Human Tibroviruses: Commensals or Lethal Pathogens?
Abstract
1. Introduction
2. Discovery of the First Tibroviruses
2.1. Bas-Congo Virus
2.2. Ekpoma Viruses 1 and 2
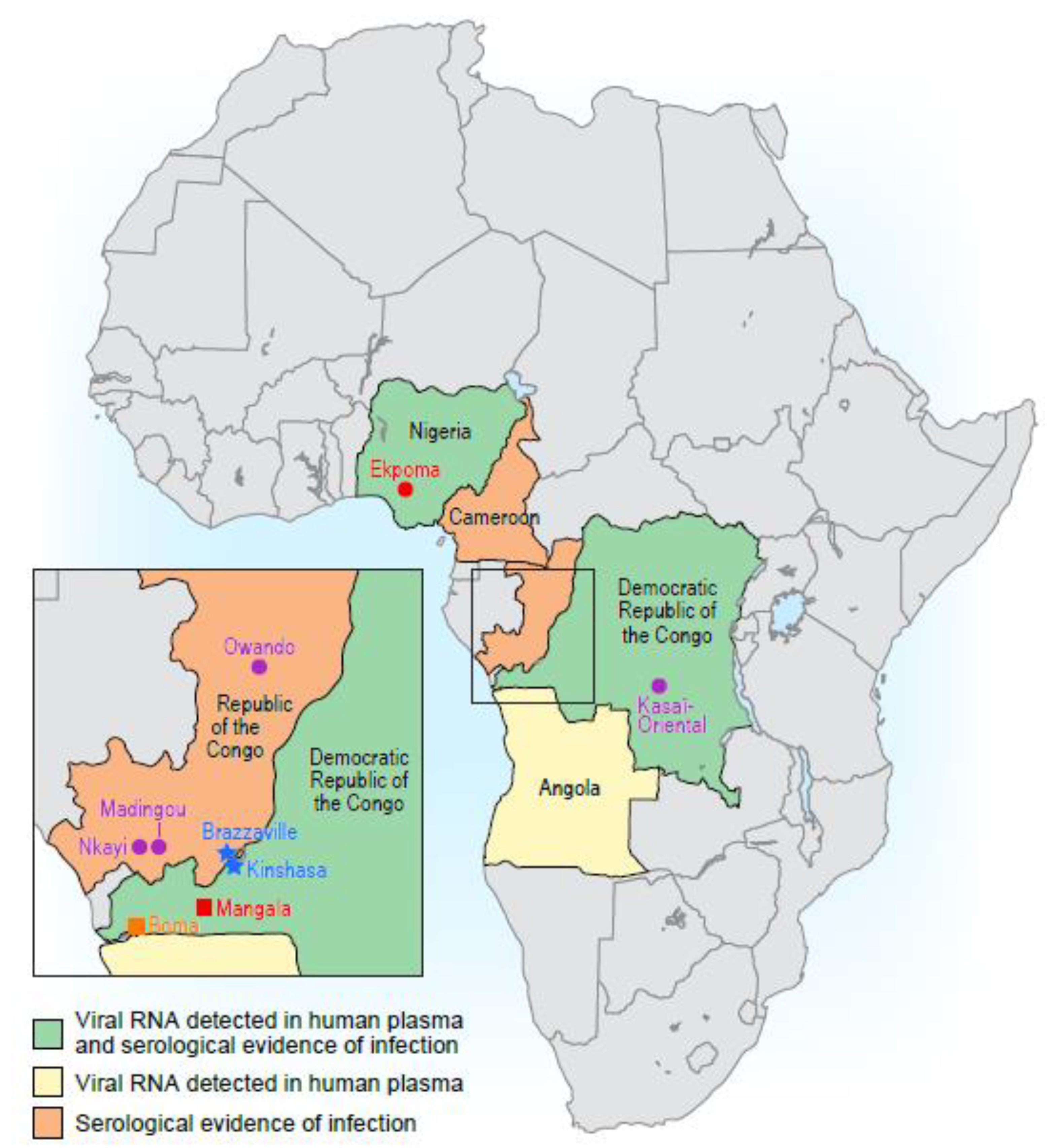
3. Exposure to Human Tibroviruses
Transmission of Human Tibroviruses
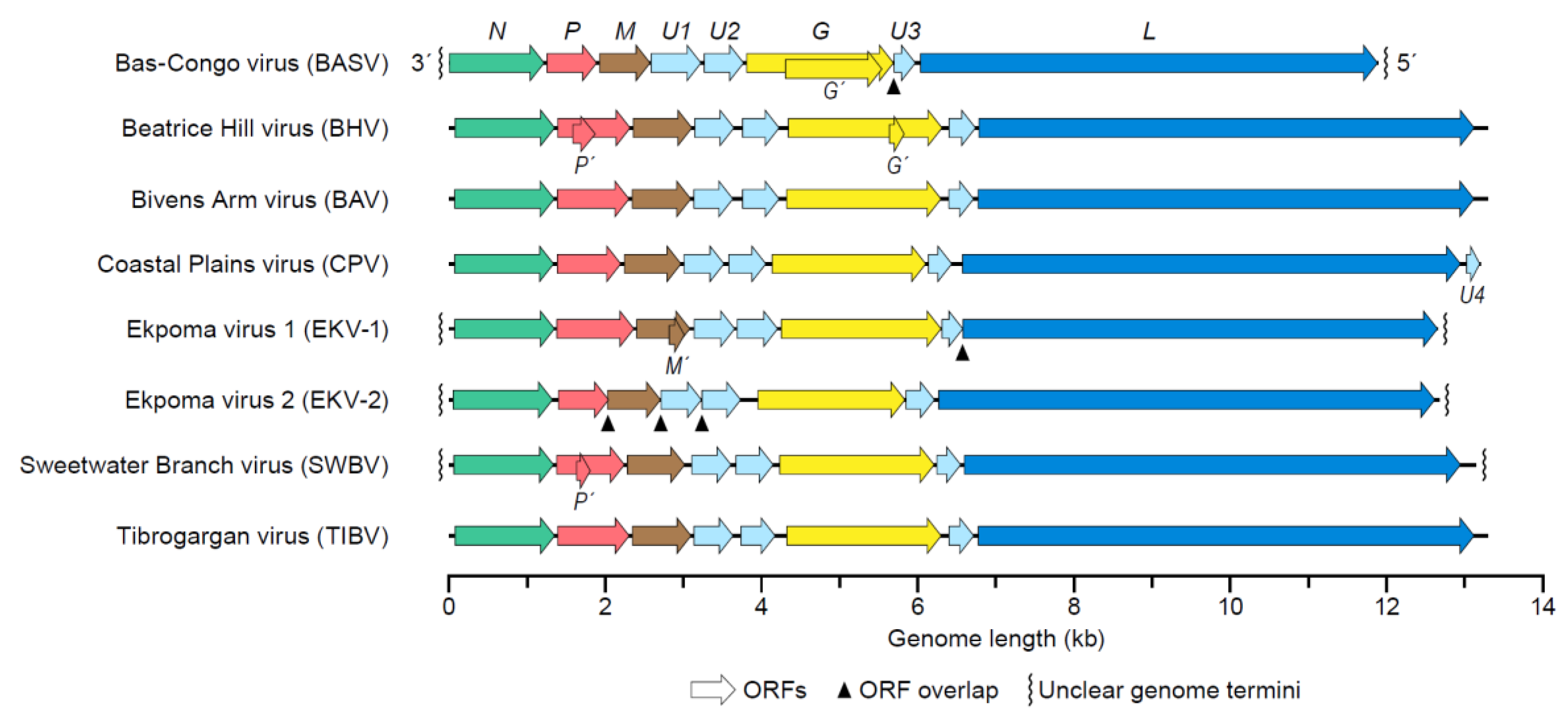
4. Tibrovirus Genome
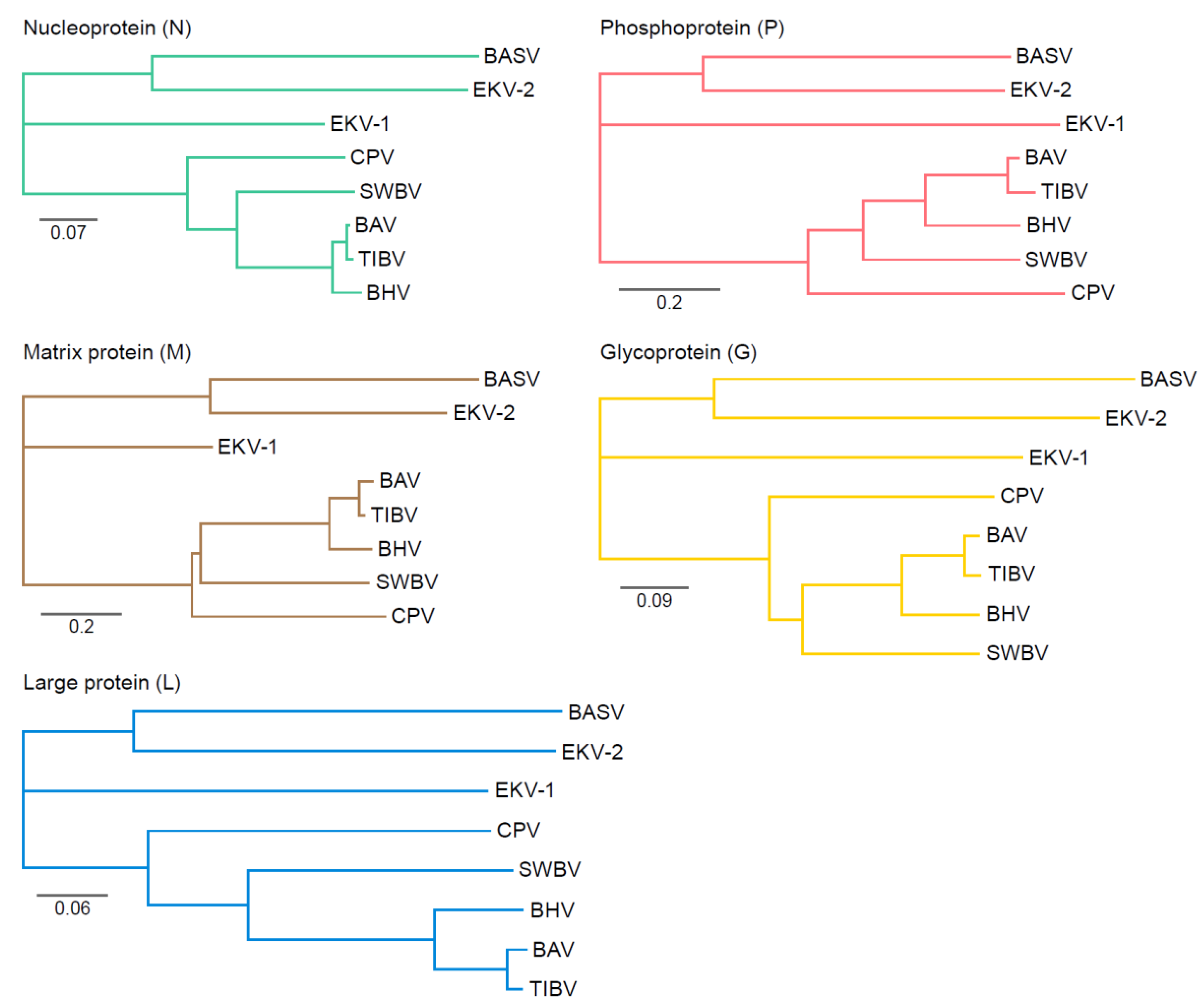
Genetic Variation of Tibroviruses
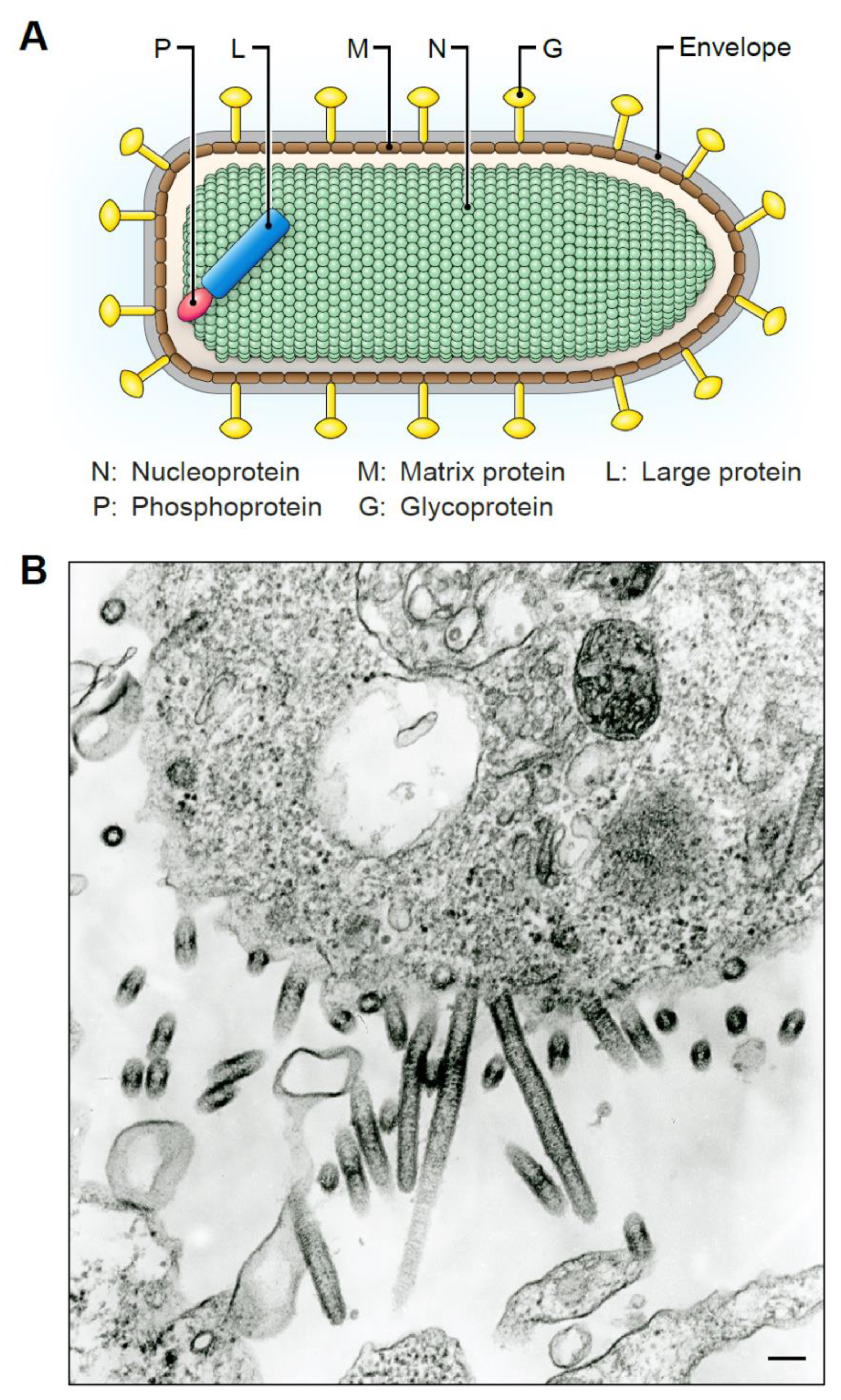
5. Tibrovirion Morphology
6. Tibrovirion Host Cell Entry and Tropism
7. In vitro Tibrovirus Replication and Cytopathic Effects
8. Implications for Human Health
