Reconstruction and Characterization of Full-Length Begomovirus and Alphasatellite Genomes Infecting Pepper through Metagenomics
Abstract
1. Introduction
2. Materials and Methods
2.1. Sampling and RCA-Seq
2.2. NGS Data Pipeline
2.3. Cloning and Sanger Sequencing of the New Begomovirus Species Identified by RCA-NGS
2.4. Cloning and Sanger Sequencing of ToYSV and a New Associated Alphasatellite from a Weed Sample
2.5. Phylogenetic Analysis of Assembled Begomovirus Sequences
2.6. Phylogenetic Analysis of Alphasatellite Sequences
2.7. Recombination Analysis
3. Results
3.1. Reconstruction of Begomovirus and Alphasatellite Genomes Infecting Pepper
3.2. Phylogenetic Analyses
3.3. Recombination Analyses
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Van Regenmortel, M.H.V.; Fauquet, C.M.; Bishop, D.H.L. Virus Taxonomy. Seventh Report of the International Committee on Taxonomy of Viruses; Academic Press: Cambridge, MA, USA, 2000; p. 1162. [Google Scholar]
- Sánchez-Campos, S.; Martinez-Ayala, A.; Marquez-Martín, B.; Aragón-Caballero, L.; Navas-Castillo, J.; Moriones, E. Fulfilling Koch’s postulates confirms the monopartite nature of Tomato leaf deformation virus: A begomovirus native to the New World. Virus Res. 2013, 173, 286–293. [Google Scholar] [CrossRef]
- Macedo, M.A.; Albuquerque, L.C.; Maliano, M.R.; Souza, J.O.; Rojas, M.R.; Inoue-Nagata, A.K.; Gilbertson, R.L. Characterization of Tomato leaf curl purple vein virus, a new monopartite New World begomovirus infecting tomato in Northeast Brazil. Arch. Virol. 2018, 163, 737–743. [Google Scholar] [CrossRef] [PubMed]
- Organización de las Naciones Unidas para La Alimentación y la Agricultura (FAO). Available online: http://faostat3.fao.org/ (accessed on 7 August 2019).
- Brown, J.K.; Poulos, B.T.; Nelson, M.R. Two whitefly-transmitted geminiviruses isolated from pepper affected with Tigré disease. Phytopathology 1989, 79, 908. [Google Scholar]
- Brown, J.K.; Poulos, B.T. Serrano golden mosaic virus: A newly identified whitefly-transmitted geminivirus of pepper and tomato in the United States and Mexico. Plant Dis. 1990, 74, 720. [Google Scholar] [CrossRef]
- Torres-Pacheco, I.; GarzonTiznado, J.A.; Brown, J.K.; Becerra, A.; Rivera-Bustamante, R.F. Detection and distribution of geminiviruses in Mexico and the southern United States. Phytopathology 1996, 86, 1186–1192. [Google Scholar] [CrossRef]
- Brown, J.K.; Idris, A.M.; Ostrow, K.M.; Goldberg, N.; French, R.; Stenger, D.C. Genetic and phenotypic variation of the Pepper golden mosaic virus complex. Phytopathology 2005, 95, 1217–1224. [Google Scholar] [CrossRef]
- McLaughlin, P.D.; McLaughlin, W.A.; Maxwell, D.P.; Roye, M.E. Identification of begomoviruses infecting crops and weeds in Belize. Plant Viruses 2008, 2, 58–63. [Google Scholar]
- Barboza, N.; Blanco-Meneses, M.; Esker, P.; Moriones, E.; Inoue-Nagata, A.K. Distribution and diversity of begomoviruses in tomato and sweet pepper plants in Costa Rica. Ann. Appl. Biol. 2018, 172, 20–32. [Google Scholar] [CrossRef]
- Torres-Pacheco, I.; Garzón-Tiznado, J.A.; Herrera-Estrella, L.; Rivera-Bustamante, R.F. Complete nucleotide sequence of Pepper huasteco virus: Analysis and comparison with bipartite geminiviruses. J. Gen. Virol. 1993, 74, 2225–2231. [Google Scholar] [CrossRef]
- Méndez-Lozano, J.; Torres-Pacheco, I.; Fauquet, C.M.; Rivera-Bustamante, R.F. Interactions between geminiviruses in a naturally occurring mixture: Pepper huasteco virus and Pepper golden mosaic virus. Phytopathology 2003, 93, 270–277. [Google Scholar] [CrossRef]
- Umaharan, P.; Padidam, M.; Phelps, R.H.; Beachy, R.N.; Fauquet, C.M. Distribution and diversity of gemminiviruses in Trinidad Tobago. Phytopathology 1998, 88, 1262–1268. [Google Scholar] [CrossRef] [PubMed]
- Castro, R.M.; Moreira, L.; Rojas, M.R.; Gilbertson, R.L.; Hernández, E.; Mora, F.; Ramírez, P. Occurrence of Squash yellow mild mottle virus and Pepper golden mosaic virus in Potential New Hosts in Costa Rica. Plant Pathol. J. 2013, 29, 285–293. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Bezerra-Agasie, I.C.; Ferreira, G.B.; de Ávila, A.C.; Inoue-Nagata, A.K. First Report of Tomato severe rugose virus in Chili Pepper in Brazil. Plant Dis. 2006, 90, 114. [Google Scholar] [CrossRef] [PubMed]
- Nozaki, D.N.; Krause-Sakate, R.; Pavan, M.A. Begomovirus infectando a cultura de pimentão no Estado de São Paulo. Summa Phytopathology 2010, 36, 244–247. [Google Scholar] [CrossRef]
- Martinez-Ayala, A.; Sánchez-Campos, S.; Cáceres, F.; Aragón-Caballero, L.; Navas-Castillo, J.; Moriones, E. Characterisation and genetic diversity of Pepper leafroll virus, a new bipartite begomovirus infecting pepper, bean and tomato in Peru. Ann. Appl. Biol. 2013, 164, 67–72. [Google Scholar] [CrossRef]
- Rojas, M.R.; Gilbertson, R.L.; Maxwell, D.P. Use of degenerate primers in the polymerase chain reaction to detect whitefly transmitted geminiviruses. Plant Dis. 1993, 77, 340–347. [Google Scholar] [CrossRef]
