Application of a Sanger-Based External Quality Assurance Strategy for the Transition of HIV-1 Drug Resistance Assays to Next Generation Sequencing
Abstract
:1. Introduction
2. What Have We Learned from Sanger-Based EQA?
3. Can We Use Existing SS EQA Programs to Evaluate NGS Data?
4. Discussion
- Quantitative measures of laboratory performance: for example, for a given sample, gene region and nucleotide position, the percent distribution for each nucleotide base may be evaluated, summary statistics computed, and outliers identified based on parametric (e.g., are the data outside a three standard deviation window?) or non-parametric (e.g., are the data below the 5th or above the 95th percentile compared to the distribution from all other laboratories?).
- Multivariate statistical techniques may be considered to evaluate the multivariate distribution of percentages of all four nucleotides. For example, the distribution of percentages may be considered in a four-dimensional space with a mean vector and correlation (or covariance) matrix. From these parameter estimates, suitable quantities to evaluate each laboratory in four-dimensional space may be computed; for example, Mahalanobis’s Distance is a measure of the “statistical distance” of each laboratory’s data point to the mean four-dimensional vector, and would quantify how far that laboratory is from the four-dimensional average in multivariate distance, accounting for correlations among the four percentages within a laboratory + nucleotide position. A suitable criterion could be developed to quantify whether a laboratory is within or outside acceptable boundaries. If traditional assumptions (e.g., Gaussian distributions) are violated, suitable transformations (e.g., natural logarithm) or alternative modelling (e.g., zero-inflated distributions) may be necessary.
- Beyond evaluation of individual laboratories, an assessment may be made of how to quantify a point estimate of what the most likely base call should be (e.g., what is the most likely nucleotide at a given location?) and the confidence in that estimate (e.g., a confidence interval using the estimated covariance matrix). Quantitative criteria would also allow for “no consensus” results, i.e., there is no clarity on what nucleotide is the best call.
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Jennings, C.; Harty, B.; Granger, S.; Wager, C.; Crump, J.A.; Fiscus, S.A.; Bremer, J.W. Cross-platform analysis of HIV-1 RNA data generated by a multicenter assay validation study with wide geographic representation. J. Clin. Microbiol. 2012, 50, 2737–2747. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lalama, C.M.; Jennings, C.; Johnson, V.A.; Coombs, R.W.; McKinnon, J.E.; Bremer, J.W.; Cobb, B.R.; Cloherty, G.A.; Mellors, J.W.; Ribaudo, H.J. Comparison of Three Different FDA-Approved Plasma HIV-1 RNA Assay Platforms Confirms the Virologic Failure Endpoint of 200 Copies per Milliliter Despite Improved Assay Sensitivity. J. Clin. Microbiol. 2015, 53, 2659–2666. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, E.R.; Parkin, N.; Jennings, C.; Brumme, C.J.; Enns, E.; Casadella, M.; Howison, M.; Coetzer, M.; Avila-Rios, S.; Capina, R.; et al. Performance comparison of next generation sequencing analysis pipelines for HIV-1 drug resistance testing. Sci. Rep. 2020, 10, 1634. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Godfrey, C.C.; Michelow, P.M.; Godard, M.; Sahasrabuddhe, V.V.; Darden, J.; Firnhaber, C.S.; Wetherall, N.T.; Bremer, J.; Coombs, R.W.; Wilkin, T. Improving diagnostic capability for HPV disease internationally within NIH-NIAID division of AIDS clinical trial networks. Am. J. Clin. Pathol. 2013, 140, 881–889. [Google Scholar] [CrossRef] [Green Version]
