicinity of Template 5′-Overhang Influence Polymerase Fidelity and Nucleoside Analog Sensitivity of HIV-1 Reverse Transcriptase. J. Biol. Chem. 2002, 277, 22345–22352. [Google Scholar] [CrossRef]
- Weiss, K.K.; Chen, R.; Skasko, M.; Reynolds, H.M.; Lee, K.; Bambara, R.A.; Mansky, L.M.; Kim, B. A Role for dNTP Binding of Human Immunodeficiency Virus Type 1 Reverse Transcriptase in Viral Mutagenesis. Biochemistry 2004, 43, 4490–4500. [Google Scholar] [CrossRef]
- Hamburgh, M.E.; Curr, K.A.; Monaghan, M.; Rao, V.R.; Tripathi, S.; Preston, B.D.; Sarafianos, S.; Arnold, E.; Darden, T.; Prasad, V.R. Structural Determinants of Slippage-mediated Mutations by Human Immunodeficiency Virus Type 1 Reverse Transcriptase. J. Biol. Chem. 2006, 281, 7421–7428. [Google Scholar] [CrossRef] [PubMed]
- Curr, K.; Tripathi, S.; Lennerstrand, J.; Larder, B.A.; Prasad, V.R. Influence of naturally occurring insertions in the fingers subdomain of human immunodeficiency virus type 1 reverse transcriptase on polymerase fidelity and mutation frequencies in vitro. J. Gen. Virol. 2006, 87, 419–428. [Google Scholar] [CrossRef] [PubMed]
- Matamoros, T.; Kim, B.; Menéndez-Arias, L. Mechanistic Insights into the Role of Val75 of HIV-1 Reverse Transcriptase in Misinsertion and Mispair Extension Fidelity of DNA Synthesis. J. Mol. Biol. 2008, 375, 1234–1248. [Google Scholar] [CrossRef] [PubMed]
- Barrioluengo, V.; Álvarez, M.; Barbieri, D.; Menéndez-Arias, L. Thermostable HIV-1 group O reverse transcriptase variants with the same fidelity as murine leukaemia virus reverse transcriptase. Biochem. J. 2011, 436, 599–607. [Google Scholar] [CrossRef] [PubMed]
- Mansky, L.M.; Temin, H.M. Lower in vivo mutation rate of human immunodeficiency virus type 1 than that predicted from the fidelity of purified reverse transcriptase. J. Virol. 1995, 69, 5087–5094. [Google Scholar] [CrossRef] [PubMed]
- Mansky, L.M. Forward mutation rate of human immunodeficiency virus type 1 in a T lymphoid cell line. AIDS Res. Hum. Retroviruses 1996, 12, 307–314. [Google Scholar] [CrossRef] [PubMed]
- Mansky, L.M. The mutation rate of human immunodeficiency virus type 1 is influenced by the vpr gene. Virology 1996, 222, 391–400. [Google Scholar] [CrossRef] [PubMed]
- Huang, K.J.; Wooley, D.P. A new cell-based assay for measuring the forward mutation rate of HIV-1. J. Virol. Methods 2005, 124, 95–104. [Google Scholar] [CrossRef] [PubMed]
- Rawson, J.M.O.; Landman, S.R.; Reilly, C.S.; Mansky, L.M. HIV-1 and HIV-2 exhibit similar mutation frequencies and spectra in the absence of G-to-A hypermutation. Retrovirology 2015, 12, 60. [Google Scholar] [CrossRef]