- Edwards, R.A.; Rohwer, F. Viral metagenomics. Nat. Rev. Microbiol. 2005, 3, 504–510. [Google Scholar] [CrossRef]
- Roossinck, M.J.; Saha, P.; Wiley, G.B.; Quan, J.; White, J.D.; Lai, H.; Chavarría, F.; Shen, G.; Roe, B.A. Ecogenomics: Using massively parallel pyrosequencing to understand virus ecology. Mol. Ecol. 2010, 1, 81–88. [Google Scholar] [CrossRef]
- Hagen, C.; Frizzi, A.; Gabriels, S.; Huang, M.; Salati, R.; Gabor, B.; Huang, S. Accurate and sensitive diagnosis of geminiviruses through enrichment, high-throughput sequencing and automated sequence identification. Arch. Virol. 2012, 157, 907–915. [Google Scholar] [CrossRef]
- Dayaram, A.; Opong, A.; Jäschke, A.; Hadfield, J.; Baschiera, M.; Dobson, R.C.; Offei, S.K.; Shepherd, D.N.; Martin, D.P.; Varsani, A. Molecular characterisation of a novel cassava associated circular ssDNA virus. Virus Res. 2012, 166, 130–135. [Google Scholar] [CrossRef]
- Idris, A.; Al-Saleh, M.; Piatek, M.J.; Al-Shahwan, I.; Ali, S.; Brown, J.K. Viral metagenomics: Analysis of begomoviruses by illumina high-throughput sequencing. Viruses 2014, 6, 1219–1236. [Google Scholar] [CrossRef] [PubMed]
- Zubair, M.; Zaidi, S.S.; Shakir, S.; Farooq, M.; Amin, I.; Scheffler, J.A.; Scheffler, B.E.; Mansoor, S. Multiple begomoviruses found associated with cotton leaf curl disease in Pakistan in early 1990 are back in cultivated cotton. Sci. Rep. 2017, 7, 680. [Google Scholar] [CrossRef] [PubMed]
- Fontenele, R.S.; Abreu, R.A.; Lamas, N.S.; Alves-Freitas, D.M.T.; Vidal, A.H.; Poppiel, R.R.; Melo, F.L.; Lacorte, C.; Martin, D.P.; Campos, M.A.; et al. Passion fruit chlorotic mottle virus: Molecular characterization of a new divergent geminivirus in Brazil. Viruses 2018, 10, e169. [Google Scholar] [PubMed]
- Wyant, P.S.; Strohmeier, S.; Schäfer, B.; Krenz, B.; Pereira Assunção, I.; de Andrade Lima, G.S.; Jeske, H. Circular DNA genomics (circomics) exemplified for geminiviruses in bean crops and weeds of northeastern Brazil. Virology 2012, 427, 151–157. [Google Scholar] [CrossRef]
- Jeske, H.; Kober, S.; Schäfer, B.; Strohmeier, S. Circomics of Cuban geminiviruses reveals the first alpha-satellite DNA in the Caribbean. Virus Genes 2014, 49, 312–324. [Google Scholar] [CrossRef]
- Ng, T.F.; Duffy, S.; Polston, J.E.; Bixby, E.; Vallad, G.E.; Breitbart, M. Exploring the diversity of plant DNA viruses and their satellites using vector-enabled metagenomics on whiteflies. PLoS ONE 2011, 6, e19050. [Google Scholar] [CrossRef]
- Rosario, K.; Seah, Y.M.; Marr, V.; Varsani, A.; Kraberger, S.; Stainton, D.; Moriones, E.; Polston, J.E.; Duffy, S.; Breitbart, M. Vector-Enabled Metagenomic (VEM) Surveys Using Whiteflies (Aleyrodidae) Reveal Novel Begomovirus Species in the New and Old Worlds. Viruses 2015, 7, 5553–5570. [Google Scholar] [CrossRef]
- Rosario, K.; Marr, C.; Varsani, A.; Kraberger, S.; Stainton, D.; Moriones, E.; Polston, J.E.; Breitbart, M. Begomovirus-Associated Satellite DNA Diversity Captured Through Vector-Enabled Metagenomic (VEM) Surveys Using Whiteflies (Aleyrodidae). Viruses 2016, 8, 36. [Google Scholar] [CrossRef]
- Candresse, T.; Filloux, D.; Muhire, B.; Julian, C.; Galzi, S.; Fort, G.; Bernardo, P.; Daugrois, J.H.; Fernandez, E.; Martin, D.P.; et al. Appearances can be deceptive: Revealing a hidden viral infection with deep sequencing in a plant quarantine context. PLoS ONE 2014, 9, 102945. [Google Scholar] [CrossRef]
- Poojari, S.; Alabi, O.J.; Fofanov, V.Y.; Naidu, R.A. A leafhopper-transmissible DNA virus with novel evolutionary lineage in the family Geminiviridae implicated in grapevine redleaf disease by next-generation sequencing. PLoS ONE 2013, 8, e64194. [Google Scholar] [CrossRef]
- Jo, Y.; Choi, H.; Kim, S.M.; Kim, S.J.; Lee, B.C.; Cho, W.K. The pepper virome: Natural co-infection of diverse viruses and their quasispecies. BMC Genom. 2017, 18, 453. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Huang, Q.; Wu, L.; Qian, Y. Identification and characterization of a maize-associated mastrevirus in China by deep sequencing small RNA populations. Virol. J. 2015, 12, 156. [Google Scholar] [CrossRef] [PubMed]
- Boukari, W.; Alcalá-Briseño, R.I.; Kraberger, S.; Fernandez, E.; Filloux, D.; Daugrois, J.H.; Comstock, J.C.; Lett, J.M.; Martin, D.P.; Varsani, A.; et al. Occurrence of a novel mastrevirus in sugarcane germplasm collections in Florida, Guadeloupe and Réunion. Virol. J 2017, 14, 146. [Google Scholar] [CrossRef]
- Posada-Céspedes, S.; Seifert, D.; Beerenwinkel, N. Recent advances in inferring viral diversity from high-throughput sequencing data. Virus Res. 2017, 239, 17–32. [Google Scholar] [CrossRef]
- Nassuth, A.; Pollari, E.; Helmeczy, K.; Stewart, S.; Kofalvi, S.A. Improved RNA extraction and one-tube RT-PCR assay for simultaneous detection of control plant RNA plus several viruses in plant extracts. J. Virol. Methods 2000, 90, 37–49. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc (accessed on 5 November 2018).