- Hong, F.; Aga, E.; Cillo, A.R.; Yates, A.L.; Besson, G.; Fyne, E.; Koontz, D.L.; Jennings, C.; Zheng, L.; Mellors, J.W. Novel assays for measurement of total cell-associated HIV-1 DNA and RNA. J. Clin. Microbiol. 2016, 54, 902–911. [Google Scholar] [CrossRef] [Green Version]
- Jennings, C.; Harty, B.; Scianna, S.R.; Granger, S.; Couzens, A.; Zaccaro, D.; Bremer, J.W. The stability of HIV-1 nucleic acid in whole blood and improved detection of HIV-1 in alternative specimen types when compared to Dried Blood Spot (DBS) specimens. J. Virol. Methods 2018, 261, 91–97. [Google Scholar] [CrossRef]
- McNulty, A.; Jennings, C.; Bennett, D.; Fitzgibbon, J.; Bremer, J.W.; Ussery, M.; Kalish, M.L.; Heneine, W.; García-Lerma, J.G. Evaluation of dried blood spots for human immunodeficiency virus type 1 drug resistance testing. J. Clin. Microbiol. 2007, 45, 517–521. [Google Scholar] [CrossRef] [Green Version]
- Parkin, N.; de Mendoza, C.; Schuurman, R.; Jennings, C.; Bremer, J.; Jordan, M.R.; Bertagnolio, S. Evaluation of in-house genotyping assay performance using dried blood spot specimens in the Global World Health Organization laboratory network. Clin. Infect. Dis. 2012, 54 (Suppl. 4), S273–S279. [Google Scholar] [CrossRef] [Green Version]
- Jennings, C.; Wager, C.G.; Scianna, S.R.; Zaccaro, D.J.; Couzens, A.; Mellors, J.W.; Coombs, R.W.; Bremer, J.W. Use of External Quality Control Material for HIV-1 RNA Testing to Assess the Comparability of Data Generated in Separate Laboratories and the Stability of HIV-1 RNA in Samples after Prolonged Storage. J. Clin. Microbiol. 2018, 56. [Google Scholar] [CrossRef] [Green Version]
- Mitchell, C.; Jennings, C.; Brambilla, D.; Aldrovandi, G.; Amedee, A.M.; Beck, I.; Bremer, J.W.; Coombs, R.; Decker, D.; Fiscus, S.; et al. Diminished human immunodeficiency virus type 1 DNA yield from dried blood spots after storage in a humid incubator at 37 °C compared to −20 °C. J. Clin. Microbiol. 2008, 46, 2945–2949. [Google Scholar] [CrossRef] [Green Version]
- Jennings, C.; Danilovic, A.; Scianna, S.; Brambilla, D.J.; Bremer, J.W. Stability of human immunodeficiency virus type 1 proviral DNA in whole-blood samples. J. Clin. Microbiol. 2005, 43, 4249–4250. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- García-Lerma, J.G.; McNulty, A.; Jennings, C.; Huang, D.; Heneine, W.; Bremer, J.W. Rapid decline in the efficiency of HIV drug resistance genotyping from dried blood spots (DBS) and dried plasma spots (DPS) stored at 37 degrees C and high humidity. J. Antimicrob. Chemother. 2009, 64, 33–36. [Google Scholar] [CrossRef] [PubMed]
- Brambilla, D.; Jennings, C.; Aldrovandi, G.; Bremer, J.; Comeau, A.M.; Cassol, S.A.; Dickover, R.; Jackson, J.B.; Pitt, J.; Sullivan, J.L.; et al. Multicenter evaluation of use of dried blood and plasma spot specimens in quantitative assays for human immunodeficiency virus RNA: Measurement, precision, and RNA stability. J. Clin. Microbiol. 2003, 41, 1888–1893. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yen-Lieberman, B.; Brambilla, D.; Jackson, B.; Bremer, J.; Coombs, R.; Cronin, M.; Herman, S.; Katzenstein, D.; Leung, S.; Lin, H.J.; et al. Evaluation of a quality assurance program for the quantitation of human immunodeficiency virus type 1 RNA in plasma by the AIDS Clinical Trials Group virology laboratories. J. Clin. Microbiol. 1996, 34, 2695–2701. [Google Scholar] [CrossRef] [Green Version]
