TSPphg Lysin from the Extremophilic Thermus Bacteriophage TSP4 as a Potential Antimicrobial Agent against Both Gram-Negative and Gram-Positive Pathogenic Bacteria
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. Bacterial Strains and Culture Conditions
2.3. Genome Sequencing and Annotation
2.4. Production and Purification of Recombinant Protein TSPphg
2.5. Assessing Thermostability of TSPphg and Effects of pH, NaCl and EDTA on Its Activity
2.6. In Vitro Antibacterial Activity Assays
2.7. Mouse Model of Skin Damage and TSPphg Treatment
2.8. Scanning Electron Microscope (SEM) Analysis
2.9. Statistical Analysis
2.10. Nucleotide and Protein Sequence Accession Number
3. Results
3.1. Identification of TSPphg by Sequencing and Annotation of Phage TSP4 Genome
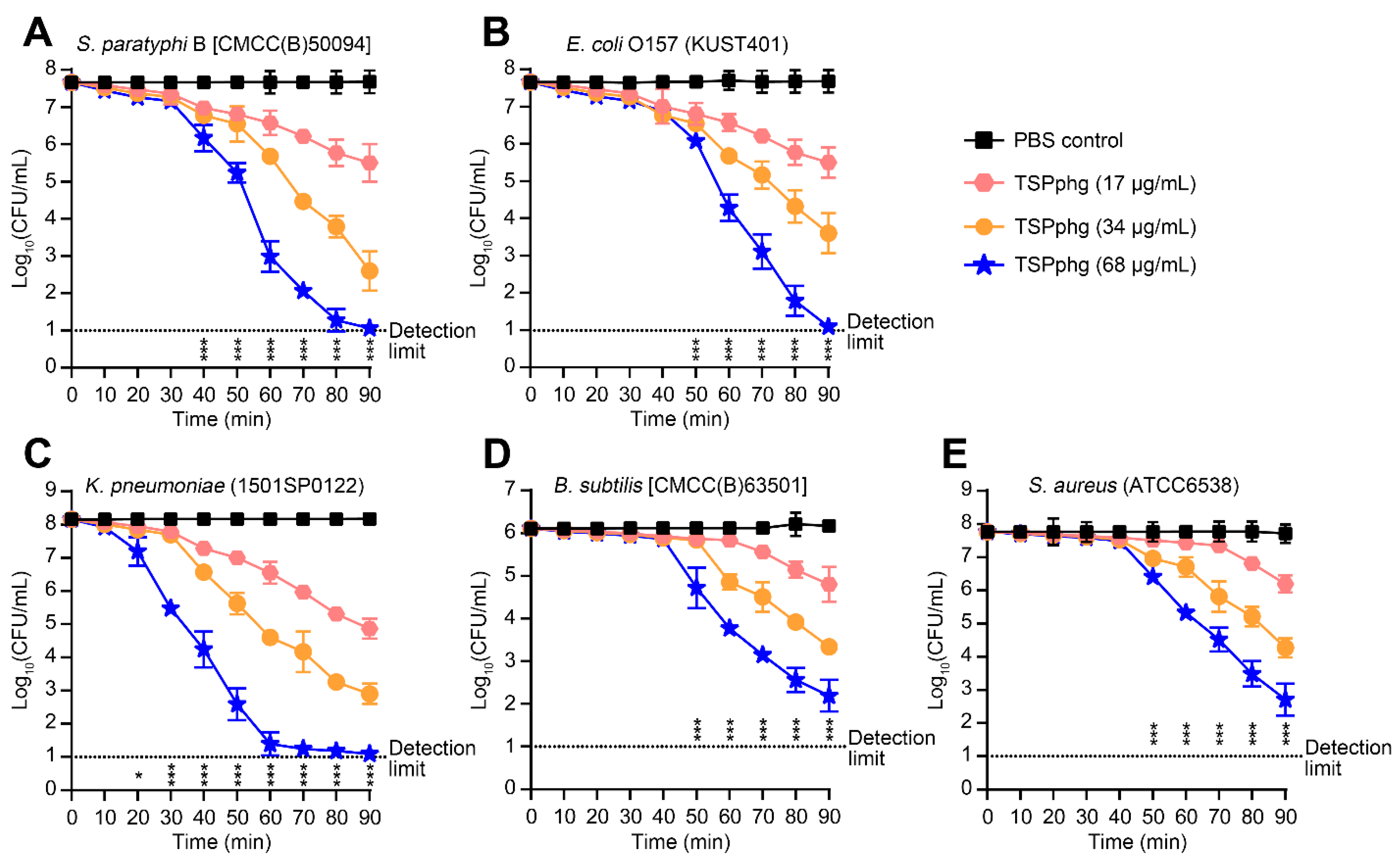
3.2. Bactericidal Activity of TSPphg against both Gram-negative and Gram-positive Bacteria
3.3. Antibacterial Activity of TSPphg against Antibiotic-Resistant Strains of Klebsiella Pneumoniae
3.4. TSPphg Accelerates Wound Healing in a Murine Skin Infection Model
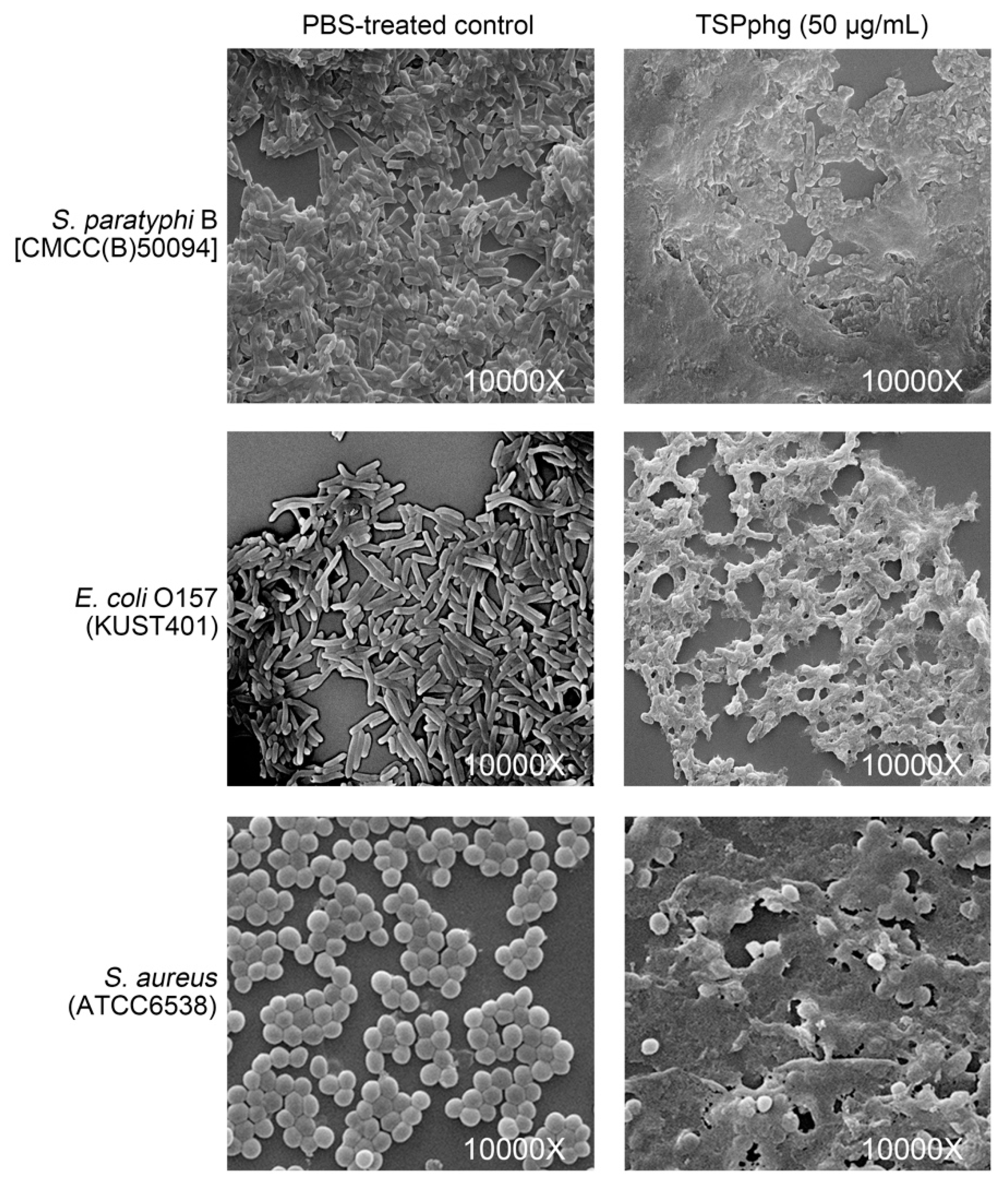
3.5. SEM Observations of the Bacteriolytic Activity of TSPphg
4. Discussion
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Mwangi, W.; de Figueiredo, P.; Criscitiello, M.F. One health: Addressing global challenges at the nexus of human, animal, and environmental health. PLoS Pathog. 2016, 12, e1005731. [Google Scholar] [CrossRef] [PubMed]
- Bordier, M.; Binot, A.; Pauchard, Q.; Nguyen, D.T.; Trung, T.N.; Fortane, N.; Goutard, F.L. Antibiotic resistance in vietnam: Moving towards a one health surveillance system. BMC Public Health 2018, 18, 1136. [Google Scholar] [CrossRef] [PubMed]
- Manyi-Loh, C.; Mamphweli, S.; Meyer, E.; Okoh, A. Antibiotic use in agriculture and its consequential resistance in environmental sources: Potential public health implications. Molecules 2018, 23, 795. [Google Scholar] [CrossRef] [PubMed]
- Blair, J.M.; Richmond, G.E.; Piddock, L.J. Multidrug efflux pumps in Gram-negative bacteria and their role in antibiotic resistance. Future Microbiol. 2014, 9, 1165–1177. [Google Scholar] [CrossRef]