- Hunt, M.; Gall, A.; Ong, S.H.; Brener, J.; Ferns, B.; Goulder, P.; Nastouli, E.; Keane, J.A.; Kellam, P.; Otto, T.D. IVA: Accurate de novo assembly of RNA virus genomes. Bioinformatics 2015, 31, 2374–2376. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Sonnhammer, E.L.; Durbin, R. A dot-matrix program with dynamic threshold control suited for genomic DNA and protein sequence analysis. Gene 1995, 167, GC1–GC10. [Google Scholar] [CrossRef]
- Rutherford, K.; Parkhill, J.; Crook, J.; Horsnell, T.; Rice, P.; Rajandream, M.A.; Barrell, B. Artemis: Sequence visualization and annotation. Bioinformatics 2000, 16, 944–945. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Muhire, B.M.; Varsani, A.; Martin, D.P. SDT: A virus classification tool based on pairwise sequence alignment and identity calculation. PLoS ONE 2014, 9, e108277. [Google Scholar] [CrossRef] [PubMed]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef]
- Stamatakis, A. “RAxML Version 8: A tool for Phylogenetic Analysis and Post-Analysis of Large Phylogenies”. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v4: Recent updates and new developments. Nucleic Acids Res. 2019, 47, W256–W259. [Google Scholar] [CrossRef]
- Darren, P.M.; Murrell, B.; Golden, M.; Khoosal, A.; Muhire, B. RDP4: Detection and analysis of recombination patterns in virus genomes. Virus Evol. 2015, 1, vev003. [Google Scholar]
- Huson, D.H.; Bryant, D. Application of phylogenetic networks in evolutionary studies. Mol. Biol. Evol. 2006, 23, 254–267. [Google Scholar] [CrossRef]
- Bruen, T.C.; Philippe, H.; Bryant, D. A simple and robust statistical test for detecting the presence of recombination. Genetics 2006, 172, 2665–2681. [Google Scholar] [CrossRef]
- Salemi, M.; Gray, R.R.; Goodenow, M.M. An exploratory algorithm to identify intra-host recombinant viral sequences. Mol. Phylogenet. Evol. 2008, 49, 618–628. [Google Scholar] [CrossRef][Green Version]
- Brown, J.K.; Zerbini, F.M.; Navas-Castillo, J.; Moriones, E.; Ramos-Sobrinho, R.; Silva, J.C.; Fiallo-Olivé, E.; Briddon, R.W.; Hernández-Zepeda, C.; Idris, A.; et al. Revision of Begomovirus taxonomy based on pairwise sequence comparisons. Arch. Virol. 2015, 160, 1593–1619. [Google Scholar] [CrossRef]
- Briddon, R.W.; Martin, D.P.; Roumagnac, P.; Navas-Castillo, J.; Fiallo-Olivé, E.; Moriones, E.; Lett, J.M.; Zerbini, F.M.; Varsani, A. Alphasatellitidae: A new family with two subfamilies for the classification of geminivirus- and nanovirus-associated alphasatellites. Arch. Virol. 2018, 163, 2587–2600. [Google Scholar] [CrossRef]
- Inoue-Nagata, A.K.; Lima, M.F.; Gilbertson, R.L. A review of geminivirus (begomovirus) diseases in vegetables and other crops in Brazil: Current status and approaches for management. Hort. Bras. 2016, 34, 008–018. [Google Scholar] [CrossRef]