- Jackson, J.B.; Drew, J.; Lin, H.J.; Otto, P.; Bremer, J.W.; Hollinger, F.B.; Wolinsky, S.M. Establishment of a quality assurance program for human immunodeficiency virus type 1 DNA polymerase chain reaction assays by the AIDS Clinical Trials Group. ACTG PCR Working Group, and the ACTG PCR Virology Laboratories. J. Clin. Microbiol. 1993, 31, 3123–3128. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parkin, N.; Bremer, J.; Bertagnolio, S. Genotyping External Quality Assurance in the World Health Organization HIV Drug Resistance Laboratory Network During 2007–2010. Clin. Infect. Dis. 2012, 54 (Suppl. 4), S266–S272. [Google Scholar] [CrossRef] [Green Version]
- Schuurman, R.; Brambilla, D.; De Groot, T.; Wang, D.; Land, S.; Bremer, J.; Benders, I.; Boucher, C.A. Underestimation of HIV type 1 drug resistance mutations: Results from the ENVA-2 genotyping proficiency program. AIDS Res. Hum. Retrovir. 2002, 18, 243–248. [Google Scholar] [CrossRef]
- Huang, D.D.; Eshleman, S.H.; Brambilla, D.J.; Palumbo, P.E.; Bremer, J.W. Evaluation of the editing process in human immunodeficiency virus type 1 genotyping. J. Clin. Microbiol. 2003, 41, 3265–3272. [Google Scholar] [CrossRef] [Green Version]
- Huang, D.D.; Bremer, J.W.; Brambilla, D.J.; Palumbo, P.E. Model for assessment of proficiency of human immunodeficiency virus type 1 sequencing-based genotypic antiretroviral assays. J. Clin. Microbiol. 2005, 43, 3963–3970. [Google Scholar] [CrossRef] [Green Version]
- Eshleman, S.H.; Crutcher, G.; Petrauskene, O.; Kunstman, K.; Cunningham, S.P.; Trevino, C.; Davis, C.; Kennedy, J.; Fairman, J.; Foley, B.; et al. Sensitivity and specificity of the ViroSeq human immunodeficiency virus type 1 (HIV-1) genotyping system for detection of HIV-1 drug resistance mutations by use of an ABI PRISM 3100 genetic analyzer. J. Clin. Microbiol. 2005, 43, 813–817. [Google Scholar] [CrossRef] [Green Version]
- Grant, R.M.; Kuritzkes, D.R.; Johnson, V.A.; Mellors, J.W.; Sullivan, J.L.; Swanstrom, R.; D’Aquila, R.T.; Van Gorder, M.; Holodniy, M.; Lloyd, R.M., Jr.; et al. Accuracy of the TRUGENE HIV-1 genotyping kit. J. Clin. Microbiol. 2003, 41, 1586–1593. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuritzkes, D.R.; Grant, R.M.; Feorino, P.; Griswold, M.; Hoover, M.; Young, R.; Day, S.; Lloyd, R.M., Jr.; Reid, C.; Morgan, G.F.; et al. Performance characteristics of the TRUGENE HIV-1 genotyping kit and the Opengene DNA sequencing system. J. Clin. Microbiol. 2003, 41, 1594–1599. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sayer, D.C.; Land, S.; Gizzarelli, L.; French, M.; Hales, G.; Emery, S.; Christiansen, F.T.; Dax, E.M. Quality assessment program for genotypic antiretroviral testing improves detection of drug resistance mutations. J. Clin. Microbiol. 2003, 41, 227–236. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Korn, K.; Reil, H.; Walter, H.; Schmidt, B. Quality control trial for human immunodeficiency virus type 1 drug resistance testing using clinical samples reveals problems with detecting minority species and interpretation of test results. J. Clin. Microbiol. 2003, 41, 3559–3565. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pandit, A.; Mackay, W.G.; Steel, C.; van Loon, A.M.; Schuurman, R. HIV-1 drug resistance genotyping quality assessment: Results of the ENVA7 genotyping proficiency programme. J. Clin. Virol. 2008, 43, 401–406. [Google Scholar] [CrossRef] [PubMed]
