Walking Together: Cross-Protection, Genome Conservation, and the Replication Machinery of Citrus tristeza virus
Abstract
:1. Brief History of Cross-Protection
2. Application of Cross-Protection against CTV
3. Terminology
4. Cross-Protection Research: A Personal Recollection
5. Cross-Protection among CTV Isolates Displaying Differential Transmission by A. gossypii
6. Failure of Cross-Protection between CTV Variants of Different Strains
7. CTV Genetics
8. Understanding the Mechanism of CTV Cross-Protection
9. Cross-Protection, Genome Conservation, and the Replication Machinery of CTV: Are These Properties Related?
10. Concluding Remarks
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Van Vuuren, S.P.; Manicom, B.Q. The response of Star Ruby grapefruit to different Citrus tristeza virus isolates. Int. Organ. Citrus Virol. Conf. Proc. 2005, 16, 112–116. [Google Scholar]
- McKinney, H.H. Mosaic diseases in the Canary Islands, West Africa and Gibraltar. J. Agric. Res. 1929, 39, 557–578. [Google Scholar]
- Salaman, R.N. Protective inoculation against a plant virus. Nature 1933, 131, 468. [Google Scholar] [CrossRef]
- Fulton, R.W. Practices and precautions in the use of cross protection for plant virus disease control. Annu. Rev. Phytopathol. 1986, 24, 67–81. [Google Scholar] [CrossRef]
- Yeh, S.D.; Gonsalves, D. Evaluation of induced mutants of papaya ringspot virus for control by cross protection. Phytopathology 1984, 74, 1086–1091. [Google Scholar] [CrossRef]
- Moreno, P.; Ambros, S.; Albiach-Martí, M.R.; Guerri, J.; Pena, L. Citrus tristeza virus: A pathogen that changed the course of the citrus industry. Mol. Plant Pathol. 2008, 9, 251–268. [Google Scholar] [CrossRef] [PubMed]
- Dawson, W.O.; Garnsey, S.M.; Tatineni, S.; Folimonova, S.Y.; Harper, S.J.; Gowda, S. Citrus tristeza virus-host interactions. Front. Microbiol. 2013, 4, 88. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fraser, L.R.; Long, K.; Cox, J. Stem pitting of grapefruit—Field protection by the use of mild virus strains. Int. Organ. Citrus Virol. Conf. Proc. 1968, 4, 27–31. [Google Scholar]
- Broadbent, P.; Bevington, K.B.; Coote, B.G. Control of stem pitting of grapefruit in Australia by mild strain cross protection. Int. Organ. Citrus Virol. Conf. Proc. 1991, 11, 64–70. [Google Scholar]
- Broadbent, P.; Dephoff, C.M.; Franks, N.; Gillings, M.; Indsto, J. Pre-immunisation of grapefruit with a mild protective isolate of Citrus tristeza virus in Australia. In Proceedings of the 3rd International Workshop on Citrus Tristeza Virus and the Brown Citrus Aphid in the Caribbean Basin: Management Strategies, Lake Alfred, FL, USA, 15–18 May 1995; pp. 163–168. [Google Scholar]
- Zhou, C.Y.; Broadbent, P.; Hailstones, D.L.; Bowyer, J.; Connor, R. Movement and titer of Citrus Tristeza Virus (pre-immunizing isolate PB61) within seedlings and field trees. Int. Organ. Citrus Virol. Conf. Proc. 2002, 15, 39–47. [Google Scholar]
- Costa, A.S.; Müller, G.W. Tristeza control by cross-protection; a U.S.-Brazil cooperative success. Plant Dis. 1980, 64, 538–541. [Google Scholar] [CrossRef]
- Costa, A.T.; Nunes, W.M.D.; Zanutto, C.A.; Müller, G.W. Stability of citrus tristeza virus protective isolates in field conditions. Pesqui. Agropecu. Bras. 2010, 45, 693–700. [Google Scholar] [CrossRef] [Green Version]