- Rodríguez-Pardina, P.E.; Hanada, K.; Laguna, G.; Zerbini, F.M.; Ducasse, D.A. Molecular characterization and relative incidence of bean and soybean infecting begomoviruses in northwestern Argentina. Ann. Appl. Biol. 2010, 158, 69–78. [Google Scholar] [CrossRef]
- Belabess, Z.; Peterschmitt, M.; Granier, M.; Tahiri, A.; Blenzar, A.; Urbino, C. The non-canonical tomato yellow leaf curl virus recombinant that displaced its parental viruses in southern Morocco exhibits a high selective advantage in experimental conditions. J. Gen. Virol. 2016, 67, 3433–3445. [Google Scholar] [CrossRef] [PubMed]
- Celli, M.G.; Perotto, M.C.; Martino, J.A.; Flores, C.R.; Conci, V.C.; Rodriguez Pardina, P. Detection and identification of the first viruses in chia (Salvia hispanica). Viruses 2014, 6, 3450–3457. [Google Scholar] [CrossRef] [PubMed]
- Calegario, R.F.; Ferreira, S.S.; Andrade, E.C.; Zerbini, F.M. Characterization of Tomato yellow spot virus, a novel tomato-infecting begomovirus in Brazil. PAB 2007, 42, 1335–1343. [Google Scholar] [CrossRef]
- Barbosa, J.C.; Eckstein, B.; Bergamin Filho, A.J.; Rezende, A.M.; Dallagnol, L.J. First Report of Tomato yellow spot virus Infecting Leonurus sibiricus in Brazil. Plant Dis. 2013, 97, 289–289. [Google Scholar] [CrossRef]
- Fernandes-Acioli, N.A.N.; Boiteux, L.S.; Fonseca, M.E.N.; Segnana, L.R.G. Report of Tomato yellow spot virus Infecting Leonurus sibiricus in Paraguay and Within Tomato Fields in Brazil. Plant Dis. 2014, 98, 1445–1445. [Google Scholar] [CrossRef]
- Andrade, E.C. Tomato yellow spot virus, a tomato-infecting begomovirus from Brazil with a closer relationship to viruses from Sida sp., forms pseudorecombinants with begomoviruses from tomato but not from Sida. J. Gen. Virol. 2006, 87, 3687–3696. [Google Scholar] [CrossRef]
- Vaghi Medina, C.G.; Lopez Lambertini, P.M. Tomato dwarf leaf virus, a New World begomovirus infecting tomato in Argentina. Arch. Virol. 2012, 157, 1975–1980. [Google Scholar] [CrossRef]
- Colariccio, A.; Eiras, M.; Chaves, A.L.R.; Bergmann, J.C.; Zerbini, F.M.; Harakava, R.; Chagas, C.M. Tomato yellow vein streak virus, a new begomovirus on tomato from Brazil: Complete DNA-A sequence and some molecular and biological features. J. Plant Pathol. 2007, 89, 385–390. [Google Scholar]
- Albuquerque, L.C.; Martin, D.P.; Avila, A.C.; Inoue-Nagata, A.K. Characterization of tomato yellow vein streak virus, a begomovirus from Brazil. Virus Genes 2010, 40, 140–147. [Google Scholar] [CrossRef] [PubMed]
- Bernis, M.E. Caracterización molecular del Tomato Yellow Vein Streak Virus (ToYVSV), un nuevo Begomovirus en la Argentina. Facultad de Ciencias Químicas; Universidad Nacional de Córdoba: Cordoba, Argentina, 2007. [Google Scholar]
- Arruabarrena, A.; Rubio, L.; González-Arcos, M.; Maeso, D.; Fiallo-Olivé, E.; Moriones, E. First Report of the Begomovirus Tomato yellow vein streak virus Infecting Tomato in Uruguay. Plant Dis. 2016, 100, 231. [Google Scholar] [CrossRef]