- Land, S.; Cunningham, P.; Zhou, J.; Frost, K.; Katzenstein, D.; Kantor, R.; Arthur Chen, Y.-M.; Oka, S.; DeLong, A.; Sayer, D.; et al. TREAT Asia quality assessment scheme (TAQAS) to standardize the outcome of HIV-1 genotypic resistnace testing in a group of Asian laboratories. J. Virol. Methods 2009, 159, 185–193. [Google Scholar] [CrossRef] [Green Version]
- Yoshida, S.; Hattori, J.; Matsuda, M.; Okada, K.; Kazuyama, Y.; Hashimoto, O.; Ibe, S.; Fujisawa, S.-I.; Chiba, H.; Tatsumi, M.; et al. Japanese external quality assessment program to standardize HIV-1 drug resistance testing (JEQS2010 program) using in vitro transcribed RNA as reference material. AIDS Res. Hum. Retrovir. 2015, 31, 318–325. [Google Scholar] [CrossRef] [Green Version]
- Saeng-aroon, S.; Saipradit, N.; Loket, R.; Klamkhai, N.; Boonmuang, R.; Kaewprommal, P.; Prommajan, K.; Takeda, N.; Sungkanuparph, S.; Shioda, T.; et al. External quality assessment scheme for HIV-1 drug-resistance genotyping in Thailand. AIDS Res. Hum. Retrovir. 2018, 34, 1028–1035. [Google Scholar] [CrossRef]
- Wensing, A.M.; Calvez, V.; Ceccherini-Silberstein, F.; Charpentier, C.; Gunthard, H.F.; Paredes, R.; Shafer, R.W.; Richman, D.D. 2019 update of the drug resistance mutations in HIV-1. Top. Antivir. Med. 2019, 27, 111–121. [Google Scholar]
- World Health Organization. WHO/HIVResNet HIV Drug Resistance Laboratory Operational Framework; WHO: Geneva, Switzerland, 2017; ISBN 978-92-4-151287-9. Available online: https://apps.who.int/iris/handle/10665/259731 (accessed on 16 December 2020).
- Ji, H.; Enns, E.; Brumme, C.J.; Parkin, N.; Howison, M.; Lee, E.R.; Capina, R.; Marinier, E.; Avila-Rios, S.; Sandstrom, P.; et al. Bioinformatic data processing pipelines in support of next-generation sequencing-based HIV-1 drug resistance testing: The Winnipeg consensus. J. Int. AIDS Soc. 2018, 21, e25193. [Google Scholar] [CrossRef] [Green Version]
- Zhou, S.; Swanstrom, R. Fact and fiction about 1%: Next generation sequencing and the detection of minor drug resistant variants in HIV-1 populations with and without unique molecular identifiers. Viruses 2020, 12, 850. [Google Scholar] [CrossRef] [PubMed]
- Becker, M.G.; Liang, D.; Cooper, B.; Le, Y.; Taylor, T.; Lee, E.R.; Wu, S.; Sandstrom, P.; Ji, H. Development and application performance assessment criteria for next-generation sequencing-based HIV drug resistance assays. Viruses 2020, 12, 627. [Google Scholar] [CrossRef] [PubMed]
- Weber, J.; Volkova, I.; Sahoo, M.K.; Tzou, P.L.; Shafer, R.W.; Pinsky, B.A. Prospective evaluation of the Vela Diagnositcs next-generation sequencing platform for HIV-1 genotypic resistance testing. J. Mol. Diag. 2019, 21, 967–970. [Google Scholar] [CrossRef] [PubMed]
- Parkin, N.; Zaccaro, D.; Avila-Rios, S.; Matias-Florentino, M.; Brumme, C.; Hunt, G.; Ledwaba, J.; Ji, H.; Lee, E.R.; Kantor, R.; et al. Multi-Laboratory comparison of next-generation to Sanger-based sequencing for HIV-1 drug resistance genotyping. Viruses 2020, 12, 694. [Google Scholar] [CrossRef] [PubMed]
- VQA HIV Gene Sequencing Proficiency Testing Scoring Criteria and Policies. 2014. Available online: https://www.hanc.info/labs/labresources/vqaResources/ptProgram/VQA%20Document%20Library/VQA%20GENO%20Scoring%20Document_v1_3.pdf (accessed on 16 December 2020).
