Selective Reactivity of Anti-Japanese Encephalitis Virus NS4B Antibody Towards Different Flaviviruses
Abstract
1. Introduction
2. Materials and Methods
2.1. Cells and Virus
2.2. Plasmid Constructs
2.3. Transient Transfections and Protein Expression
2.4. Antibodies
2.5. Indirect Immunofluorescence Test
2.6. Cell Lysis
2.7. Western Blotting Assay
2.8. Hydrophobicity and Protein Sequence Alignment Analyses
3. Results
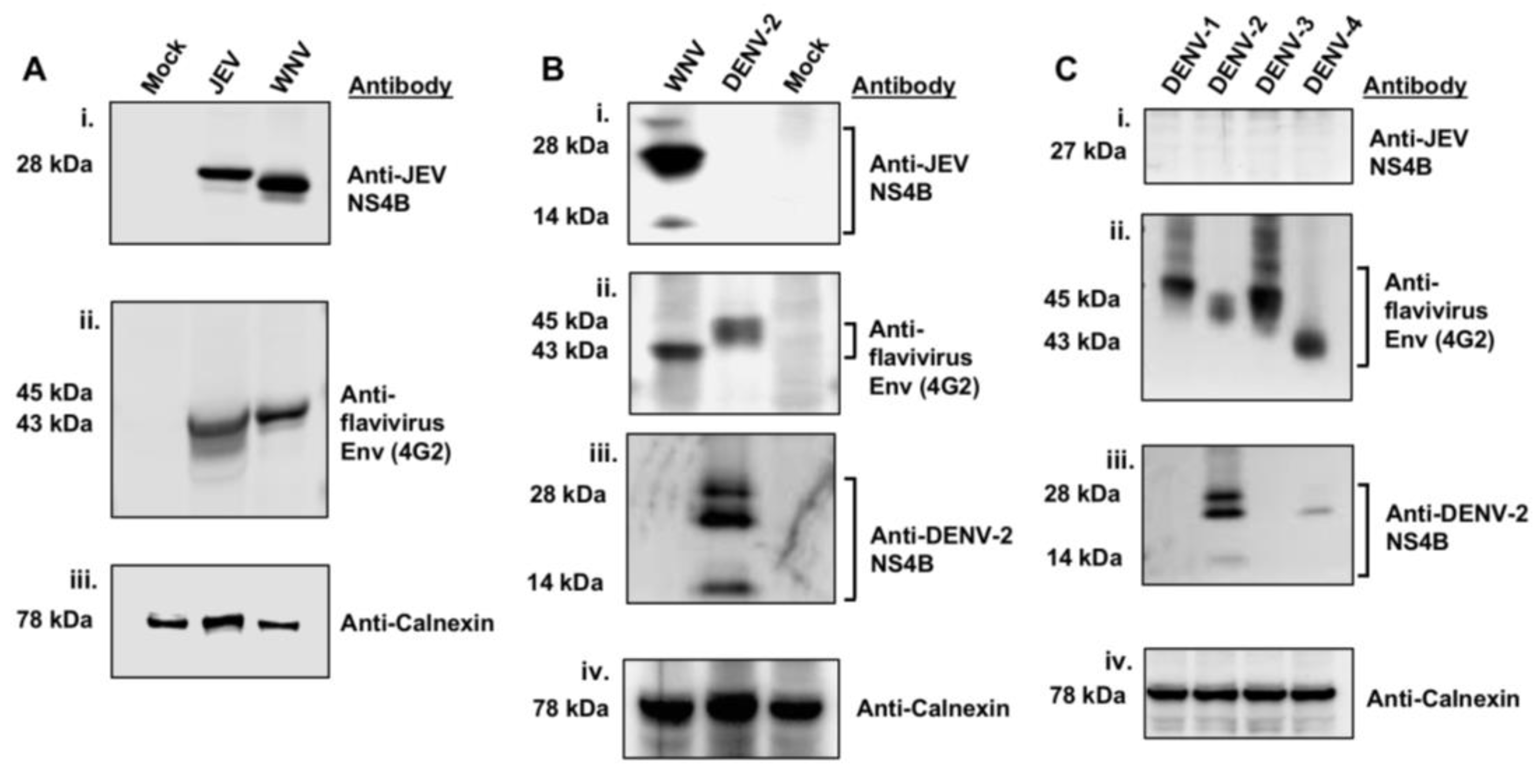
3.1. JEV NS4B Antibody Detects NS4B Protein of WNVNY99 but Not of the Four DENV Serotypes
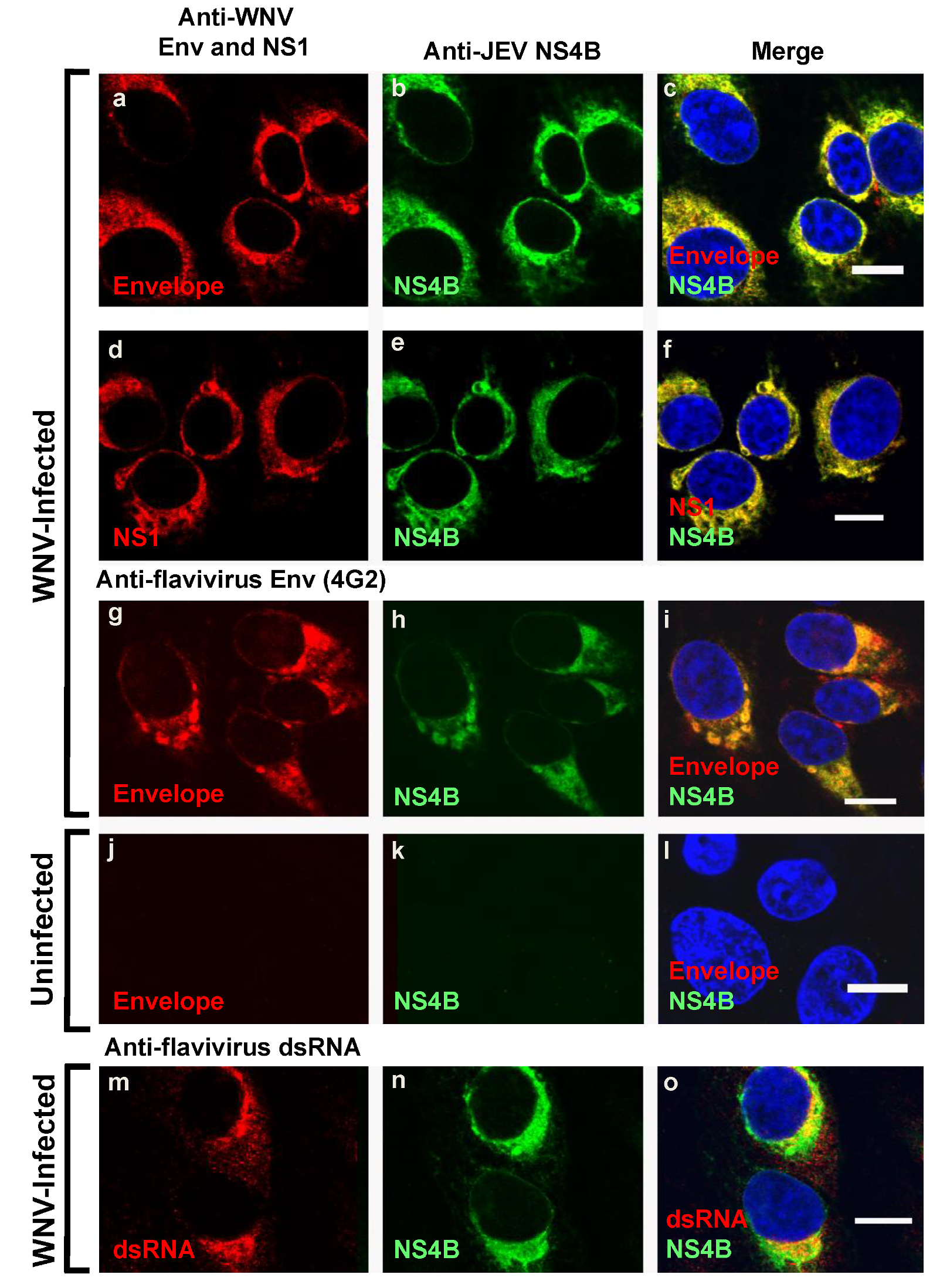
3.2. Assessment of the Distribution and Localization of Intracellular WNVNY99 NS4B Protein to Other Viral Replication Components
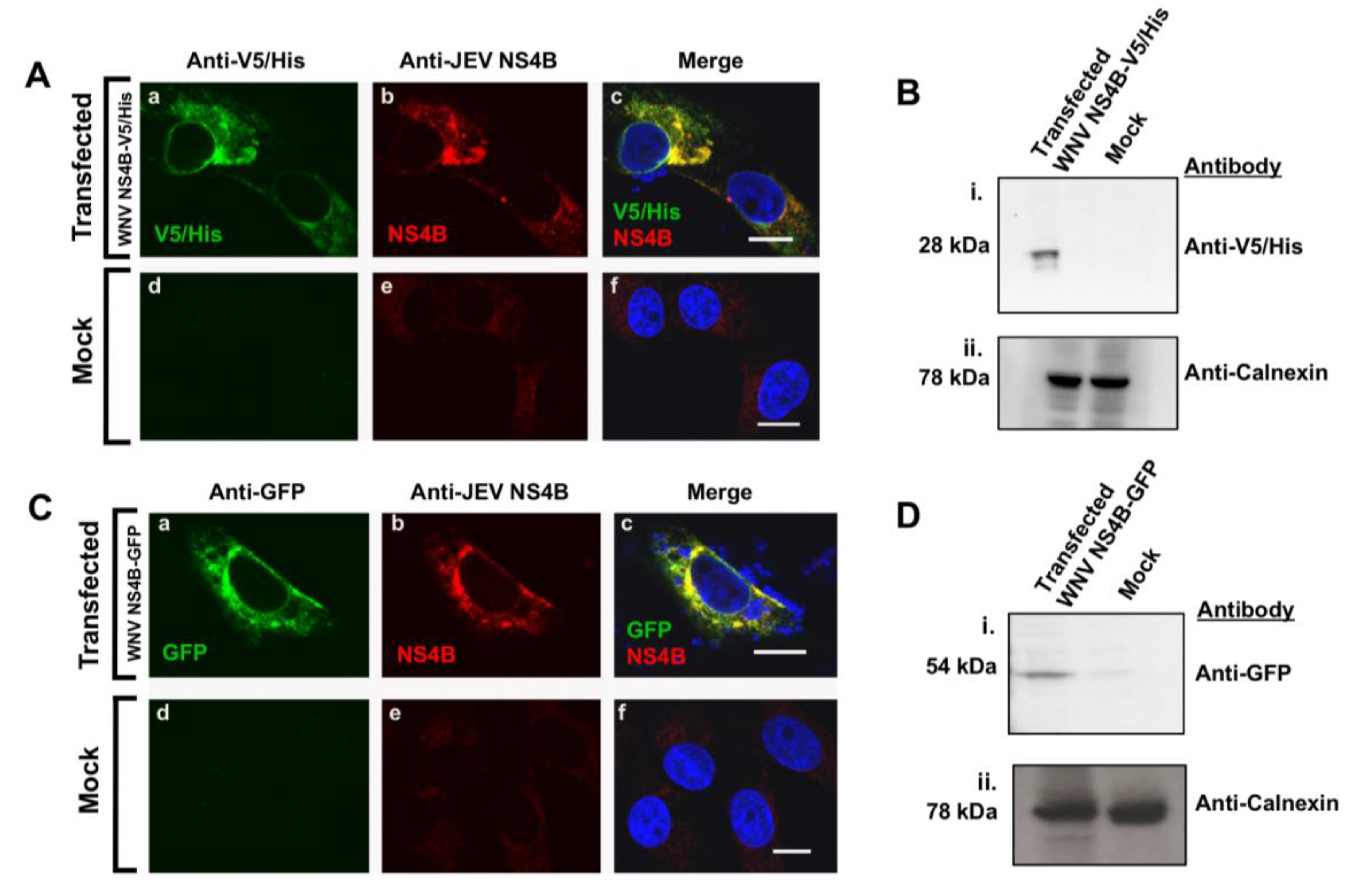
3.3. Detection of NS4B Protein Harbored in the NS4B C-Terminal Tagged Plasmids with JEV NS4B Antibody
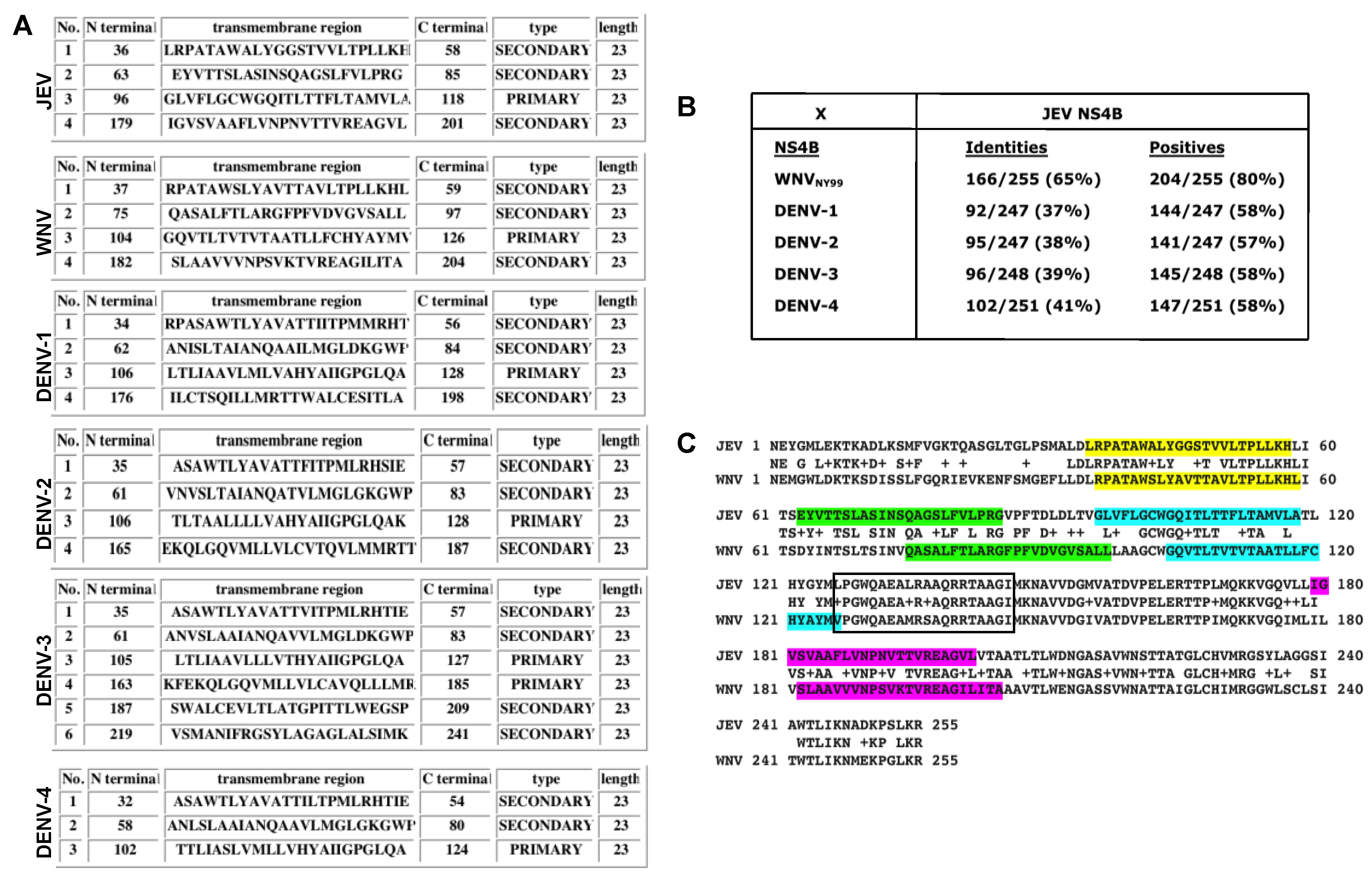
3.4. In Silico Analyses of Genus Flavivirus NS4B Protein
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Stiasny, K.; Kiermayr, S.; Holzmann, H.; Heinz, F.X. Cryptic properties of a cluster of dominant flavivirus cross-reactive antigenic sites. J. Virol. 2006, 80, 9557–9568. [Google Scholar] [CrossRef]
- Gould, E.A. Evolution of the Japanese encephalitis serocomplex viruses. Curr. Top. Microbiol. Immunol. 2002, 267, 391–404. [Google Scholar] [CrossRef]