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Dietzgen, R.G.; Kondo, H.; Goodin, M.M.; Kurath, G.; Vasilakis, N. The family Rhabdoviridae: Mono- and bipartite negative-sense RNA viruses with diverse genome organization and common evolutionary origins. Virus Res. 2017, 227, 158–170. [Google Scholar] [CrossRef]
- Kuzmin, I.V.; Walker, P.J. Vector-borne rhabdoviruses. In Arboviruses: Molecular Biology, Evolution and Control; Vasilakis, N., Gubler, D.J., Eds.; Caister Academic Press: Wymondham, UK, 2016; pp. 71–88. [Google Scholar]
- Walker, P.J.; Firth, C.; Widen, S.G.; Blasdell, K.R.; Guzman, H.; Wood, T.G.; Paradkar, P.N.; Holmes, E.C.; Tesh, R.B.; Vasilakis, N. Evolution of genome size and complexity in the Rhabdoviridae. PLoS Pathog. 2015, 11, e1004664. [Google Scholar] [CrossRef]
- Walker, P.J.; Blasdell, K.R.; Calisher, C.H.; Dietzgen, R.G.; Kondo, H.; Kurath, G.; Longdon, B.; Stone, D.M.; Tesh, R.B.; Tordo, N.; et al. ICTV virus taxonomy profile: Rhabdoviridae. J. Gen. Virol. 2018, 99, 447–448. [Google Scholar] [CrossRef] [PubMed]
- Amarasinghe, G.K.; Ayllón, M.A.; Bào, Y.; Basler, C.F.; Bavari, S.; Blasdell, K.R.; Briese, T.; Brown, P.A.; Bukreyev, A.; Balkema-Buschmann, A.; et al. Taxonomy of the order Mononegavirales: Update 2019. Arch. Virol. 2019, 164, 1967–1980. [Google Scholar] [CrossRef] [PubMed]
- Internationcal Committee on Taxonomy of Viruses. ICTV. 2019. Available online: https://talk.ictvonline.org/ (accessed on 21 February 2020).