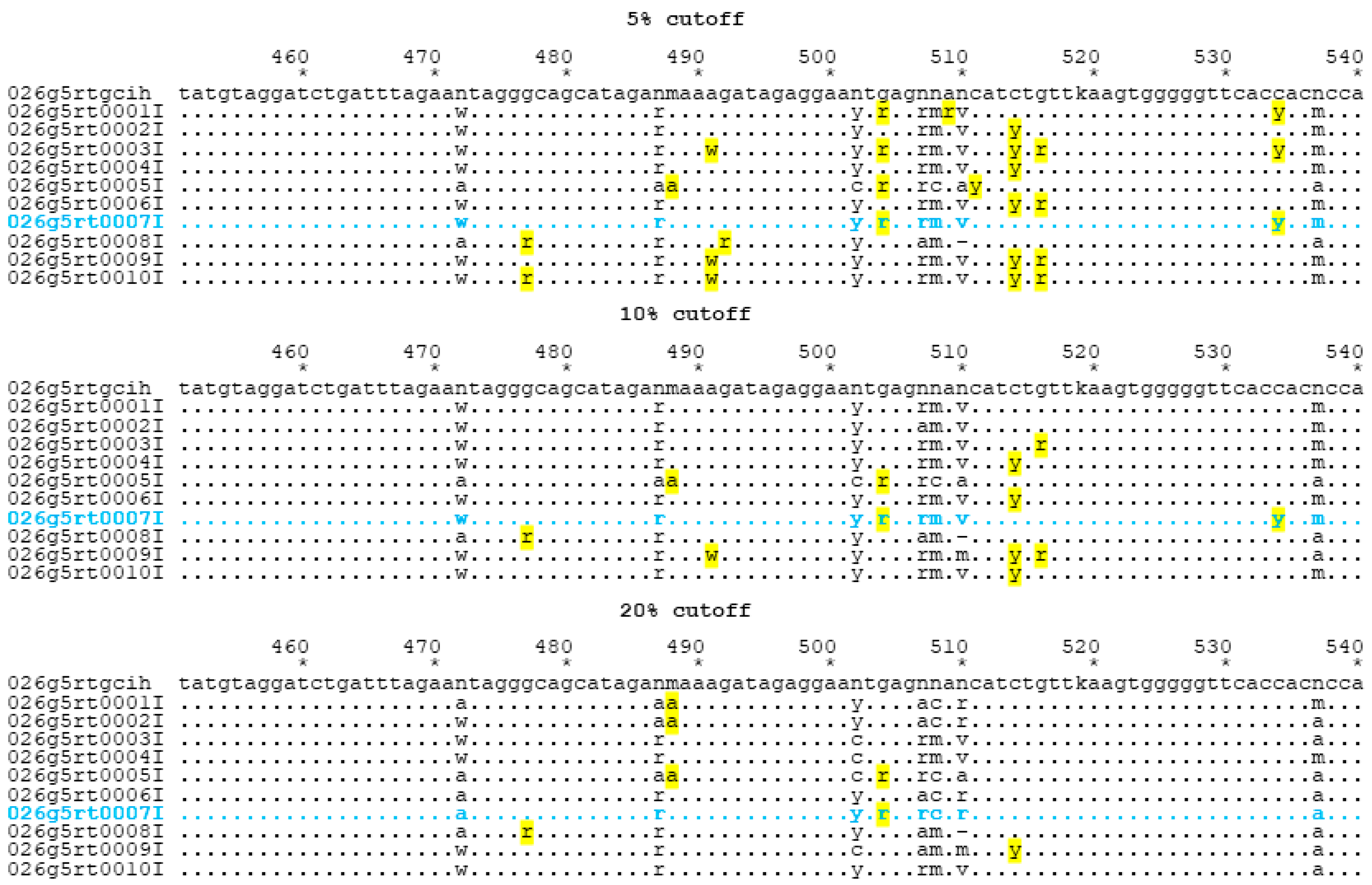
| Laboratory | 5% Cutoff | 10% Cutoff | 20% Cutoff | ||||||
|---|---|---|---|---|---|---|---|---|---|
| % Homology * | Stage 1 Errors | Poisson p-Value | % Homology * | Stage 1 Errors | Poisson p-Value | % Homology * | Stage 1 Errors | Poisson p-Value | |
| 1 | 99.0 | 5 | 99.6 | 2 | 99.4 | 3 | |||
| 2 | 98.3 | 10 | 0.021 | 99.1 | 5 | 99.1 | 5 | ||
| 3 | 97.6 | 14 | 0.0004 | 99.0 | 6 | 100 | 0 | ||
| 4 | 98.1 | 11 | 0.009 | 99.3 | 4 | 99.8 | 1 | ||
| 5 | 97.2 | 16 | <0.0001 | 97.2 | 16 | <0.0001 | 97.4 | 15 | 0.00011 |
| 6 | 97.9 | 12 | 0.003 | 99.3 | 4 | 100 | 0 | ||
| 7 | 98.3 | 10 | 0.021 | 99.1 | 5 | 99.5 | 3 | ||
| 8 | 98.1 | 11 | 0.009 | 98.3 | 10 | 0.021 | 98.3 | 10 | 0.02138 |
| 9 | 98.1 | 11 | 0.009 | 98.1 | 11 | 0.009 | 98.3 | 10 | 0.02138 |
| 10 | 97.4 | 15 | 0.0001 | 99.1 | 6 | 100 | 0 | ||
| Position | Depth | A | C | G | T |
|---|---|---|---|---|---|
| 472 | 23,583 | 87 | 13 | ||
| 487 | 19,894 | 75 | 24 | ||
| 488 | 19,907 | 76 | 24 | ||
| 502 | 21,915 | 78 | 22 | ||
| 504 | 20,530 | 29 | 71 | ||
| 507 | 21,464 | 59 | 41 | ||
| 508 | 21,617 | 16 | 84 | ||
| 510 | 20,543 | 57 | 17 | 25 | |
| 519 | 26,631 | 61 | 39 | ||
| 537 | 27,318 | 86 | 13 |
| Lab | 24 g Total Points, Score | 26 g Total Points, Score | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 5% | 10% | 20% | 5% | 10% | 20% | |||||||
| Points | Score | Points | Score | Points | Score | Points | Score | Points | Score | Points | Score | |
| 1 | 13 | PC | 4 | C | 2 | C | 9 | PC | 1 | C | 2 | C |
| 2 | 8 | PC | 0 | C | 0 | C | 14 | PC | 2 | C | 2 | C |
| 3 | 6 | C | 2 | C | 0 | C | 6 | C | 3 | C | 0 | C |
| 4 | 9 | PC | 2 | C | 5 | C | 8 | PC | 0 | C | 0 | C |