- Paprotka, T.; Metzler, V.; Jeske, H. The first DNA 1-like alpha satellites in association with New World begomoviruses in natural infections. Virology 2010, 404, 148–157. [Google Scholar] [CrossRef]
- Varela, G.; Ávalos, V.; Reyna, P.; Laguna, G.; Rodriguez-Pardina, P. Identification, molecular characterization and relative incidence of begomoviruses infecting bean crops in northwestern Argentina: An update. Aust. Plant Pathol. 2018, 47, 343–350. [Google Scholar] [CrossRef]
- Simmonds, P.; Adams, M.J.; Benkő, M.; Breitbart, M.; Brister, J.R.; Carstens, E.B.; Davison, A.J.; Delwart, E.; Gorbalenya, A.E.; Harrach, B.; et al. Consensus statement: Virus taxonomy in the age of metagenomics. Nat. Rev. Microbiol. 2017, 15, 161–168. [Google Scholar] [CrossRef]
- Ferro, C.G.; Silva, J.P.; Xavier, C.A.D.; Godinho, M.T.; Lima, A.T.M.; Mar, T.B.; Lau, D.; Zerbini, F.M. The ever increasing diversity of begomoviruses infecting non-cultivated hosts: New species from Sida spp. and Leonurus sibiricus, plus two New World alphasatellites. Ann. Appl. Biol. 2017, 170, 204–218. [Google Scholar] [CrossRef]
- Instituto de Botánica Darwinion (IBOMA). Available online: http://www2.darwin.edu.ar/Proyectos/FloraArgentina/DetalleEspecie.asp?forma=&variedad=&subespecie=&especie=japonicus&genero=Leonurus&espcod=9285 (accessed on 10 September 2019).
- Mar, T.B.; Mendes, I.R.; Lau, D.; Fiallo-Olivé, E.; Navas-Castillo, J.; Alves, M.S.; Murilo Zerbini, F. Interaction between the New World begomovirus Euphorbia yellow mosaic virus and its associated alphasatellite: Effects on infection and transmission by the whitefly Bemisia tabaci. J. Gen. Virol. 2017, 98, 1552–1562. [Google Scholar] [CrossRef]
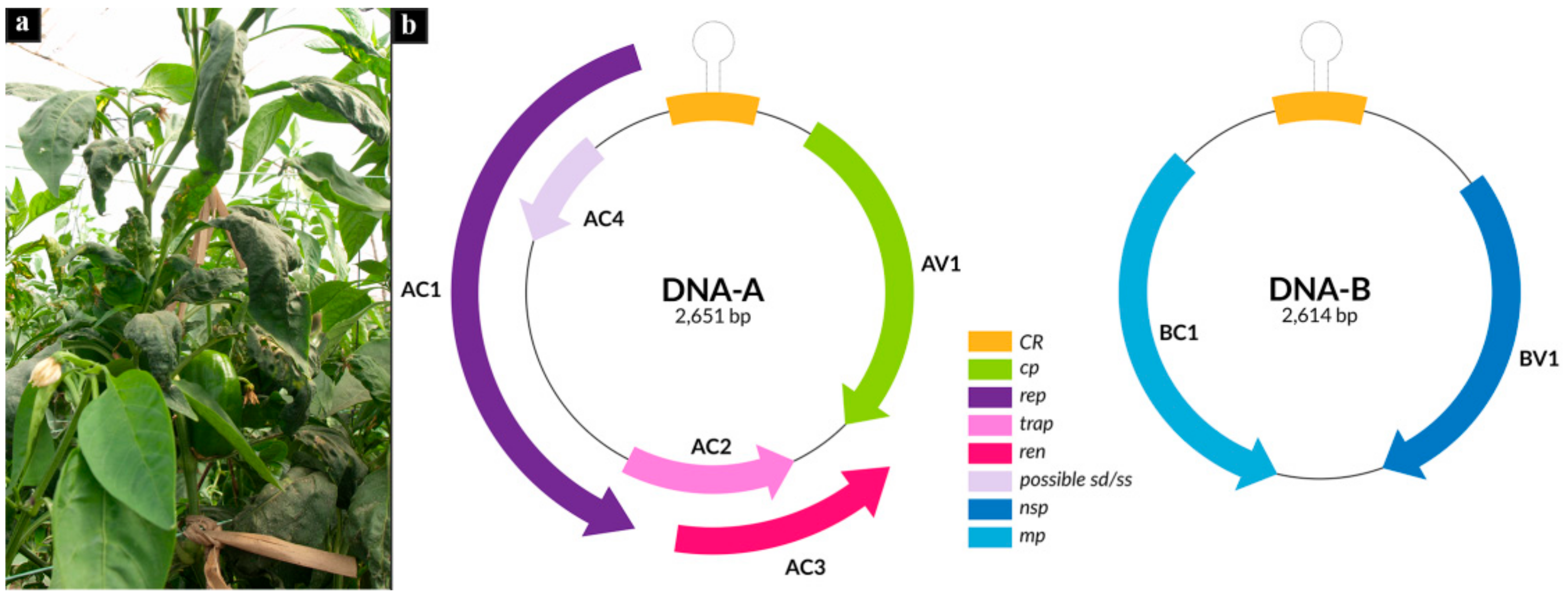
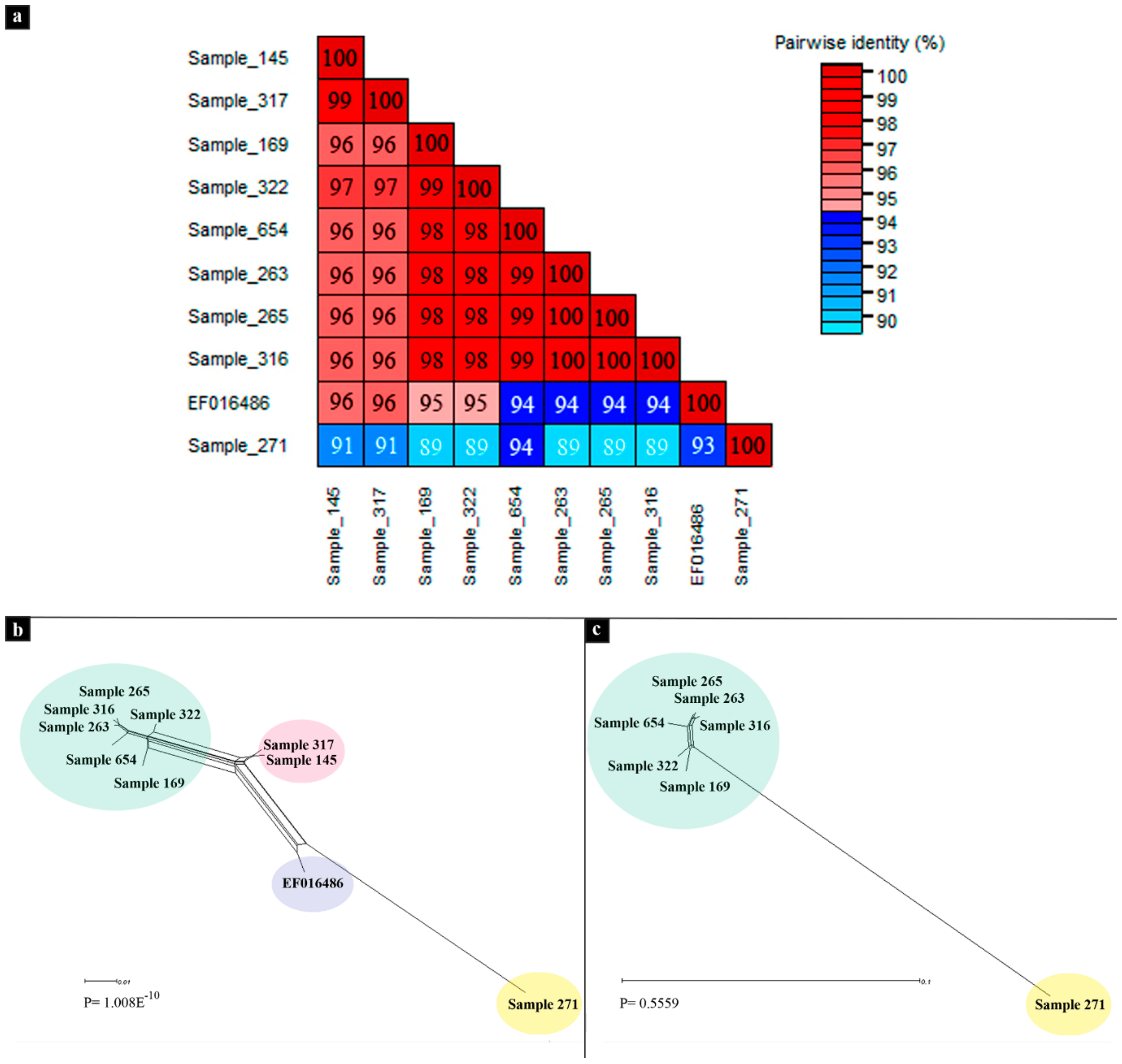
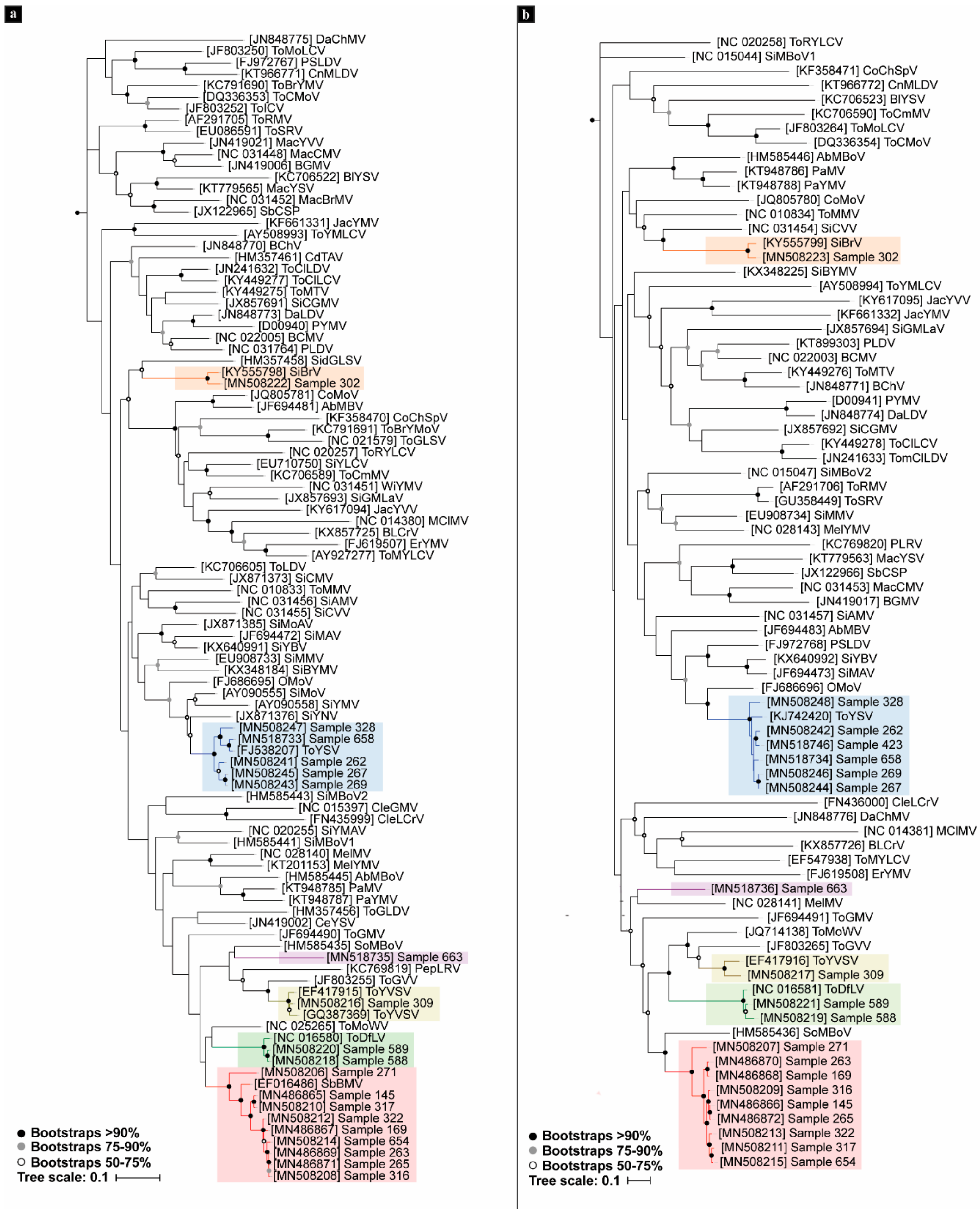
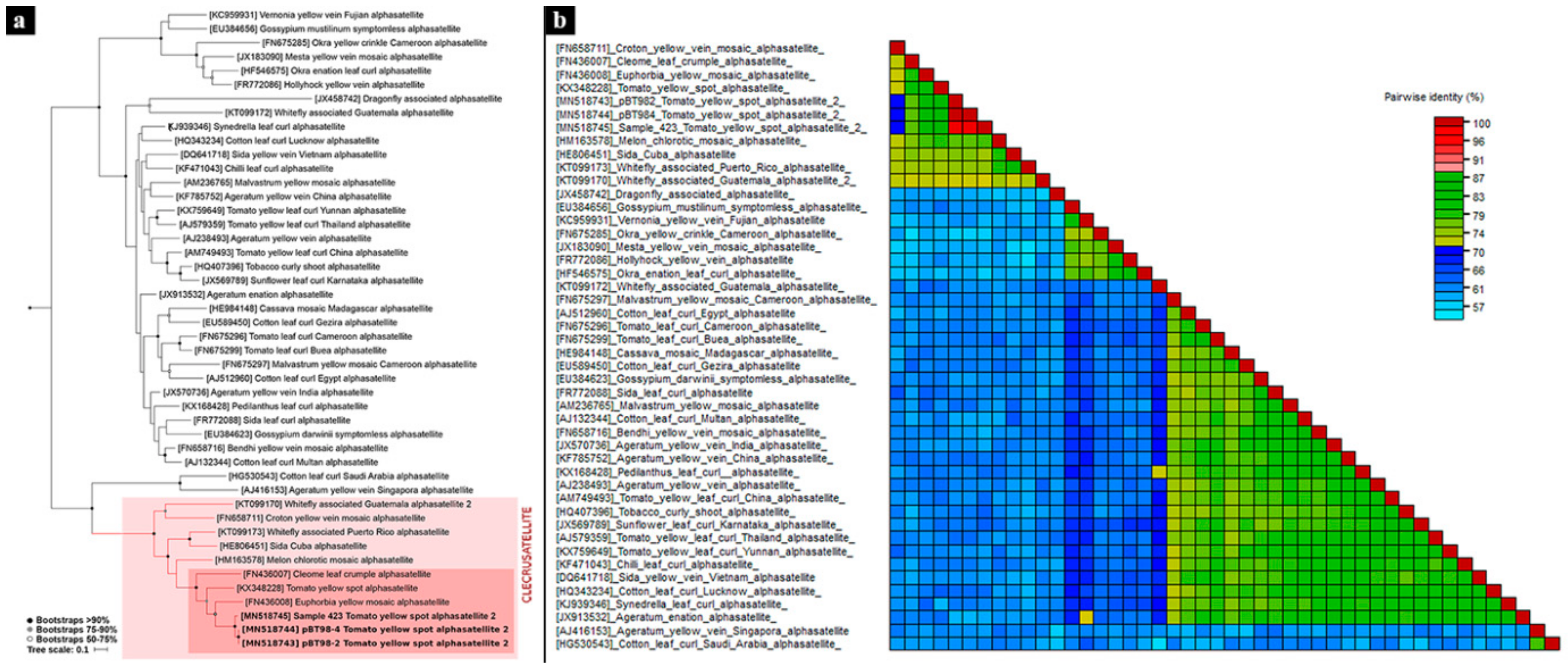
| Sample | Isolate | Raw Paired Reads | Reads Post Trimming | % C. annum | Viral Contigs |
|---|---|---|---|---|---|
| 145 | AR:Salta:Pichanal:Pepper145:2005 | 4,612,436 | 2,469,680 | 46.64 | 2 |
| 169 | AR:Salta:Pichanal:Pepper169:2006 | 10,002,748 | 4,604,741 | 66.78 | 2 |
| 262 | AR:Salta:Pichanal:Pepper262:2007 | 8,874,712 | 4,060,381 | 76.03 | 2 |
| 263 | AR:Salta:Pichanal:Pepper263:2007 | 10,935,998 | 4,851,060 | 44.78 | 2 |
| 265 | AR:Salta:Pichanal:Pepper265:2007 | 4,846,478 | 2,218,946 | 32.66 | 2 |
| 267 | AR:Salta:Pichanal:Pepper267:2007 | 10,927,593 | 4,694,431 | 54.31 | 2 |
| 269 | AR:Salta:Pichanal:Pepper269:2007 | 9,735,179 | 4,060,166 | 77.69 | 2 |
| 271 | AR:Salta:Pichanal:Pepper271:2007 | 10,869,197 | 4,959,285 | 54.16 | 2 |
| 302 | AR:Salta:Orán: Pepper302:2007 | 5,122,034 | 2,563,396 | 78.54 | 1 |
| 309 | AR:Salta:Pichanal:Pepper309:2007 | 9,401,380 | 4,537,658 | 85.43 | 2 |
| 316 | AR:Salta:Pichanal:Pepper316:2008 | 9,626,908 | 5,106,324 | 25.19 | 2 |
| 317 | AR:Salta:Pichanal:Pepper317:2008 | 3,952,259 | 1,675,427 | 32.37 | 2 |
| 322 | AR:Salta:Pichanal:Pepper322:2008 | 10,178,983 | 4,918,919 | 12.87 | 2 |