- Kuno, G. Serodiagnosis of flaviviral infections and vaccinations in humans. Adv. Virus Res. 2003, 61, 3–65. [Google Scholar] [CrossRef] [PubMed]
- Hua, R.H.; Liu, L.K.; Huo, H.; Li, Y.N.; Guo, L.P.; Wang, X.L.; Qin, C.F.; Bu, Z.G. Comprehensive mapping of a novel NS1 epitope conserved in flaviviruses within the Japanese encephalitis virus serocomplex. Virus Res. 2014, 185, 103–109. [Google Scholar] [CrossRef] [PubMed]
- Konishi, E.; Konishi, M. Nonstructural protein 1 antibody-based epitope-blocking enzyme-linked immunosorbent assay to differentiate Japanese encephalitis virus from dengue virus infections in humans. Jpn. J. Infect. Dis. 2011, 64, 284–291. [Google Scholar] [PubMed]
- Hua, R.H.; Chen, N.S.; Qin, C.F.; Deng, Y.Q.; Ge, J.Y.; Wang, X.J.; Qiao, Z.J.; Chen, W.Y.; Wen, Z.Y.; Liu, W.X.; et al. Identification and characterization of a virus-specific continuous B-cell epitope on the PrM/M protein of Japanese Encephalitis Virus: Potential application in the detection of antibodies to distinguish Japanese Encephalitis Virus infection from West Nile Virus and Dengue Virus infections. Virol. J. 2010, 7, 249. [Google Scholar] [CrossRef]
- Brinton, M.A. The molecular biology of West Nile Virus: A new invader of the western hemisphere. Annu. Rev. Microbiol. 2002, 56, 371–402. [Google Scholar] [CrossRef]