- Baba, V.Y.; Giampani, J.S.; Tazima, Z.H.; Yada, I.F.U.; Paccola-Meirelles, L.D.; Leite, R.P. Agronomic performance of pera and related sweet orange accessions naturally infected with citrus tristeza virus in Northern Parana State, Brazil. Trop. Plant Pathol. 2014, 39, 442–448. [Google Scholar] [CrossRef] [Green Version]
- Van Vuuren, S.P.; Collins, R.P.; da Graça, J.V. The performance of exotic citrus tristeza virus isolates as preimmunizing agents for sweet orange on sour orange rootstock under natural disease pressure in South Africa. Int. Organ. Citrus Virol. Conf. Proc. 1991, 11, 60–63. [Google Scholar]
- Da Graça, J.V.; van Vuuren, S.P. Managing Citrus tristeza virus losses using cross protection. In Citrus Tristeza Virus Complex and Tristeza Diseases; Karasev, A.V., Hilf, M.E., Eds.; APS Press: Eagan, MN, USA, 2010; pp. 247–260. [Google Scholar]
- Bederski, K.; Roistacher, C.N.; Müller, G.W. Cross protection against severe Citrus tristeza virus stem pitting in Peru. Int. Organ. Citrus Virol. Conf. Proc. 2006, 16, 117–126. [Google Scholar]
- Bederski, K.; Roistacher, C.N.; Silvestre, O.P.; Müller, G.W. Long-term cross-protection of severe stem pitting citrus tristeza virus in Peru. Int. Organ. Citrus Virol. Conf. Proc. 2010, 17, 67–79. [Google Scholar]
- Satyanarayana, T.; Gowda, S.; Boyko, V.P.; Albiach-Marti, M.R.; Mawassi, M.; Navas-Castillo, J.; Karasev, A.V.; Dolja, V.; Hilf, M.E.; Lewandowski, D.J.; et al. An engineered closterovirus RNA replicon and analysis of heterologous terminal sequences for replication. Proc. Natl. Acad. Sci. USA 1999, 96, 7433–7438. [Google Scholar] [CrossRef] [Green Version]
- Folimonov, A.S.; Folimonova, S.Y.; Bar-Joseph, M.; Dawson, W.O. A stable RNA virus-based vector for citrus trees. Virology 2007, 368, 205–216. [Google Scholar] [CrossRef] [Green Version]
- Folimonova, S.Y.; Robertson, C.J.; Shilts, T.; Folimonov, A.S.; Hilf, M.E.; Garnsey, S.M.; Dawson, W.O. Infection with strains of Citrus tristeza virus does not exclude superinfection by other strains of the virus. J. Virol. 2010, 84, 1314–1325. [Google Scholar] [CrossRef] [Green Version]
- Folimonova, S.Y. Developing an understanding of cross-protection by citrus tristeza virus. Front. Microbiol. 2013, 4, 76. [Google Scholar] [CrossRef] [Green Version]
- Mawassi, M.; Gafny, R.; Bar-Joseph, M. Nucleotide sequence of the coat protein gene of Citrus tristeza virus: Comparison of biologically diverse isolates collected in Israel. Virus Genes 1993, 73, 265–275. [Google Scholar] [CrossRef] [PubMed]
- Cook, G.; van Vuuren, S.P.; Breytenbach, J.H.J.; Steyn, C.; Burger, J.T.; Maree, H.J. Characterization of Citrus tristeza virus single-variant sources in grapefruit in greenhouse and field trials. Plant Dis. 2016, 100, 2251–2256. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bar-Joseph, M. A historical note on two unreported obstacles for cross-protecting mature citrus trees against severe Citrus tristeza virus isolates. J. Cit. Pathol. 2015, 2, 1–4. [Google Scholar]
- Bar-Joseph, M. Cross protection incompleteness-possible cause for natural spread of citrus tristeza virus after a prolonged lag period in Israel. Phytopathology 1978, 68, 1110–1111. [Google Scholar] [CrossRef]
- Rosner, A.; Ginzburg, I.; Bar-Joseph, M. Molecular cloning of complementary DNA sequences of citrus tristeza virus RNA. J. Gen. Virol. 1983, 64, 1757–1763. [Google Scholar] [CrossRef]
- Rosner, A.; Bar-Joseph, M. Diversity of citrus tristeza virus strains indicated by hybridization with cloned cDNA sequences. Virology 1984, 139, 189–193. [Google Scholar] [CrossRef]