- Abram, M.E.; Ferris, A.L.; Shao, W.; Alvord, W.G.; Hughes, S.H. Nature, Position, and Frequency of Mutations Made in a Single Cycle of HIV-1 Replication. J. Virol. 2010, 84, 9864–9878. [Google Scholar] [CrossRef]
- Abram, M.E.; Ferris, A.L.; Das, K.; Quinoñes, O.; Shao, W.; Tuske, S.; Alvord, W.G.; Arnold, E.; Hughes, S.H. Mutations in HIV-1 Reverse Transcriptase Affect the Errors Made in a Single Cycle of Viral Replication. J. Virol. 2014, 88, 7589–7601. [Google Scholar] [CrossRef] [PubMed]
- Geller, R.; Domingo-Calap, P.; Cuevas, J.M.; Rossolillo, P.; Negroni, M.; Sanjuán, R. The external domains of the HIV-1 envelope are a mutational cold spot. Nat. Commun. 2015, 6, 8571. [Google Scholar] [CrossRef] [PubMed]
- Mansky, L.M.; Rouzic, E.L.; Benichou, S.; Gajary, L.C. Influence of Reverse Transcriptase Variants, Drugs, and Vpr on Human Immunodeficiency Virus Type 1 Mutant Frequencies. J. Virol. 2003, 77, 2071–2080. [Google Scholar] [CrossRef] [PubMed]
- Su, C.T.-T.; Koh, D.W.-S.; Gan, S.K.-E. Reviewing HIV-1 Gag Mutations in Protease Inhibitors Resistance: Insights for Possible Novel Gag Inhibitor Designs. Molecules 2019, 24, 3243. [Google Scholar] [CrossRef] [PubMed]
- Eckert, K.A.; Kunkel, T.A. Fidelity of DNA synthesis catalyzed by human DNA polymerase alpha and HIV-1 reverse transcriptase: Effect of reaction pH. Nucleic Acids Res. 1993, 21, 5212–5220. [Google Scholar] [CrossRef] [PubMed]
- Bhatti, A.B.; Usman, M.; Kandi, V. Current Scenario of HIV/AIDS, Treatment Options, and Major Challenges with Compliance to Antiretroviral Therapy. Cureus 2016, 8, e515. [Google Scholar] [CrossRef]
- Wensing, A.M.; Calvez, V.; Ceccherini-Silberstein, F.; Charpentier, C.; Günthard, H.F.; Paredes, R.; Shafer, R.W.; Richman, D.D. 2019 Update of the Drug Resistance Mutations in HIV-1. Top. Antivir. Med. 2019, 27, 111–121. [Google Scholar]
- Seckler, J.M.; Barkley, M.D.; Wintrode, P.L. Allosteric suppression of HIV-1 reverse transcriptase structural dynamics upon inhibitor binding. Biophys. J. 2011, 100, 144–153. [Google Scholar] [CrossRef]
- Patel, D.; Antwi, J.; Koneru, P.C.; Serrao, E.; Forli, S.; Kessl, J.J.; Feng, L.; Deng, N.; Levy, R.M.; Fuchs, J.R.; et al. A New Class of Allosteric HIV-1 Integrase Inhibitors Identified by Crystallographic Fragment Screening of the Catalytic Core Domain. J. Biol. Chem. 2016, 291, 23569–23577. [Google Scholar] [CrossRef]
- Ung, P.M.-U.; Dunbar, J.B.; Gestwicki, J.E.; Carlson, H.A. An allosteric modulator of HIV-1 protease shows equipotent inhibition of wild-type and drug-resistant proteases. J. Med. Chem. 2014, 57, 6468–6478. [Google Scholar] [CrossRef]
- Chan, K.-F.; Phua, S.-X.; Su, C.T.-T.; Gan, S.K.-E. Inhibiting HIV-1 and MMLV Reverse Transcriptase: The potential of an Allosteric Broad-Spectrum Inhibitor. Manuscript in preparation.