- Abedon, S.T.; Garcia, P.; Mullany, P.; Aminov, R. Editorial: Phage therapy: Past, present and future. Front. Microbiol. 2017, 8, 981. [Google Scholar] [CrossRef]
- Abedon, S.T.; Kuhl, S.J.; Blasdel, B.G.; Kutter, E.M. Phage treatment of human infections. Bacteriophage 2011, 1, 66–85. [Google Scholar] [CrossRef]
- Barrow, P.; Lovell, M.; Berchieri, A., Jr. Use of lytic bacteriophage for control of experimental Escherichia coli septicemia and meningitis in chickens and calves. Clin. Diagn. Lab. Immunol. 1998, 5, 294–298. [Google Scholar] [CrossRef]
- Knoll, B.M.; Mylonakis, E. Antibacterial bioagents based on principles of bacteriophage biology: An overview. Clin. Infect. Dis. 2014, 58, 528–534. [Google Scholar] [CrossRef]
- Yang, H.; Yu, J.; Wei, H. Engineered bacteriophage lysins as novel anti-infectives. Front. Microbiol. 2014, 5, 542. [Google Scholar] [CrossRef]
- Schuch, R.; Nelson, D.; Fischetti, V.A. A bacteriolytic agent that detects and kills Bacillus anthracis. Nature 2002, 418, 884–889. [Google Scholar] [CrossRef]
- Loessner, M.J. Bacteriophage endolysins—Current state of research and applications. Curr. Opin. Microbiol. 2005, 8, 480–487. [Google Scholar] [CrossRef] [PubMed]
- Love, M.J.; Bhandari, D.; Dobson, R.C.J.; Billington, C. Potential for bacteriophage endolysins to supplement or replace antibiotics in food production and clinical care. Antibiotics 2018, 7, 17. [Google Scholar] [CrossRef] [PubMed]
- Trudil, D. Phage lytic enzymes: A history. Virol. Sin. 2015, 30, 26–32. [Google Scholar] [CrossRef]
- Lin, L.; Hong, W.; Ji, X.; Han, J.; Huang, L.; Wei, Y. Isolation and characterization of an extremely long tail Thermus bacteriophage from tengchong hot springs in china. J. Basic Microbiol. 2010, 50, 452–456. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Ji, X.; Chen, M.; Guo, J.; Deng, X.; Lin, L. Rapid purification of bacteriophage endolysin TSPphg and its exogenous treatment could act as an alternative bacterial cell disruption method. Protein Expr. Purif. 2018, 148, 54–58. [Google Scholar] [CrossRef] [PubMed]