- Sekiya, M.E.; Lawrence, S.D.; McCaffery, M.; Cline, K. Molecular cloning and nucleotide sequencing of the coat protein gene of citrus tristeza virus. J. Gen. Virol. 1991, 72, 1013–1020. [Google Scholar] [CrossRef]
- Bar-Joseph, M.; Garnsey, S.M.; Gonsalves, D. The closteroviruses: A distinct group of elongated plant viruses. Adv. Virus Res. 1979, 25, 93–168. [Google Scholar]
- Dolja, V.V.; Karasev, A.V.; Koonin, E.V. Molecular biology and evolution of closteroviruses: Sophisticated build-up of large RNA genomes. Annu. Rev. Phytopathol. 1994, 32, 261–285. [Google Scholar] [CrossRef]
- Dolja, V.V.; Kreuze, J.F.; Valkonen, J.P.T. Comparative and functional genomics of closteroviruses. Virus Res. 2006, 117, 38–51. [Google Scholar] [CrossRef]
- Agranovsky, A.A. Principles of molecular organization, expression, and evolution of closteroviruses: Over the barriers. Adv. Virus Res. 1996, 47, 119–158. [Google Scholar] [PubMed]
- Agranovsky, A.A. Plant Viruses: Evolution and Management. In Closteroviruses:Molecular Biology, Evolution and Interactions with Cells; Springer: Berlin/Heidelberg, Germany, 2016; pp. 231–252. [Google Scholar]
- Karasev, A.V. Genetic diversity and evolution of closteroviruses. Annu. Rev. Phytopathol. 2000, 38, 293–324. [Google Scholar] [CrossRef] [PubMed]
- Folimonova, S.Y. Citrus tristeza virus: A large RNA virus with complex biology turned into a valuable tool for crop protection. PLoS Pathog. 2020, 16, e1008416. [Google Scholar] [CrossRef] [PubMed]
- Karasev, A.V.; Boyko, V.; Gowda, S.; Nikolaeva, O.V.; Hilf, M.E.; Koonin, E.V.; Niblett, C.L.; Cline, K.; Gumpf, D.J.; Lee, R.F.; et al. Complete sequence of the citrus tristeza virus RNA genome. Virology 1995, 208, 511–520. [Google Scholar] [CrossRef] [Green Version]
- Satyanarayana, T.; Gowda, S.; Mawassi, M.; Albiach-Martí, M.R.; Ayllón, M.A.; Robertson, C.; Garnsey, S.M.; Dawson, W.O. Closterovirus encoded HSP70 homolog and p61 in addition to both coat proteins function in efficient virion assembly. Virology 2000, 278, 253–265. [Google Scholar] [CrossRef] [Green Version]
- Tatineni, S.; Robertson, C.J.; Garnsey, S.M.; Bar-Joseph, M.; Gowda, S.; Dawson, W.O. Three genes of citrus tristeza virus are dispensable for infection and movement throughout some varieties of citrus trees. Virology 2008, 376, 297–307. [Google Scholar] [CrossRef] [Green Version]
- Lu, R.; Folimonov, A.; Shintaku, M.; Li, W.-X.; Falk, B.W.; Dawson, W.O.; Ding, S.-W. Three distinct suppressors of RNA silencing encoded by a 20-kb viral RNA genome. Proc. Natl. Acad. Sci. USA 2004, 101, 15742–15747. [Google Scholar] [CrossRef] [Green Version]
- Tatineni, S.; Robertson, C.J.; Garnsey, S.M.; Dawson, W.O. A plant virus evolved by acquiring multiple nonconserved genes to extend its host range. Proc. Natl. Acad. Sci. USA 2011, 108, 17366–17371. [Google Scholar] [CrossRef] [Green Version]
- Gowda, S.; Ayllon, M.A.; Satyanarayana, T.; Bar-Joseph, M.; Dawson, W.O. Transcription strategy in a Closterovirus: A novel 5′-proximal controller element of citrus tristeza virus produces 5′- and 3′-terminal subgenomic RNAs and differs from 3′ open reading frame controller elements. J. Virol. 2003, 77, 340–352. [Google Scholar] [CrossRef] [Green Version]
- Mawassi, M.; Karasev, A.V.; Mietkiewska, E.; Gafny, R.; Lee, R.F.; Dawson, W.O.; Bar-Joseph, M. Defective RNA molecules associated with citrus tristeza virus. Virology 1995, 208, 383–387. [Google Scholar] [CrossRef] [Green Version]