| 5 | 9 | PC | 6 | C | 3 | C | 9 | PC | 9 | PC | 8 | PC |
| 6 | 6 | C | 2 | C | 0 | C | 12 | PC | 6 | C | 3 | C |
| 7 | 3 | C | 2 | C | 0 | C | 3 | C | 2 | C | 0 | C |
| 8 | 3 | C | 3 | C | 4 | C | 4 | C | 3 | C | 3 | C |
| 9 | 9 | PC | 5 | C | 1 | C | 12 | PC | 8 | PC | 7 | C |
| 10 | 10 | PC | 1 | C | 0 | C | 9 | PC | 1 | C | 0 | C |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jennings, C.; Parkin, N.T.; Zaccaro, D.J.; Capina, R.; Sandstrom, P.; Ji, H.; Brambilla, D.J.; Bremer, J.W. Application of a Sanger-Based External Quality Assurance Strategy for the Transition of HIV-1 Drug Resistance Assays to Next Generation Sequencing. Viruses 2020, 12, 1456. https://doi.org/10.3390/v12121456
Jennings C, Parkin NT, Zaccaro DJ, Capina R, Sandstrom P, Ji H, Brambilla DJ, Bremer JW. Application of a Sanger-Based External Quality Assurance Strategy for the Transition of HIV-1 Drug Resistance Assays to Next Generation Sequencing. Viruses. 2020; 12(12):1456. https://doi.org/10.3390/v12121456
Chicago/Turabian StyleJennings, Cheryl, Neil T. Parkin, Daniel J. Zaccaro, Rupert Capina, Paul Sandstrom, Hezhao Ji, Donald J. Brambilla, and James W. Bremer. 2020. "Application of a Sanger-Based External Quality Assurance Strategy for the Transition of HIV-1 Drug Resistance Assays to Next Generation Sequencing" Viruses 12, no. 12: 1456. https://doi.org/10.3390/v12121456
APA StyleJennings, C., Parkin, N. T., Zaccaro, D. J., Capina, R., Sandstrom, P., Ji, H., Brambilla, D. J., & Bremer, J. W. (2020). Application of a Sanger-Based External Quality Assurance Strategy for the Transition of HIV-1 Drug Resistance Assays to Next Generation Sequencing. Viruses, 12(12), 1456. https://doi.org/10.3390/v12121456