- Ballinger, M.J.; Bruenn, J.A.; Taylor, D.J. Phylogeny, integration and expression of sigma virus-like genes in Drosophila. Mol. Phylogenet. Evol. 2012, 65, 251–258. [Google Scholar] [CrossRef] [PubMed]
- Geisler, C.; Jarvis, D.L. Rhabdovirus-like endogenous viral elements in the genome of Spodoptera frugiperda insect cells are actively transcribed: Implications for adventitious virus detection. Biologicals 2016, 44, 219–225. [Google Scholar] [CrossRef] [PubMed]
- Longdon, B.; Murray, G.G.R.; Palmer, W.J.; Day, J.P.; Parker, D.J.; Welch, J.J.; Obbard, D.J.; Jiggins, F.M. The evolution, diversity, and host associations of rhabdoviruses. Virus Evol. 2015, 1, vev014. [Google Scholar] [CrossRef] [PubMed]
- Hampson, K.; Coudeville, L.; Lembo, T.; Sambo, M.; Kieffer, A.; Attlan, M.; Barrat, J.; Blanton, J.D.; Briggs, D.J.; Cleaveland, S.; et al. Estimating the global burden of endemic canine rabies. PLoS Negl. Trop. Dis. 2015, 9, e0003709. [Google Scholar] [CrossRef]
- Rupprecht, C.; Kuzmin, I.; Meslin, F. Lyssaviruses and rabies: Current conundrums, concerns, contradictions and controversies. F1000Research 2017, 6, 184. [Google Scholar] [CrossRef]
- Merritt, T.; Taylor, K.; Cox-Witton, K.; Field, H.; Wingett, K.; Mendez, D.; Power, M.; Durrheim, D. Australian bat lyssavirus. Aust. J. Gen. Pract. 2018, 47, 93–96. [Google Scholar] [CrossRef]
- van Thiel, P.-P.A.M.; de Bie, R.M.A.; Eftimov, F.; Tepaske, R.; Zaaijer, H.L.; van Doornum, G.J.J.; Schutten, M.; Osterhaus, A.D.M.E.; Majoie, C.B.L.M.; Aronica, E.; et al. Fatal human rabies due to Duvenhage virus from a bat in Kenya: Failure of treatment with coma-induction, ketamine, and antiviral drugs. PLoS Negl. Trop. Dis. 2009, 3, e428. [Google Scholar] [CrossRef] [PubMed]
- McElhinney, L.M.; Marston, D.A.; Wise, E.L.; Freuling, C.M.; Bourhy, H.; Zanoni, R.; Moldal, T.; Kooi, E.A.; Neubauer-Juric, A.; Nokireki, T.; et al. Molecular epidemiology and evolution of European bat lyssavirus 2. Int. J. Mol. Sci. 2018, 19, 156. [Google Scholar] [CrossRef] [PubMed]
- Leonova, G.N.; Somova, L.M.; Belikov, S.I.; Kondratov, I.G.; Plekhova, N.G.; Krylova, N.V.; Pavlenko, E.V.; Tiunov, M.P.; Tkachev, S.E. The fatal case of lyssavirus encephalitis in the Russian Far East. In Encephalitis; Tkachev, S., Ed.; IntechOpen: London, UK, 2013; pp. 231–250. [Google Scholar]
- Kgaladi, J.; Wright, N.; Coertse, J.; Markotter, W.; Marston, D.; Fooks, A.R.; Freuling, C.M.; Müller, T.F.; Sabeta, C.T.; Nel, L.H. Diversity and epidemiology of Mokola virus. PLoS Negl. Trop. Dis. 2013, 7, e2511. [Google Scholar] [CrossRef] [PubMed]
- Menghani, S.; Chikhale, R.; Raval, A.; Wadibhasme, P.; Khedekar, P. Chandipura virus: An emerging tropical pathogen. Acta Trop. 2012, 124, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Sapkal, G.N.; Sawant, P.M.; Mourya, D.T. Chandipura viral encephalitis: A brief review. Open Virol. J. 2018, 12, 44–51. [Google Scholar] [CrossRef] [PubMed]
- Rao, B.L.; Basu, A.; Wairagkar, N.S.; Gore, M.M.; Arankalle, V.A.; Thakare, J.P.; Jadi, R.S.; Rao, K.A.; Mishra, A.C. A large outbreak of acute encephalitis with high fatality rate in children in Andhra Pradesh, India, in 2003, associated with Chandipura virus. Lancet 2004, 364, 869–874. [Google Scholar] [CrossRef]
- Dietzgen, R.G.; Kuzmin, I.V. Rhabdoviruses: Molecular Taxonomy, Evolution, Genomics, Ecology, Host-Vector Interactions, Cytopathology, and Control; Caister Academic Press: Wymondham, UK, 2012; pp. 23–26. [Google Scholar]
- Quiroz, E.; Moreno, N.; Peralta, P.H.; Tesh, R.B. A human case of encephalitis associated with vesicular stomatitis virus (Indiana serotype) infection. Am. J. Trop. Med. Hyg. 1988, 39, 312–314. [Google Scholar] [CrossRef]
- Brody, J.A.; Fischer, G.F.; Peralta, P.H. Vesicular stomatitis virus in Panama. Human serologic patterns in a cattle raising area. Am. J. Epidemiol. 1967, 86, 158–161. [Google Scholar] [CrossRef]
- Shelokov, A.; Peralta, P.H. Vesicular stomatitis virus, Indiana type: An arbovirus infection of tropical sandflies and humans? Am. J. Epidemiol. 1967, 86, 149–157. [Google Scholar] [CrossRef]
- Tesh, R.B.; Peralta, P.H.; Johnson, K.M. Ecologic studies of vesicular stomatitis virus. I. Prevalence of infection among animals and humans living in an area of endemic VSV activity. Am. J. Epidemiol. 1969, 90, 255–261. [Google Scholar] [CrossRef]