- Miller, S.; Sparacio, S.; Bartenschlager, R. Subcellular localization and membrane topology of the Dengue virus type 2 Non-structural protein 4B. J. Biol. Chem. 2006, 281, 8854–8863. [Google Scholar] [CrossRef]
- Kaufusi, P.H.; Kelley, J.F.; Yanagihara, R.; Nerurkar, V.R. Induction of endoplasmic reticulum-derived replication-competent membrane structures by West Nile virus non-structural protein 4B. PLoS ONE 2014, 9, e84040. [Google Scholar] [CrossRef]
- Liu, W.J.; Wang, X.J.; Mokhonov, V.V.; Shi, P.Y.; Randall, R.; Khromykh, A.A. Inhibition of interferon signaling by the New York 99 strain and Kunjin subtype of West Nile virus involves blockage of STAT1 and STAT2 activation by nonstructural proteins. J. Virol. 2005, 79, 1934–1942. [Google Scholar] [CrossRef]
- Munoz-Jordan, J.L.; Laurent-Rolle, M.; Ashour, J.; Martinez-Sobrido, L.; Ashok, M.; Lipkin, W.I.; Garcia-Sastre, A. Inhibition of alpha/beta interferon signaling by the NS4B protein of flaviviruses. J. Virol. 2005, 79, 8004–8013. [Google Scholar] [CrossRef]
- Kelley, J.F.; Kaufusi, P.H.; Volper, E.M.; Nerurkar, V.R. Maturation of dengue virus nonstructural protein 4B in monocytes enhances production of dengue hemorrhagic fever-associated chemokines and cytokines. Virology 2011, 418, 27–39. [Google Scholar] [CrossRef]