- Swift, S.M.; Seal, B.S.; Garrish, J.K.; Oakley, B.B.; Hiett, K.; Yeh, H.-Y.; Woolsey, R.; Schegg, K.M.; Line, J.E.; Donovan, D.M. A thermophilic phage endolysin fusion to a Clostridium perfringens-specific cell wall binding domain creates an anti-clostridium antimicrobial with improved thermostability. Viruses 2015, 7, 3019–3034. [Google Scholar] [CrossRef]
- Plotka, M.; Kaczorowska, A.K.; Morzywolek, A.; Makowska, J.; Kozlowski, L.P.; Thorisdottir, A.; Skirnisdottir, S.; Hjorleifsdottir, S.; Fridjonsson, O.H.; Hreggvidsson, G.O.; et al. Biochemical characterization and validation of a catalytic site of a highly thermostable Ts2631 endolysin from the Thermus scotoductus phage vB_Tsc2631. PLoS ONE 2015, 10, e0137374. [Google Scholar] [CrossRef]
- Plotka, M.; Kapusta, M.; Dorawa, S.; Kaczorowska, A.K.; Kaczorowski, T. Ts2631 endolysin from the extremophilic Thermus scotoductus bacteriophage vB_Tsc2631 as an antimicrobial agent against Gram-negative multidrug-resistant bacteria. Viruses 2019, 11, 657. [Google Scholar] [CrossRef]
- Yang, H.; Wang, M.; Yu, J.; Wei, H. Antibacterial activity of a novel peptide-modified lysin against Acinetobacter baumannii and Pseudomonas aeruginosa. Front. Microbiol. 2015, 6, 1471. [Google Scholar] [CrossRef]
- Kugelberg, E.; Norstrom, T.; Petersen, T.K.; Duvold, T.; Andersson, D.I.; Hughes, D. Establishment of a superficial skin infection model in mice by using Staphylococcus aureus and Streptococcus pyogenes. Antimicrob. Agents Chemother. 2005, 49, 3435–3441. [Google Scholar] [CrossRef]
- Pastagia, M.; Euler, C.; Chahales, P.; Fuentes-Duculan, J.; Krueger, J.G.; Fischetti, V.A. A novel chimeric lysin shows superiority to mupirocin for skin decolonization of methicillin-resistant and -sensitive Staphylococcus aureus strains. Antimicrob. Agents Chemother. 2011, 55, 738–744. [Google Scholar] [CrossRef] [PubMed]