- Tesh, R.; Saidi, S.; Javadian, E.; Loh, P.; Nadim, A. Isfahan virus, a new Vesiculovirus infecting humans, gerbils, and sandflies in Iran. Am. J. Trop. Med. Hyg. 1977, 26, 299–306. [Google Scholar] [CrossRef] [PubMed]
- Saidi, S.; Tesh, R.; Javadian, E.; Sahabi, Z.; Nadim, A. Studies on the epidemiology of sandfly fever in Iran. II. The prevalence of human and animal infection with five phlebotomus fever virus serotypes in Isfahan province. Am. J. Trop. Med. Hyg. 1977, 26, 288–293. [Google Scholar] [CrossRef]
- Tesh, R.B.; Boshell, J.; Modi, G.B.; Morales, A.; Young, D.G.; Corredor, A.; Ferro de Carrasquilla, C.; de Rodriguez, C.; Walters, L.L.; Gaitan, M.O. Natural infection of humans, animals, and phlebotomine sand flies with the Alagoas serotype of vesicular stomatitis virus in Colombia. Am. J. Trop. Med. Hyg. 1987, 36, 653–661. [Google Scholar] [CrossRef]
- Figueiredo, L.T.M.; da Rosa, A.P.A.T.; Fiorillo, A.M. Prevalência de anticorpos neutralizantes para o arbovirus Piry em individuos da região de Ribeirão Preto, Estado de São Paulo. Rev. Inst. Med. Trop. São Paulo 1985, 27, 157–161. [Google Scholar] [CrossRef] [PubMed]
- Pinheiro, F.P.; Bensabath, G.; Andrade, A.H.P.; Lins, Z.C.; Fraiha, H.; Tang, A.T.; Lainson, R.; Shaw, J.J.; Azevedo, M.C. Infectious diseases along Brazil’s trans-Amazon highway: Surveillance and research. Bull. Pan Am. Health Organ. 1974, 8, 111–122. [Google Scholar] [PubMed]
- Centers for Disease Control and Prevention. Arbovirus Catalog. 1985. Available online: https://wwwn.cdc.gov/Arbocat/Default.aspx (accessed on 21 February 2020).
- Berge, T.O. International Catalogue of Arboviruses including certain other Viruses of Vertebrates; US Department of Health, Education and Welfare Publication No. (CDC) 75-8301; US Department of Health, Education and Welfare: Washington, DC, USA, 1975.
- Woodruff, A.W.; Ansdell, V.E.; Bowen, E.T.W. Le Dantec virus infection in a patient who had not been to West Africa. Br. Med. J. 1977, 2, 1632–1633. [Google Scholar] [CrossRef] [PubMed]
- Grard, G.; Fair, J.N.; Lee, D.; Slikas, E.; Steffen, I.; Muyembe, J.-J.; Sittler, T.; Veeraraghavan, N.; Ruby, J.G.; Wang, C.; et al. A novel rhabdovirus associated with acute hemorrhagic fever in Central Africa. PLoS Pathog. 2012, 8, e1002924. [Google Scholar] [CrossRef]
- Stremlau, M.H.; Andersen, K.G.; Folarin, O.A.; Grove, J.N.; Odia, I.; Ehiane, P.E.; Omoniwa, O.; Omoregie, O.; Jiang, P.-P.; Yozwiak, N.L.; et al. Discovery of novel rhabdoviruses in the blood of healthy individuals from West Africa. PLoS Negl. Trop. Dis. 2015, 9, e0003631. [Google Scholar] [CrossRef]
- Cybinski, D.H.; Gard, G.P. Isolation of a new rhabdovirus in Australia related to Tibrogargan virus. Aust. J. Biol. Sci. 1986, 39, 225–232. [Google Scholar] [CrossRef]
- Cybinski, D.H.; St. George, T.D.; Standfast, H.A.; McGregor, A. Isolation of Tibrogargan virus, a new Australian rhabdovirus, from Culicoides brevitaris. Vet. Microbiol. 1980, 5, 301–308. [Google Scholar] [CrossRef]
- Gibbs, E.P.; Calisher, C.H.; Tesh, R.B.; Lazuick, J.S.; Bowen, R.; Greiner, E.C. Bivens Arm virus: A new rhabdovirus isolated from Culicoides insignis in Florida and related to Tibrogargan virus of Australia. Vet. Microbiol. 1989, 19, 141–150. [Google Scholar] [CrossRef]
- Standfast, H.A.; Dyce, A.L.; St. George, T.D.; Muller, M.J.; Doherty, R.L.; Carley, J.G.; Filippich, C. Isolation of arboviruses from insects collected at Beatrice Hill, Northern Territory of Australia, 1974-1976. Aust. J. Biol. Sci. 1984, 37, 351–366. [Google Scholar] [CrossRef] [PubMed]
- Gubala, A.; Davis, S.; Weir, R.; Melville, L.; Cowled, C.; Boyle, D. Tibrogargan and Coastal Plains rhabdoviruses: Genomic characterization, evolution of novel genes and seroprevalence in Australian livestock. J. Gen. Virol. 2011, 92, 2160–2170. [Google Scholar] [CrossRef] [PubMed]
- Lauck, M.; Yú, S.; Caì, Y.; Hensley, L.E.; Chiu, C.Y.; O’Connor, D.H.; Kuhn, J.H. Genome sequence of Bivens Arm virus, a tibrovirus belonging to the species Tibrogargan virus (Mononegavirales: Rhabdoviridae). Genome Announc. 2015, 3, e00089-15. [Google Scholar] [CrossRef]