- Blaney, J.E., Jr.; Manipon, G.G.; Firestone, C.Y.; Johnson, D.H.; Hanson, C.T.; Murphy, B.R.; Whitehead, S.S. Mutations which enhance the replication of dengue virus type 4 and an antigenic chimeric dengue virus type 2/4 vaccine candidate in Vero cells. Vaccine 2003, 21, 4317–4327. [Google Scholar] [CrossRef]
- Ni, H.; Chang, G.J.; Xie, H.; Trent, D.W.; Barrett, A.D. Molecular basis of attenuation of neurovirulence of wild-type Japanese encephalitis virus strain SA14. J. Gen. Virol. 1995, 76 Pt 2, 409–413. [Google Scholar] [CrossRef]
- Pletnev, A.G.; Putnak, R.; Speicher, J.; Wagar, E.J.; Vaughn, D.W. West Nile virus/dengue type 4 virus chimeras that are reduced in neurovirulence and peripheral virulence without loss of immunogenicity or protective efficacy. Proc. Natl. Acad. Sci. USA 2002, 99, 3036–3041. [Google Scholar] [CrossRef]
- Wang, E.; Ryman, K.D.; Jennings, A.D.; Wood, D.J.; Taffs, F.; Minor, P.D.; Sanders, P.G.; Barrett, A.D. Comparison of the genomes of the wild-type French viscerotropic strain of yellow fever virus with its vaccine derivative French neurotropic vaccine. J. Gen. Virol. 1995, 76 Pt 11, 2749–2755. [Google Scholar] [CrossRef]