- Che, X.; Piestun, D.; Mawassi, M.; Yang, G.; Satyanarayana, T.; Gowda, S.; Dawson, W.O.; Bar-Joseph, M. 5′-coterminal subgenomic RNAs in citrus tristeza virus-infected cells. Virology 2001, 283, 374–381. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gowda, S.; Tatineni, S.; Folimonova, S.Y.; Hilf, M.E.; Dawson, W.O. Accumulation of a 5′ proximal subgenomic RNA of citrus tristeza virus is correlated with encapsidation by the minor coat protein. Virology 2009, 389, 122–131. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hilf, M.E.; Karasev, A.V.; Pappu, H.R.; Gumpf, D.J.; Niblett, C.L.; Garnsey, S.M. Characterization of citrus tristeza virus subgenomic RNAs in infected tissue. Virology 1995, 208, 576–582. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karasev, A.V.; Hilf, M.E.; Garnsey, S.M.; Dawson, W.O. Transcriptional strategy of closteroviruses: Mapping the 5′ termini of the citrus tristeza virus subgenomic RNAs. J. Virol. 1997, 71, 6233–6236. [Google Scholar] [CrossRef] [Green Version]
- Bar-Joseph, M.; Mawassi, M. The defective RNAs of Closteroviridae. Front. Microbiol. 2013, 4, 1–6. [Google Scholar] [CrossRef] [Green Version]
- Hilf, M.E.; Mavrodieva, V.A.; Garnsey, S.M. Genetic marker analysis of a global collection of isolates of Citrus tristeza virus: Characterization and distribution of CTV genotypes and association with symptoms. Phytopathology 2005, 95, 909–917. [Google Scholar] [CrossRef] [Green Version]
- Harper, S.J. Citrus tristeza virus: Evolution of complex and varied genotypic groups. Front. Microbiol. 2013, 4, 93. [Google Scholar] [CrossRef] [Green Version]
- Mawassi, M.; Mietkiewska, E.; Gofman, R.; Yang, G.; Bar-Joseph, M. Unusual sequence relationships between two isolates of Citrus tristeza virus. J. Gen. Virol. 1996, 77, 2359–2364. [Google Scholar] [CrossRef]
- López, C.; Ayllón, M.A.; Navas-Castillo, J.; Guerri, J.; Moreno, P.; Flores, R. Molecular variability of the 5′- and 3′-terminal regions of Citrus tristeza virus RNA. Phytopathology 1998, 88, 685–691. [Google Scholar] [CrossRef]
- Kong, P.; Rubio, L.; Polek, M.; Falk, B.W. Population structure and genetic diversity within California Citrus tristeza virus (CTV) field isolates. Virus Genes 2000, 21, 139–145. [Google Scholar] [CrossRef]
- Rubio, L.; Ayllón, M.A.; Kong, P.; Fernández, A.; Polek, M.; Guerri, J.; Moreno, P.; Falk, B.W. Genetic variation of Citrus tristeza virus isolates from California and Spain: Evidence for mixed infections and recombination. J. Virol. 2001, 75, 8054–8062. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roy, A.; Manjunath, K.L.; Brlansky, R.H. Assessment of sequence diversity in the 5′-terminal region of Citrus tristeza virus from India. Virus Res. 2005, 113, 132–142. [Google Scholar] [CrossRef] [PubMed]
- Silva, G.; Marques, N.; Nolasco, G. The evolutionary rate of citrus tristeza virus ranks among the rates of the slowest RNA viruses. J. Gen. Virol. 2012, 93, 419–429. [Google Scholar] [CrossRef] [PubMed]
- Albiach-Martí, M.R.; Mawassi, M.; Gowda, S.; Satyanarayana, T.; Hilf, M.E.; Shanker, S.; Almira, E.C.; Vives, M.C.; Lopez, C.; Guerri, J.; et al. Sequences of Citrus tristeza virus separated in time and space are essentially identical. J. Virol. 2000, 74, 6856–6865. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lbida, B.; Fonseca, F.; Santos, C.; Zemzami, M.; Bennani, A.; Nolasco, G. Genomic variability of Citrus tristeza virus (CTV) isolates introduced into Morocco. Phytopathol. Mediterr. 2004, 43, 205–210. [Google Scholar]