- Cheng, M.; Zhang, L.; Zhang, H.; Li, X.; Wang, Y.; Xia, F.; Wang, B.; Cai, R.; Guo, Z.; Zhang, Y.; et al. An ointment consisting of the phage lysin LysGH15 and apigenin for decolonization of methicillin-resistant Staphylococcus aureus from skin wounds. Viruses 2018, 10, 244. [Google Scholar] [CrossRef] [PubMed]
- Yoong, P.; Schuch, R.; Nelson, D.; Fischetti, V.A. Identification of a broadly active phage lytic enzyme with lethal activity against antibiotic-resistant Enterococcus faecalis and Enterococcus faecium. J. Bacteriol. 2004, 186, 4808–4812. [Google Scholar] [CrossRef] [PubMed]
- Larpin, Y.; Oechslin, F.; Moreillon, P.; Resch, G.; Entenza, J.M.; Mancini, S. In vitro characterization of PlyE146, a novel phage lysin that targets Gram-negative bacteria. PLoS ONE 2018, 13, e0192507. [Google Scholar] [CrossRef] [PubMed]
- Candan, E.D.; Aksoz, N. Klebsiella pneumoniae: Characteristics of carbapenem resistance and virulence factors. Acta Biochim. Pol. 2015, 62, 867–874. [Google Scholar] [CrossRef] [PubMed]
- Martin, R.M.; Bachman, M.A. Colonization, infection, and the accessory genome of Klebsiella pneumoniae. Front. Cell. Infect. Microbiol. 2018, 8, 4. [Google Scholar] [CrossRef]
- Wei, J.; Wenjie, Y.; Ping, L.; Na, W.; Haixia, R.; Xuequn, Z. Antibiotic resistance of Klebsiella pneumoniae through beta-arrestin recruitment-induced beta-lactamase signaling pathway. Exp. Ther. Med. 2018, 15, 2247–2254. [Google Scholar]
- Maciejewska, B.; Olszak, T.; Drulis-Kawa, Z. Applications of bacteriophages versus phage enzymes to combat and cure bacterial infections: An ambitious and also a realistic application? Appl. Microbiol. Biotechnol. 2018, 102, 2563–2581. [Google Scholar] [CrossRef]
- Allen, H.K.; Trachsel, J.; Looft, T.; Casey, T.A. Finding alternatives to antibiotics. Ann. N. Y. Acad. Sci. 2014, 1323, 91–100. [Google Scholar] [CrossRef]
- Plotka, M.; Sancho-Vaello, E.; Dorawa, S.; Kaczorowska, A.K.; Kozlowski, L.P.; Kaczorowski, T.; Zeth, K. Structure and function of the Ts2631 endolysin of Thermus scotoductus phage vB_Tsc2631 with unique N-terminal extension used for peptidoglycan binding. Sci. Rep. 2019, 9, 1261. [Google Scholar] [CrossRef]