- Su, C.T.-T.; Lua, W.-H.; Ling, W.-L.; Gan, S.K.-E. Allosteric Effects between the Antibody Constant and Variable Regions: A Study of IgA Fc Mutations on Antigen Binding. Antibodies 2018, 7, 20. [Google Scholar] [CrossRef] [PubMed]
- Chiang, R.Z.-H.; Gan, S.K.-E.; Su, C.T.-T. A computational study for rational HIV-1 non-nucleoside reverse transcriptase inhibitor selection and the discovery of novel allosteric pockets for inhibitor design. Biosci. Rep. 2018, 38, BSR20171113. [Google Scholar] [CrossRef]
- Freed, E.O. HIV-1 assembly, release and maturation. Nat. Rev. Microbiol. 2015, 13, 484–496. [Google Scholar] [CrossRef] [PubMed]
- Su, C.T.-T.; Kwoh, C.-K.; Verma, C.S.; Gan, S.K.-E. Modeling the full length HIV-1 Gag polyprotein reveals the role of its p6 subunit in viral maturation and the effect of non-cleavage site mutations in protease drug resistance. J. Biomol. Struct. Dyn. 2018, 36, 4366–4377. [Google Scholar] [CrossRef] [PubMed]
- Gatanaga, H.; Suzuki, Y.; Tsang, H.; Yoshimura, K.; Kavlick, M.F.; Nagashima, K.; Gorelick, R.J.; Mardy, S.; Tang, C.; Summers, M.F. Amino acid substitutions in Gag protein at non-cleavage sites are indispensable for the development of a high multitude of HIV-1 resistance against protease inhibitors. J. Biol. Chem. 2002, 277, 5952–5961. [Google Scholar] [CrossRef]
- Guarnera, E.; Berezovsky, I.N. Structure-Based Statistical Mechanical Model Accounts for the Causality and Energetics of Allosteric Communication. PLoS Comput. Biol. 2016, 12, e1004678. [Google Scholar] [CrossRef]
- Guarnera, E.; Tan, Z.W.; Zheng, Z.; Berezovsky, I.N. AlloSigMA: Allosteric signaling and mutation analysis server. Bioinformatics 2017, 33, 3996–3998. [Google Scholar] [CrossRef]
- Guarnera, E.; Berezovsky, I.N. On the perturbation nature of allostery: Sites, mutations, and signal modulation. Curr. Opin. Struct. Biol. 2019, 56, 18–27. [Google Scholar] [CrossRef]
- Guarnera, E.; Berezovsky, I.N. Toward Comprehensive Allosteric Control over Protein Activity. Structure 2019, 27, 866–878. [Google Scholar] [CrossRef]
- Tan, Z.W.; Tee, W.-V.; Guarnera, E.; Booth, L.; Berezovsky, I.N. AlloMAPS: Allosteric mutation analysis and polymorphism of signaling database. Nucleic Acids Res. 2018, 47, D265–D270. [Google Scholar] [CrossRef] [PubMed]
- Chung, S.; Miller, J.T.; Lapkouski, M.; Tian, L.; Yang, W.; Le Grice, S.F.J. Examining the Role of the HIV-1 Reverse Transcriptase p51 Subunit in Positioning and Hydrolysis of RNA/DNA Hybrids. J. Biol. Chem. 2013, 288, 16177–16184. [Google Scholar] [CrossRef] [PubMed]
- Appadurai, R.; Senapati, S. Dynamical Network of HIV-1 Protease Mutants Reveals the Mechanism of Drug Resistance and Unhindered Activity. Biochemistry 2016, 55, 1529–1540. [Google Scholar] [CrossRef] [PubMed]
- Su, C.T.-T.; Ling, W.-L.; Lua, W.-H.; Haw, Y.-X.; Gan, S.K.-E. Structural analyses of 2015-updated drug-resistant mutations in HIV-1 protease: An implication of protease inhibitor cross-resistance. BMC Bioinformatics 2016, 17, 500. [Google Scholar] [CrossRef] [PubMed]
- Maguire, M.F.; Guinea, R.; Griffin, P.; Macmanus, S.; Elston, R.C.; Wolfram, J.; Richards, N.; Hanlon, M.H.; Porter, D.J.T.; Wrin, T.; et al. Changes in Human Immunodeficiency Virus Type 1 Gag at Positions L449 and P453 Are Linked to I50V Protease Mutants In Vivo and Cause Reduction of Sensitivity to Amprenavir and Improved Viral Fitness In Vitro. J. Virol. 2002, 76, 7398. [Google Scholar] [CrossRef] [PubMed]
- Doyon, L.; Croteau, G.; Thibeault, D.; Poulin, F.; Pilote, L.; Lamarre, D. Second locus involved in human immunodeficiency virus type 1 resistance to protease inhibitors. J. Virol. 1996, 70, 3763. [Google Scholar] [CrossRef]
- Turner, D.; Brenner, B.; Wainberg, M.A. Multiple Effects of the M184V Resistance Mutation in the Reverse Transcriptase of Human Immunodeficiency Virus Type 1. Clin. Diagn. Lab. Immunol. 2003, 10, 979–981. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).