| 328 | AR:Jujuy:Yuto: Pepper328:2008 | 5,322,366 | 2,446,806 | 21.44 | 3 |
| 423 | AR:Jujuy:Yuto: Pepper423:2008 | 4,965,540 | 2,438,148 | 87.37 | 0 |
| 588 | AR:Jujuy:Yuto: Pepper588:2011 | 3,955,481 | 1,903,671 | 59.05 | 2 |
| 589 | AR:Jujuy:Yuto: Pepper589:2011 | 10,554,833 | 4,753,535 | 59.32 | 2 |
| 654 | AR:Salta:Orán: Pepper654:2014 | 10,858,704 | 4,626,688 | 75.22 | 1 |
| 658 | AR:Salta:Orán: Pepper658:2014 | 4,316,078 | 2,066,084 | 86.22 | 2 |
| 663 | AR:Salta:Orán: Pepper663:2014 | 10,342,387 | 4,946,984 | 83.71 | 2 |
| Sample | Assembly Method | Coverage | Genome Length | Genomic Component | Virus Acronym | GenBank Accession Number | SDT % Identity | Best BLAST Hit |
|---|---|---|---|---|---|---|---|---|
| 145 | de novo | 7831.28 | 2603 | DNA-A | SbBMV | MN486865 | 95.4 | Soybean blistering mosaic virus DNA-A (EF016486) |
| 7838.68 | 2551 | DNA-B | SbBMV | MN486866 | 82.8 | Solanum mosaic Bolivia virus DNA-B (HM585436) | ||
| 169 | de novo | 7836.68 | 2586 | DNA-A | SbBMV | MN486867 | 94 | Soybean blistering mosaic virus DNA-A (EF016486) |
| 7848.72 | 2589 | DNA-B | SbBMV | MN486868 | 58.7 | Solanum mosaic Bolivia virus DNA-B (HM585436) | ||
| 262 | de novo | 7437.22 | 2466 | DNA-A | ToYSV | MN508241 | 98.2 | Tomato yellow spot virus DNA-A (KJ742419) |
| 7742.25 | 2602 | DNA-B | ToYSV | MN508242 | 95.5 | Tomato yellow spot virus DNA-B (KJ742420) | ||
| 263 | de novo | 7859.79 | 2603 | DNA-A | SbBMV | MN486869 | 93.4 | Soybean blistering mosaic virus DNA-A (EF016486) |
| 7855.25 | 2551 | DNA-B | SbBMV | MN486870 | 83 | Solanum mosaic Bolivia virus DNA-B (HM585436) | ||
| 265 | de novo | 7803.97 | 2603 | DNA-A | SbBMV | MN486871 | 93.4 | Soybean blistering mosaic virus DNA-A (EF016486) |
| 7855.33 | 2552 | DNA-B | SbBMV | MN486872 | 82.8 | Solanum mosaic Bolivia virus DNA-B (HM585436) | ||
| 267 | de novo | 7790.12 | 2632 | DNA-A | ToYSV | MN508243 | 97.6 | Tomato yellow spot virus DNA-A (KJ742419) |
| 7836.16 | 2594 | DNA-B | ToYSV | MN508244 | 96 | Tomato yellow spot virus DNA-B (KJ742420) | ||
| 269 | de novo | 6504.38 | 2632 | DNA-A | ToYSV | MN508245 | 97.6 | Tomato yellow spot virus DNA-A (KJ742419) |
| 7627.65 | 2594 | DNA-B | ToYSV | MN508246 | 97.6 | Tomato yellow spot virus DNA-B (KJ742420) | ||
| 271 | de novo | 7809.05 | 2592 | DNA-A | SbBMV | MN508206 | 92.6 | Soybean blistering mosaic virus DNA-A (EF016486) |
| 7859.85 | 2578 | DNA-B | SbBMV | MN508207 | 83.1 | Solanum mosaic Bolivia virus DNA-B (HM585436) | ||
| 302 | filtering + reference | 307.31 | 2653 | DNA-A | SiGMBRV | MN508222 | 95.4 | Sida Brazil virus DNA A (KY555798) |
| 221.93 | 2399 | DNA-B | SiGMBRV | MN508223 | 94.9 | Sida Brazil virus DNA B (KY555799) | ||
| 309 | de novo | 2396.62 | 2564 | DNA-A | ToYVSV | MN508216 | 98.7 | Tomato yellow vein streak virus DNA-A (KC136337) |
| 5370.95 | 2560 | DNA-B | ToYVSV | MN508217 | 95.8 | Tomato yellow vein streak virus DNA-B (KC136338) | ||