- Wicker, J.A.; Whiteman, M.C.; Beasley, D.W.; Davis, C.T.; Zhang, S.; Schneider, B.S.; Higgs, S.; Kinney, R.M.; Barrett, A.D. A single amino acid substitution in the central portion of the West Nile virus NS4B protein confers a highly attenuated phenotype in mice. Virology 2006, 349, 245–253. [Google Scholar] [CrossRef]
- Evans, J.D.; Seeger, C. Differential effects of mutations in NS4B on West Nile virus replication and inhibition of interferon signaling. J. Virol. 2007, 81, 11809–11816. [Google Scholar] [CrossRef]
- Sambrook, J.; Russell, D.W. Molecular Cloning: A Laboratory Manual; Cold Spring Harbor Laboratory Press: New York, NY, USA, 2001. [Google Scholar]
- Lu, C.Y.; Hour, M.J.; Wang, C.Y.; Huang, S.H.; Mu, W.X.; Chang, Y.C.; Lin, C.W. Single-Round Infectious Particle Antiviral Screening Assays for the Japanese Encephalitis Virus. Viruses 2017, 9, 76. [Google Scholar] [CrossRef]
- Wicker, J.A.; Whiteman, M.C.; Beasley, D.W.; Davis, C.T.; McGee, C.E.; Lee, J.C.; Higgs, S.; Kinney, R.M.; Huang, C.Y.; Barrett, A.D. Mutational analysis of the West Nile virus NS4B protein. Virology 2012, 426, 22–33. [Google Scholar] [CrossRef]
- Nybakken, G.E.; Nelson, C.A.; Chen, B.R.; Diamond, M.S.; Fremont, D.H. Crystal structure of the West Nile virus envelope glycoprotein. J. Virol. 2006, 80, 11467–11474. [Google Scholar] [CrossRef]