- Jenkins, G.M.; Rambaut, A.; Pybus, O.G.; Holmes, E.C. Rates of molecular evolution in RNA viruses: A quantitative phylogenetic analysis. J. Mol. Evol. 2002, 54, 156–165. [Google Scholar] [CrossRef]
- Sanjuán, R.; Nebot, M.R.; Chirico, N.; Mansky, L.M.; Belshaw, R. Viral mutation rates. J. Virol. 2010, 84, 9733–9748. [Google Scholar] [CrossRef] [Green Version]
- Minskaia, E.; Hertzig, T.; Gorbalenya, A.E.; Campanacci, V.; Cambillau, C.; Canard, B.; Ziebuhr, J. Discovery of an RNA virus 3′→5′ exoribonuclease that is critically involved in coronavirus RNA synthesis. Proc. Natl. Acad. Sci. USA 2006, 103, 5108–5113. [Google Scholar] [CrossRef] [Green Version]
- Eckerle, L.D.; Becker, M.M.; Halpin, R.A.; Li, K.; Venter, E.; Lu, X.; Scherbakova, S.; Graham, R.L.; Baric, R.S.; Stockwell, T.B.; et al. Infidelity of SARS-CoV Nsp14-exonuclease mutant virus replication is revealed by complete genome sequencing. PLoS Pathog. 2010, 6, e1000896. [Google Scholar] [CrossRef] [Green Version]
- Gorbalenya, A.E.; Enjuanes, L.; Ziebuhr, J.; Snijder, E.J. Nidovirales: Evolving the largest RNA virus genome. Virus Res. 2006, 117, 17–37. [Google Scholar] [CrossRef]
- Batuman, O.; Mawassi, M.; Bar-Joseph, M. Transgenes consisting of a dsRNA of an RNAi suppressor plus the 3′ UTR provide resistance to Citrus tristeza virus sequences in Nicotiana benthamiana but not in citrus. Virus Genes 2006, 33, 319–327. [Google Scholar] [PubMed]
- Peña, L.; Fagoaga, C.; Lopez, C.; Dominguez, A.; Ghorbel, R.; de Mendoza, A.H.; Moreno, P.; Navarro, L.; Flores, R. Pathogen-derived resistance to citrus tristeza virus in transgenic citrus plants. In Citrus Tristeza Virus Complex and Tristeza Diseases; Karasev, A.V., Hilf, M.E., Eds.; The American Phytopathological Society Press: Saint Paul, MN, USA, 2010; pp. 203–216. [Google Scholar]
- Cillo, F.; Palukaitis, P. Transgenic resistance. Adv. Virus Res. 2014, 90, 35–146. [Google Scholar] [PubMed]
- Lee, R.F.; Keremane, M.L. Mild strain cross protection of tristeza: A review of research to protect against decline on sour orange in Florida. Front. Microbiol. 2013, 4, 259. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grant, T.J.; Higgins, R.P. Occurrence of mixtures of tristeza virus strains in citrus. Phytopathology 1957, 47, 272–276. [Google Scholar]
- Vives, M.C.; Rubio, L.; Sambade, A.; Mirkov, T.E.; Moreno, P.; Guerri, J. Evidence of multiple recombination events between two RNA sequence variants within a Citrus tristeza virus isolate. Virology 2005, 331, 232–237. [Google Scholar] [CrossRef] [PubMed]
- Roy, A.; Brlansky, R.H. Population dynamics of a Florida Citrus trisreza virus isolate and aphid-transmitted sub-isolates: Identification of three genotypic groups and recombinants after aphid transmission. Phytopathology 2009, 11, 1297–1306. [Google Scholar] [CrossRef]
- Scott, K.A.; Hlela, Q.; Zablocki, O.; Read, D.; van Vuuren, S.; Pietersen, G. Genotype composition of population of grapefruit-cross-protecting citrus tristeza virus strain GFMS12 in different host plants and aphid-transmitted sub-isolates. Arch. Virol. 2013, 158, 27–37. [Google Scholar] [CrossRef] [Green Version]
- Bergua, M.; Kang, S.-H.; Folimonova, S.Y. Understanding superinfection exclusion by complex populations of Citrus tristeza virus. Virology 2016, 499, 331–339. [Google Scholar] [CrossRef]