- Huang, B.; Allcock, R.; Warrilow, D. Newly characterized arboviruses of northern Australia. Virol. Rep. 2016, 6, 11–17. [Google Scholar] [CrossRef]
- Buchko, G.W.; Clifton, M.C.; Wallace, E.G.; Atkins, K.A.; Myler, P.J. Backbone chemical shift assignments and secondary structure analysis of the U1 protein from the Bas-Congo virus. Biomol. NMR Assign. 2017, 11, 51–56. [Google Scholar] [CrossRef] [PubMed]
- Calisher, C.H.; Borde, G.E.N.; Tuekam, T.; Gibbs, E.P.J. Antibody to Bivens Arm virus in Trinidadian buffalo: A temporal and evolutionary link between Australia, Asia and the Caribbean? In Bovine Ephemeral Fever and Related Rhabdoviruses. 1st International Symposium, Bovine Ephemeral Fever and Related rhabdovIruses. ACIAR Proceedings; ACIAR: Canberra, Australia, 1993; Volume 44, pp. 80–83. [Google Scholar]
- St George, T.D. Studies on the pathogenesis of bovine ephemeral fever in sentinel cattle. I. Virology and serology. Vet. Microbiol. 1985, 10, 493–504. [Google Scholar] [CrossRef]
- Gard, G.P.; Shorthose, J.E.; Weir, R.P.; Walsh, S.J.; Melville, L.F. Arboviruses recovered from sentinel livestock in northern Australia. Vet. Microbiol. 1988, 18, 109–118. [Google Scholar] [CrossRef]
- Tuekam, T.; Greiner, E.C.; Gibbs, E.P.J. Seroepidemiology of Bivens Arm virus infections of cattle in Florida, St Croix and Puerto Rico. Vet. Microbiol. 1991, 28, 121–127. [Google Scholar] [CrossRef]
- Chiu, C.; Fair, J.; Leroy, E.M. Bas-Congo virus: Another deadly virus? Future Microbiol. 2013, 8, 139–141. [Google Scholar] [CrossRef]
- Grard, G.; Fair, J.; Chiu, C.; Leroy, E. Bas-Congo virus: A novel rhabdovirus associated with acute hemorrhagic fever. In Emerging Infectious Diseases; Ergönül, Ö., Can, F., Madoff, L., Akova, M., Eds.; Academic Press: Amsterdam, The Netherlands, 2014; pp. 13–24. [Google Scholar]
- Chiu, C.Y. Viral pathogen discovery. Curr. Opin. Microbiol. 2013, 16, 468–478. [Google Scholar] [CrossRef] [PubMed]
- Branco, L.M.; Garry, R.F. Bas-Congo Virus—Not an Established Pathogen. 2018. Available online: https://science.sciencemag.org/content/362/6414/577/tab-e-letters (accessed on 21 February 2020).
- Vu, D.-L.; Cordey, S.; Simonetta, F.; Brito, F.; Docquier, M.; Turin, L.; van Delden, C.; Boely, E.; Dantin, C.; Pradier, A.; et al. Human pegivirus persistence in human blood virome after allogeneic haematopoietic stem-cell transplantation. Clin. Microbiol. Infect. 2019, 25, 225–232. [Google Scholar] [CrossRef] [PubMed]
- Moustafa, A.; Xie, C.; Kirkness, E.; Biggs, W.; Wong, E.; Turpaz, Y.; Bloom, K.; Delwart, E.; Nelson, K.E.; Venter, J.C.; et al. The blood DNA virome in 8,000 humans. PLoS Pathog. 2017, 13, e1006292. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Li, L.; Deng, X.; Blümel, J.; Nübling, C.M.; Hunfeld, A.; Baylis, S.A.; Delwart, E. Viral nucleic acids in human plasma pools. Transfusion 2016, 56, 2248–2255. [Google Scholar] [CrossRef]
- Ngoi, C.N.; Siqueira, J.; Li, L.; Deng, X.; Mugo, P.; Graham, S.M.; Price, M.A.; Sanders, E.J.; Delwart, E. The plasma virome of febrile adult Kenyans shows frequent parvovirus B19 infections and a novel arbovirus (Kadipiro virus). J. Gen. Virol. 2016, 97, 3359–3367. [Google Scholar] [CrossRef]
- Manso, C.F.; Bibby, D.F.; Mbisa, J.L. Efficient and unbiased metagenomic recovery of RNA virus genomes from human plasma samples. Sci. Rep. 2017, 7, 4173. [Google Scholar] [CrossRef]
- Bai, H.L.; Jin, B.H.; He, Y.; Leng, P.E.; Chen, R.; Zhu, Y.Y.; Zhang, C.Z.; Xu, F.; Pan, H.; Wu, H.Y. Research development of Ekpoma virus and its biological characterization. J. Microbes Infect. 2017, 12, 66–69. [Google Scholar]
- Kemp, G.E.; Lee, V.H.; Moore, D.L.; Shope, R.E.; Causey, O.R.; Murphy, F.A. Kotonkan, a new rhabdovirus related to Mokola virus of the rabies serogroup. Am. J. Epidemiol. 1973, 98, 43–49. [Google Scholar] [CrossRef]
- Babayan, S.A.; Orton, R.J.; Streicker, D.G. Predicting reservoir hosts and arthropod vectors from evolutionary signatures in RNA virus genomes. Science 2018, 362, 577–580. [Google Scholar] [CrossRef]
- Vasilakis, N.; Weaver, S.C. The history and evolution of human dengue emergence. Adv. Virus Res. 2008, 72, 1–76. [Google Scholar] [CrossRef]