- Roosendaal, J.; Westaway, E.G.; Khromykh, A.; Mackenzie, J.M. Regulated cleavages at the West Nile virus NS4A-2K-NS4B junctions play a major role in rearranging cytoplasmic membranes and Golgi trafficking of the NS4A protein. J. Virol. 2006, 80, 4623–4632. [Google Scholar] [CrossRef]
- Westaway, E.G.; Khromykh, A.A.; Kenney, M.T.; Mackenzie, J.M.; Jones, M.K. Proteins C and NS4B of the flavivirus Kunjin translocate independently into the nucleus. Virology 1997, 234, 31–41. [Google Scholar] [CrossRef]
- Miller, S.; Kastner, S.; Krijnse-Locker, J.; Buhler, S.; Bartenschlager, R. The non-structural protein 4A of dengue virus is an integral membrane protein inducing membrane alterations in a 2K-regulated manner. J. Biol. Chem. 2007, 282, 8873–8882. [Google Scholar] [CrossRef]
- Mackenzie, J.M.; Khromykh, A.A.; Jones, M.K.; Westaway, E.G. Subcellular localization and some biochemical properties of the flavivirus Kunjin nonstructural proteins NS2A and NS4A. Virology 1998, 245, 203–215. [Google Scholar] [CrossRef]
- Westaway, E.G.; Mackenzie, J.M.; Kenney, M.T.; Jones, M.K.; Khromykh, A.A. Ultrastructure of Kunjin virus-infected cells: Colocalization of NS1 and NS3 with double-stranded RNA, and of NS2B with NS3, in virus-induced membrane structures. J. Virol. 1997, 71, 6650–6661. [Google Scholar] [CrossRef]