- Stothard, P. The sequence manipulation suite: Javascript programs for analyzing and formatting protein and DNA sequences. Biotechniques 2000, 28, 1102–1104. [Google Scholar] [CrossRef] [PubMed]
- Brogden, K.A. Antimicrobial peptides: Pore formers or metabolic inhibitors in bacteria? Nat. Rev. Microbiol. 2005, 3, 238–250. [Google Scholar] [CrossRef] [PubMed]
- Mihajlovic, M.; Lazaridis, T. Antimicrobial peptides in toroidal and cylindrical pores. Biochim. Biophys. Acta 2010, 1798, 1485–1493. [Google Scholar] [CrossRef]
- Zhang, Y.; Shi, W.; Tang, S.; Li, J.; Yin, S.; Gao, X.; Wang, L.; Zou, L.; Zhao, J.; Huang, Y.; et al. The influence of cathelicidin LL37 in human anti-neutrophils cytoplasmic antibody (anca)-associated vasculitis. Arthritis Res. Ther. 2013, 15, R161. [Google Scholar] [CrossRef] [PubMed]
- Briers, Y.; Lavigne, R. Breaking barriers: Expansion of the use of endolysins as novel antibacterials against Gram-negative bacteria. Future Microbiol. 2015, 10, 377–390. [Google Scholar] [CrossRef] [PubMed]
- Deslouches, B.; Steckbeck, J.D.; Craigo, J.K.; Doi, Y.; Burns, J.L.; Montelaro, R.C. Engineered cationic antimicrobial peptides to overcome multidrug resistance by ESKAPE pathogens. Antimicrob. Agents Chemother. 2015, 59, 1329–1333. [Google Scholar] [CrossRef] [PubMed]


| Strain | Antibiotic Resistant a | Starting Bacterial Count (Log10 CFU/mL) | Log10 of (Starting Count/Final Count) b |
|---|---|---|---|
| S. paratyphi B | |||
| CMCC(B)50094 | None | 7.66 ± 0.09 | 3.91 ± 0.25 |
| E. coli O157 | |||
| KUST401 | STR, TET and AMP | 7.63 ± 0.20 | 3.03 ± 0.34 |
| K. pneumoniae | |||
| 14V0622 | CRO, AMP, CFZ, ATM, FOX and FEP | 8.31 ± 0.10 | 6.12 ± 0.10 |
| 1501SP0122 | AMP, CFZ, ATM, AMX and FEP | 8.16 ± 0.14 | 6.41 ± 0.07 |
| 13A14918 | CRO, AMP, CFZ, NIT, AMP, AMX and GEN | 7.48 ± 0.16 | > 6.48 (*) |
| 13A15188 | AMP and NIT | 7.42 ± 0.18 | 2.45 ± 0.01 |
| 13V1837 | CRO, AMP, CFZ, ATM, NIT and FEP | 8.05 ± 0.04 | 1.09 ± 0.40 |