- Gal-On, A.; Shiboleth, Y.M. Cross protection. In Natural Resistance Mechanisms of Plants to Viruses; Loebenstein, G., Carr, J.P., Eds.; Springer: Dordrecht, The Netherlands, 2006; pp. 261–288. [Google Scholar]
- Folimonova, S.Y.; Harper, S.J.; Leonard, M.T.; Triplett, E.W.; Shilts, T. Superinfection exclusion by Citrus tristeza virus does not correlate with the production of viral small RNAs. Virology 2014, 468, 462–471. [Google Scholar] [CrossRef] [Green Version]
- Folimonova, S.Y. Superinfection exclusion is an active virus–controlled function that requires a specific viral protection. J. Virol. 2012, 86, 5554–5561. [Google Scholar] [CrossRef] [Green Version]
- Atallah, O.O.; Kang, S.-H.; El-Mohtar, C.A.; Shilts, T.; Bergua, M.; Folimonova, S.Y. A 5′-proximal region of the Citrus tristeza virus genome encoding two leader proteases is involved in virus superinfection exclusion. Virology 2016, 489, 108–115. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Domingo, E. Molecular basis of genetic variation of viruses: Error-prone replication. Virus Popul. 2020, 35–71. [Google Scholar]
- Adams, R.H.; Brown, D.T. BHK cells expressing Sindbis virus-induced homologous interference allow the translation of nonstructural genes of superinfecting virus. J. Virol. 1985, 54, 351–357. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Johnston, R.E.; Wan, K.; Bose, H.R. Homologous interference induced by Sindbis virus. J. Virol. 1974, 14, 1076–1082. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karpf, A.R.; Lenches, E.; Strauss, E.G.; Strauss, J.H.; Brown, D.T. Superinfection exclusion of alphaviruses in three mosquito cell lines persistently infected with Sindbis virus. J. Virol. 1997, 71, 7119–7123. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, Y.M.; Tscherne, D.M.; Yun, S.I.; Frolov, I.; Rice, C.M. Dual mechanisms of pestiviral superinfection exclusion at entry and RNA replication. J. Virol. 2005, 79, 3231–3242. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schaller, T.; Appel, N.; Koutsoudakis, G.; Kallis, S.; Lohmann, V.; Pietschmann, T.; Bartenschlager, R. Analysis of hepatitis C virus superinfection exclusion by using novel fluorochrome gene-tagged viral genomes. J. Virol. 2007, 81, 4591–4603. [Google Scholar] [CrossRef] [Green Version]
- Zou, G.; Zhang, B.; Lim, P.Y.; Yuan, Z.; Bernard, K.A.; Shi, P.Y. Exclusion of West Nile virus superinfection through RNA replication. J. Virol. 2009, 83, 11765–11776. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.F.; Sun, R.; Guo, Q.; Zhang, S.; Meulia, T.; Halfmann, R.; Li, D.; Qu, F. A self-perpetuating repressive state of a viral replication protein blocks superinfection by the same virus. PLoS Pathog. 2017, 13, e1006253. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.F.; Zhang, S.; Guo, Q.; Sun, R.; Wei, T.; Qu, F. A new mechanistic model for viral cross protection and superinfection exclusion. Front. Plant Sci. 2018, 9, 40. [Google Scholar] [CrossRef] [Green Version]
- Nagy, P.D.; Strating, J.R.P.M.; van Kuppeveld, F.J.M. Building Viral Replication Organelles: Close Encounters of the Membrane Types. PLoS Pathog. 2016, 12, e1005912. [Google Scholar] [CrossRef] [PubMed]
- Jin, X.; Cao, X.; Wang, X.; Jiang, J.; Wan, J.; Laliberté, J.F.; Zhang, Y. Three-dimensional architecture and biogenesis of membrane structures associated with plant virus replication. Front. Plant Sci. 2018, 9, 57. [Google Scholar] [CrossRef] [PubMed]