- Wiley, M.R.; Prieto, K.; Blasdell, K.R.; Caì, Y.; Campos Lawson, C.; Walker, P.J.; Chiu, C.Y.; Palacios, G.; Kuhn, J.H. Beatrice Hill virus represents a novel species in the genus Tibrovirus (Mononegavirales: Rhabdoviridae). Genome Announc. 2017, 5, e01485-16. [Google Scholar] [CrossRef]
- Walker, P.J.; Dietzgen, R.G.; Joubert, D.A.; Blasdell, K.R. Rhabdovirus accessory genes. Virus Res. 2011, 162, 110–125. [Google Scholar] [CrossRef] [PubMed]
- Devaux, P.; Songsungthong, W.; Springfeld, C.; von Messling, V.; Cattaneo, R. Measles virus host defense evasion proteins V and C have also transcription modulation and infectivity factor functions. Mol. Ther. 2005, 11, S43. [Google Scholar] [CrossRef]
- Nasar, F.; Matassov, D.; Seymour, R.L.; Latham, T.; Gorchakov, R.V.; Nowak, R.M.; Leal, G.; Hamm, S.; Eldridge, J.H.; Tesh, R.B.; et al. Recombinant Isfahan virus and vesicular stomatitis virus vaccine vectors provide durable, multivalent, single-dose protection against lethal alphavirus challenge. J. Virol. 2017, 91, e01729-16. [Google Scholar] [CrossRef]
- Rieder, M.; Conzelmann, K.-K. Interferon in rabies virus infection. Adv. Virus Res. 2011, 79, 91–114. [Google Scholar] [CrossRef] [PubMed]
- Brown, J.C.; Newcomb, W.W.; Wertz, G.W. Helical virus structure: The case of the rhabdovirus bullet. Viruses 2010, 2, 995–1001. [Google Scholar] [CrossRef] [PubMed]
- Ge, P.; Tsao, J.; Schein, S.; Green, T.J.; Luo, M.; Zhou, Z.H. Cryo-EM model of the bullet-shaped vesicular stomatitis virus. Science 2010, 327, 689–693. [Google Scholar] [CrossRef]
- Jackson, A.O.; Dietzgen, R.G.; Goodin, M.M.; Bragg, J.N.; Deng, M. Biology of plant rhabdoviruses. Annu Rev. Phytopathol. 2005, 43, 623–660. [Google Scholar] [CrossRef]
- Steffen, I.; Liss, N.M.; Schneider, B.S.; Fair, J.N.; Chiu, C.Y.; Simmons, G. Characterization of the Bas-Congo virus glycoprotein and its function in pseudotyped viruses. J. Virol. 2013, 87, 9558–9568. [Google Scholar] [CrossRef]
- Albertini, A.A.; Baquero, E.; Ferlin, A.; Gaudin, Y. Molecular and cellular aspects of rhabdovirus entry. Viruses 2012, 4, 117–139. [Google Scholar] [CrossRef]
- Roche, S.; Bressanelli, S.; Rey, F.A.; Gaudin, Y. Crystal structure of the low-pH form of the vesicular stomatitis virus glycoprotein G. Science 2006, 313, 187–191. [Google Scholar] [CrossRef] [PubMed]
- Roche, S.; Rey, F.A.; Gaudin, Y.; Bressanelli, S. Structure of the prefusion form of the vesicular stomatitis virus glycoprotein G. Science 2007, 315, 843–848. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, Y.; Bowden, T.A.; Wilson, I.A.; Crispin, M. Exploitation of glycosylation in enveloped virus pathobiology. Biochim. Biophys. Acta Gen. Subj. 2019, 1863, 1480–1497. [Google Scholar] [CrossRef] [PubMed]
- Caì, Y.; Yú, S.; Jangra, R.K.; Postnikova, E.N.; Wada, J.; Tesh, R.B.; Whelan, S.P.J.; Lauck, M.; Wiley, M.R.; Finch, C.L.; et al. Human, nonhuman primate, and bat cells are broadly susceptible to tibrovirus particle cell entry. Front. Microbiol. 2019, 10, 856. [Google Scholar] [CrossRef]
- Gramberg, T.; Soilleux, E.; Fisch, T.; Lalor, P.F.; Hofmann, H.; Wheeldon, S.; Cotterill, A.; Wegele, A.; Winkler, T.; Adams, D.H.; et al. Interactions of LSECtin and DC-SIGN/DC-SIGNR with viral ligands: Differential pH dependence, internalization and virion binding. Virology 2008, 373, 189–201. [Google Scholar] [CrossRef]
- Gillies, S.; Stollar, V. Generation of defective interfering particles of vesicular stomatitis virus in Aedes albopictus cells. Virology 1980, 107, 497–508. [Google Scholar] [CrossRef]
- Mudd, J.A.; Leavitt, R.W.; Kingsbury, D.T.; Holland, J.J. Natural selection of mutants of vesicular stomatitis virus by cultured cells of Drosophila melanogaster. J. Gen. Virol. 1973, 20, 341–351. [Google Scholar] [CrossRef]
- Seganti, L.; Superti, F.; Girmenia, C.; Melucci, L.; Orsi, N. Study of receptors for vesicular stomatitis virus in vertebrate and invertebrate cells. Microbiologica 1986, 9, 259–267. [Google Scholar]
- Lyles, D.S. Cytopathogenesis of rhabdoviruses. In Biology and Pathogenesis of Rhabdo- and Filoviruses; Pattnaik, A.K., Ed.; World Scientific: Singapore, 2015; pp. 141–169. [Google Scholar]
- Brun, J.; McManus, D.; Lefebvre, C.; Hu, K.; Falls, T.; Atkins, H.; Bell, J.C.; McCart, J.A.; Mahoney, D.; Stojdl, D.F. Identification of genetically modified Maraba virus as an oncolytic rhabdovirus. Mol. Ther. 2010, 18, 1440–1449. [Google Scholar] [CrossRef]