| 316 | de novo | 7863.88 | 2603 | DNA-A | SbBMV | MN508208 | 93.4 | Soybean blistering mosaic virus DNA-A (EF016486) |
| 7882.99 | 2549 | DNA-B | SbBMV | MN508209 | 82.8 | Solanum mosaic Bolivia virus DNA-B (HM585436) | ||
| 317 | de novo | 7803.6 | 2603 | DNA-A | SbBMV | MN508210 | 95.5 | Soybean blistering mosaic virus DNA-A (EF016486) |
| 7833.51 | 2584 | DNA-B | SbBMV | MN508211 | 58.3 | Solanum mosaic Bolivia virus DNA-B (HM585436) | ||
| 322 | de novo | 7865.51 | 2605 | DNA-A | SbBMV | MN508212 | 94 | Soybean blistering mosaic virus DNA-A (EF016486) |
| 7885.42 | 2550 | DNA-B | SbBMV | MN508213 | 82.9 | Solanum mosaic Bolivia virus DNA-B (HM585436) | ||
| 328 | de novo | 7787.35 | 2641 | DNA-A | ToYSV | MN508247 | 95.8 | Tomato yellow spot virus DNA-A (KJ742419) |
| 7846.58 | 2609 | DNA-B | ToYSV | MN508248 | 94.5 | Tomato yellow spot virus DNA-B (KJ742420) | ||
| 423 | filtering + de novo | 938.35 | 1350 | Alphasastellite | ToYSA 2 | MN518745 | 85.1 | Tomato yellow spot alphasatellite (KX348228) |
| 432.04 | 2594 | DNA-B | ToYSV | MN518746 | 95.9 | Tomato yellow spot virus DNA-B (KJ742420) | ||
| 588 | filtering + de novo | 7253.73 | 2540 | DNA-A | ToDfLV | MN508218 | 97.4 | Tomato dwarf leaf virus DNA-A (JN564749) |
| 7777.29 | 2501 | DNA-B | ToDfLV | MN508219 | 97.2 | Tomato dwarf leaf virus DNA-B (JN564750) | ||
| 589 | de novo | 7834.03 | 2539 | DNA-A | ToDfLV | MN508220 | 98.1 | Tomato dwarf leaf virus DNA-A (JN564749) |
| 7818.73 | 2501 | DNA-B | ToDfLV | MN508221 | 99.2 | Tomato dwarf leaf virus DNA-B (JN564750) | ||
| 654 | filtering + reference | 7767.16 | 2603 | DNA-A | SbBMV | MN508214 | 93.4 | Soybean blistering mosaic virus DNA-A (EF016486) |
| 7808 | 2641 | DNA-B | SbBMV | MN508215 | 82.9 | Solanum mosaic Bolivia virus DNA-B (HM585436) | ||
| 658 | de novo | 3845.53 | 2630 | DNA-A | ToYSV | MN518733 | 98.5 | Tomato yellow spot virus DNA-A (FJ538207) |
| 5562.73 | 2591 | DNA-B | ToYSV | MN518734 | 98.5 | Tomato yellow spot virus DNA-B (KJ742420) | ||
| 663 | filtering + de novo | 4660.23 | 2651 | DNA-A | PepBLV | MN518735 | 83.1 | Solanum mosaic Bolivia virus DNA-A (HM585435) |
| 1256.06 | 2614 | DNA-B | PepBLV | MN518736 | 83.7 | Sida mosaic Bolivia virus DNA-B (HM585442) |
| Sample | Isolate (Country: Province: Location: Host Species-Sample Number: Year of Collection) | GenBank Accession Number | Viral Sequence Length | Virus Acronym/Genomic Component | SDT % Identity of BLAST Hit/Acronym of Begomovirus Identified /GenBank Accession Number |
|---|---|---|---|---|---|
| 417 | AR: Jujuy:Yuto: Leonurus417:2014 | MN518741 | 2632 | ToYSV/DNA-A | 98.9% ToYSV DNA-A (KJ742419) |
| MN518742 | 2592 | ToYSV/DNA-B | 96% ToYSV DNA-B (KJ742420) | ||
| MN518743 | 1350 | ToYSA 2 | 84.2% Tomato yellow spot alphasatellite (KX348228) | ||
| MN518744 | 1350 | ToYSA 2 | 84.2% Tomato yellow spot alphasatellite (KX348228) | ||