| 1501SP0351 | CRO, AMP, CFZ, ATM, AMX and FEP | 8.47 ± 0.01 | 1.90 ± 0.01 |
| 13A14165 | AMP, NIT and CFP | 8.57 ± 0.05 | 3.05 ± 0.20 |
| 14A0287 | CRO, AMP, TZB, CFZ, ATM, NIT, FOX and AMK | 7.74 ± 0.03 | > 6.74 (*) |
| 1412SP0200 | AMP, NIT and CIP | 7.82 ± 0.05 | > 6.82 (*) |
| B. subtilis | |||
| CMCC(B)63501 | None | 6.11 ± 0.08 | 1.99 ± 0.22 |
| Staphylococcus epidermidis | |||
| ATCC12228 | None | 5.57 ± 0.12 | 2.77 ± 0.26 |
| Micrococcus luteus | |||
| ATCC4698 | None | 7.81 ± 0.09 | > 6.81 (*) |
| S. aureus | |||
| 1606BL1486 | CIP, CLI, ERY, GEN, LVX, LZD, MXF, NIT, OXA, PEN and RIF | 6.67 ± 0.06 | 2.96 ± 0.43 |
| ATCC6538 | None | 7.76 ± 0.05 | 2.11 ± 0.12 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, F.; Ji, X.; Li, Q.; Zhang, G.; Peng, J.; Hai, J.; Zhang, Y.; Ci, B.; Li, H.; Xiong, Y.; et al. TSPphg Lysin from the Extremophilic Thermus Bacteriophage TSP4 as a Potential Antimicrobial Agent against Both Gram-Negative and Gram-Positive Pathogenic Bacteria. Viruses 2020, 12, 192. https://doi.org/10.3390/v12020192
Wang F, Ji X, Li Q, Zhang G, Peng J, Hai J, Zhang Y, Ci B, Li H, Xiong Y, et al. TSPphg Lysin from the Extremophilic Thermus Bacteriophage TSP4 as a Potential Antimicrobial Agent against Both Gram-Negative and Gram-Positive Pathogenic Bacteria. Viruses. 2020; 12(2):192. https://doi.org/10.3390/v12020192
Chicago/Turabian StyleWang, Feng, Xinyu Ji, Qiupeng Li, Guanling Zhang, Jiani Peng, Jun Hai, Yao Zhang, Baiquan Ci, Hongwei Li, Yan Xiong, and et al. 2020. "TSPphg Lysin from the Extremophilic Thermus Bacteriophage TSP4 as a Potential Antimicrobial Agent against Both Gram-Negative and Gram-Positive Pathogenic Bacteria" Viruses 12, no. 2: 192. https://doi.org/10.3390/v12020192
APA StyleWang, F., Ji, X., Li, Q., Zhang, G., Peng, J., Hai, J., Zhang, Y., Ci, B., Li, H., Xiong, Y., Deng, X., & Lin, L. (2020). TSPphg Lysin from the Extremophilic Thermus Bacteriophage TSP4 as a Potential Antimicrobial Agent against Both Gram-Negative and Gram-Positive Pathogenic Bacteria. Viruses, 12(2), 192. https://doi.org/10.3390/v12020192