- Beckham, J.D.; Tyler, K.L. Arbovirus infections. Continuum (Minneap Minn) 2015, 21, 1599–1611. [Google Scholar] [CrossRef]
- Burne, J.C. Viræmia in rabies. Lancet 1970, 295/i, 195–196. [Google Scholar] [CrossRef]
- Constantine, D.G. Absence of prenatal infection of bats with rabies virus. J. Wildl. Dis. 1986, 22, 249–250. [Google Scholar] [CrossRef] [PubMed]
- Sitprija, V.; Sriaroon, C.; Lumlertdaecha, B.; Wacharapluesadee, S.; Phumesin, P.; Khawplod, P.; Wilde, H.; Hemachudha, T. Does contact with urine and blood from a rabid dog represent a rabies risk? Clin. Infect. Dis. 2003, 37, 1399–1400. [Google Scholar] [CrossRef] [PubMed]
- Johnson, J.E.; Nasar, F.; Coleman, J.W.; Price, R.E.; Javadian, A.; Draper, K.; Lee, M.; Reilly, P.A.; Clarke, D.K.; Hendry, R.M.; et al. Neurovirulence properties of recombinant vesicular stomatitis virus vectors in non-human primates. Virology 2007, 360, 36–49. [Google Scholar] [CrossRef] [PubMed]
- Reis, J.L., Jr. Pathogenesis of Acute Vesicular Stomatitis Infection in Experimentally Infected Cattle. Ph.D. Thesis, University of Georgia, Athens, GA, USA, 2010. [Google Scholar]
- Lin, W.-H.W.; Kouyos, R.D.; Adams, R.J.; Grenfell, B.T.; Griffin, D.E. Prolonged persistence of measles virus RNA is characteristic of primary infection dynamics. Proc. Natl. Acad. Sci. USA 2012, 109, 14989–14994. [Google Scholar] [CrossRef] [PubMed]
- Saxena, V.; Xie, G.; Li, B.; Farris, T.; Welte, T.; Gong, B.; Boor, P.; Wu, P.; Tang, S.-J.; Tesh, R.; et al. A hamster-derived West Nile virus isolate induces persistent renal infection in mice. PLoS Negl. Trop. Dis. 2013, 7, e2275. [Google Scholar] [CrossRef] [PubMed]
- Simon, I.D.; van Rooijen, N.; Rose, J.K. Vesicular stomatitis virus genomic RNA persists in vivo in the absence of viral replication. J. Virol. 2010, 84, 3280–3286. [Google Scholar] [CrossRef]
| Virus | Country | Year | Clinical Status | Sequence (nt) | Genome Status | GenBank |
|---|---|---|---|---|---|---|
| BASV | Democratic Republic of the Congo | 2009 | Febrile | 11,892 | Near-complete | JX297815.1 |
| EKV-1 | Nigeria | 2013 | Healthy | 13,158 | Coding-complete | KP324827.1 |
| EKV-2 | Nigeria | 2013 | Healthy | 12,707 | Coding-complete | KP324828.1 |
| EKV-2 | Democratic Republic of the Congo | 2014 | Healthy | 1228 | Partial | KY766247.1–KY766250.1 |
| EKV-2 | Angola | 2016 | Febrile | 12,638 | Coding-complete | MF079256.1 |
| Virus Pair | N | P | M | U1 | U2 | G | U3 | L |
|---|---|---|---|---|---|---|---|---|
| BASV × EKV-1 | 41 | 19 | 23 | 13 | 15 | 30 | 13 | 43 |
| BASV × EKV-2 | 46 | 30 | 28 | 20 | 17 | 34 | 13 | 49 |
| EKV1 × EKV-2 | 41 | 17 | 22 | 18 | 18 | 26 | 20 | 43 |
| EKV-2 (Nigeria) × EKV-2 (Angola) | 99 | 100 | 98 | 99 | 99 | 98 | 90 | 98 |
| Virus | Leader (nt) | N | P | M | U1 | U2 | G | U3 | L | Trailer (nt) |
|---|---|---|---|---|---|---|---|---|---|---|
| EKV-1 | 71 | 429 | 332 | 230 | 176 | 176 | 685 | 92 | 2166 | 80 |
| EKV-2 | 69 | 427 | 217 | 229 | 175 | 168 | 630 | 125 | 2120 | 60 |
| BASV | ND | 429 | 215 | 218 | 216 | 171 | 629 | 94 | 2123 | ND |
| TIBV | 72 | 428 | 308 | 253 | 170 | 149 | 662 | 109 | 2119 | 170 |
| CPV | 67 | 425 | 274 | 244 | 173 | 161 | 658 | 103 | 2128 | 250 |
| BHV | 72 | 428 | 312 | 253 | 170 | 159 | 659 | 117 | 2116 | 168 |
| SWBV | ND | 428 | 292 | 248 | 170 | 162 | 662 | 105 | 2120 | ND |
| BAV | ND | 428 | 309 | 253 | 170 | 159 | 662 | 109 | 2119 | ND |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kuhn, J.H.; Pān, H.; Chiu, C.Y.; Stremlau, M. Human Tibroviruses: Commensals or Lethal Pathogens? Viruses 2020, 12, 252. https://doi.org/10.3390/v12030252
Kuhn JH, Pān H, Chiu CY, Stremlau M. Human Tibroviruses: Commensals or Lethal Pathogens? Viruses. 2020; 12(3):252. https://doi.org/10.3390/v12030252
Chicago/Turabian StyleKuhn, Jens H., Hào Pān, Charles Y. Chiu, and Matthew Stremlau. 2020. "Human Tibroviruses: Commensals or Lethal Pathogens?" Viruses 12, no. 3: 252. https://doi.org/10.3390/v12030252
APA StyleKuhn, J. H., Pān, H., Chiu, C. Y., & Stremlau, M. (2020). Human Tibroviruses: Commensals or Lethal Pathogens? Viruses, 12(3), 252. https://doi.org/10.3390/v12030252