| 663 | AR:Salta:Orán: Pepper663:2014 | MN518737 MN518738 | 2651 | PepBLV/DNA-A | 83% SoMBoV DNA-A (HM585435) |
| MN518739 MN518740 | 2614 | PepBLV/DNA-B | 78% SiMBV1 DNA-B (NC_015044) |
| Recombinant Sequence | Major Parental | Minor Parental | Methods | Av. p-Value | Beginning Breakpoint | Ending Breakpoint |
|---|---|---|---|---|---|---|
| DNA-A | ||||||
| ToYSV (DQ336350, Sample 658, 328, 269, 267, 262, NC007726, KX348179, KX348178, KX348177, KX348176, KX348175, KX348174, KX348173, KX348172, KX348171, KX348170, KX348169, KX348168, KX348167, KX348166, KX348165, (KJ742419, KC706628, JX513952, FJ538207) | ToDfLV (Sample 589) | SiYMV (AY090558) | RGBMCST | 1.057 × 10−12 | 2664 | 2029 |
| SoMBoV (HM585435) | PepBLV (Sample 663) | SbBMV (EF016486) | RBMCST | 2.432 × 10−18 | 30 | 1948 |
| SbBMV (Sample 317, 145) | SbBMV (Sample 322) | SbBMV (Sample 271) | RGBMCST | 5.130 × 10−17 | 201 | 584 |
| CeYSV (JN419002) | PepBLV (Sample 663) | CleGMV (NC_015397) | RGBMCST | 2.312 × 10−9 | 2001 | 2222 |
| SbBMV (EF016486) | SbBMV (Sample 322, 654, 316, 265, 263, 169) | SbBMV (Sample 271) | RGBMCST | 3.971 × 10−11 | 867 | 176 |
| DNA-B | ||||||
| ToYSV (KX348212) | ToYSV (KX34820) | ToYSV (KX348216) | RMCST | 9.303 × 10−17 | 58 | 1373 |
| ToYSV (KX348211) | ToYSV (KX348213) | ToYSV (KX348222) | RGBMCST | 3.369 × 10−7 | 437 | 1257 |
| ToYSV (KX348205) | ToYSV (KX348223) | ToYSV (KX348206) | RGMCST | 7.057 × 10−7 | 982 | 2510 |
| ToYSV (Sample 269) | ToYSV (KJ742420) | ToYSV (Sample 328) | RGBMCST | 1.856 × 10−6 | 2203 | 2406 |
| SbBMV (Sample 263) | SbBMV (Sample 654) | SbBMV (Sample 271) | RGBMCST | 5.872 × 10−7 | 2256 | 2336 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bornancini, V.A.; Irazoqui, J.M.; Flores, C.R.; Vaghi Medina, C.G.; Amadio, A.F.; López Lambertini, P.M. Reconstruction and Characterization of Full-Length Begomovirus and Alphasatellite Genomes Infecting Pepper through Metagenomics. Viruses 2020, 12, 202. https://doi.org/10.3390/v12020202
Bornancini VA, Irazoqui JM, Flores CR, Vaghi Medina CG, Amadio AF, López Lambertini PM. Reconstruction and Characterization of Full-Length Begomovirus and Alphasatellite Genomes Infecting Pepper through Metagenomics. Viruses. 2020; 12(2):202. https://doi.org/10.3390/v12020202
Chicago/Turabian StyleBornancini, Verónica A., José M. Irazoqui, Ceferino R. Flores, Carlos G. Vaghi Medina, Ariel F. Amadio, and Paola M. López Lambertini. 2020. "Reconstruction and Characterization of Full-Length Begomovirus and Alphasatellite Genomes Infecting Pepper through Metagenomics" Viruses 12, no. 2: 202. https://doi.org/10.3390/v12020202
APA StyleBornancini, V. A., Irazoqui, J. M., Flores, C. R., Vaghi Medina, C. G., Amadio, A. F., & López Lambertini, P. M. (2020). Reconstruction and Characterization of Full-Length Begomovirus and Alphasatellite Genomes Infecting Pepper through Metagenomics. Viruses, 12(2), 202. https://doi.org/10.3390/v12020202