- Anwar, A.; Hosoya, T.; Leong, K.M.; Onogi, H.; Okuno, Y.; Hiramatsu, T.; Koyama, H.; Suzuki, M.; Hagiwara, M.; Garcia-Blanco, M.A. The kinase inhibitor SFV785 dislocates dengue virus envelope protein from the replication complex and blocks virus assembly. PLoS ONE 2011, 6, e23246. [Google Scholar] [CrossRef]
- Hirokawa, T.; Boon-Chieng, S.; Mitaku, S. SOSUI: Classification and secondary structure prediction system for membrane proteins. Bioinformatics 1998, 14, 378–379. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kaufusi, P.H.; Tseng, A.C.; Kelley, J.F.; Nerurkar, V.R. Selective Reactivity of Anti-Japanese Encephalitis Virus NS4B Antibody Towards Different Flaviviruses. Viruses 2020, 12, 212. https://doi.org/10.3390/v12020212
Kaufusi PH, Tseng AC, Kelley JF, Nerurkar VR. Selective Reactivity of Anti-Japanese Encephalitis Virus NS4B Antibody Towards Different Flaviviruses. Viruses. 2020; 12(2):212. https://doi.org/10.3390/v12020212
Chicago/Turabian StyleKaufusi, Pakieli H., Alanna C. Tseng, James F. Kelley, and Vivek R. Nerurkar. 2020. "Selective Reactivity of Anti-Japanese Encephalitis Virus NS4B Antibody Towards Different Flaviviruses" Viruses 12, no. 2: 212. https://doi.org/10.3390/v12020212
APA StyleKaufusi, P. H., Tseng, A. C., Kelley, J. F., & Nerurkar, V. R. (2020). Selective Reactivity of Anti-Japanese Encephalitis Virus NS4B Antibody Towards Different Flaviviruses. Viruses, 12(2), 212. https://doi.org/10.3390/v12020212