- Wolff, G.; Melia, C.E.; Snijder, E.J.; Bárcena, M. Double-membrane vesicles as platforms for viral replication. Trends Microbiol. 2020. [Google Scholar] [CrossRef] [PubMed]
- Esau, K.; Hoefert, L.L. Cytology of beet yellows virus infection in Tetragonia. I. Parenchyma cells in infected leaf. Protoplasma 1971, 72, 255–273. [Google Scholar] [CrossRef]
- Wang, J.; Stewart, L.R.; Kiss, Z.; Falk, B.W. Lettuce infectious yellows virus (LIYV) RNA 1-encoded P34 is an RNA-binding protein and exhibits perinuclear localization. Virology 2010, 403, 67–77. [Google Scholar] [CrossRef] [Green Version]
- Erokhina, T.N.; Vitushkina, M.V.; Zinovkin, R.A.; Lesemann, D.E.; Jelkmann, W.; Koonin, E.V.; Agranovsky, A.A. Ultrastructural localization and epitope mapping of the methyltransferase-like and helicase-like proteins of Beet yellows virus. J. Gen. Virol. 2001, 82, 1983–1994. [Google Scholar] [CrossRef]
- Zinovkin, R.A.; Erokhina, T.N.; Lesemann, D.E.; Jelkmann, W.; Agranovsky, A.A. Processing and subcellular localization of the leader papain-like proteinase of Beet yellows closterovirus. J. Gen. Virol. 2003, 84, 2265–2270. [Google Scholar] [CrossRef]
- Gushchin, V.A.; Solovyev, A.G.; Erokhina, T.N.; Morozov, S.Y.; Agranovsky, A.A. Beet yellows virus replicase and replicative compartments: Parallels with other RNA viruses. Front. Microbiol. 2013, 4, 38. [Google Scholar] [CrossRef] [Green Version]
- Tilsner, J.; Oparka, K.J. Missing links?—The connection between replication and movement of plant RNA viruses. Curr. Opin. Virol. 2012, 2, 705–711. [Google Scholar] [CrossRef]
- Tilsner, J.; Linnik, O.; Louveaux, M.; Roberts, I.M.; Chapman, S.N.; Oparka, K.J. Replication and trafficking of a plant virus are coupled at the entrances of plasmodesmata. J. Cell Biol. 2013, 201, 981–995. [Google Scholar] [CrossRef] [Green Version]
- Grangeon, R.; Jian, J.; Wan, J.; Agbeci, M.; Zheng, H.; Laliberté, J.F. 6K2-induced vesicles can move cell to cell during turnip mosaic virus infection. Front. Microbiol. 2013, 4, 351. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wan, J.; Cabanillas, D.G.; Zheng, H.; Laliberté, J.F. Turnip mosaic virus moves systemically through both phloem and xylem as membrane-associated complexes. Plant Physiol. 2015, 167, 1374–1388. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wan, J.; Laliberté, J.F. Membrane-associated virus replication complexes locate to plant conducting tubes. Plant Signal. Behav. 2015, 10, e1042639. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Folimonova, S.Y.; Folimonov, A.S.; Tatineni, S.; Dawson, W.O. Citrus tristeza virus: Survival at the edge of the movement continuum. J. Virol. 2008, 82, 6546–6556. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bergua, M.; Zwart, M.P.; El-Mohtar, C.; Shilts, T.; Elena, S.F.; Folimonova, S.Y. A viral protein mediates superinfection exclusion at the whole-organism level but is not required for exclusion at the cellular level. J. Virol. 2014, 88, 11327–11338. [Google Scholar] [CrossRef] [Green Version]
- Tennant, P.F.; Gonsalves, C.; Ling, K.S.; Fitch, M.; Manshardt, R.; Slightom, J.L.; Gonsalves, D. Differential protection against papaya ringspot virus isolates in coat protein gene transgenic papaya and classically cross-protected papaya. Phytopathology 1994, 84, 1359–1365. [Google Scholar] [CrossRef] [Green Version]
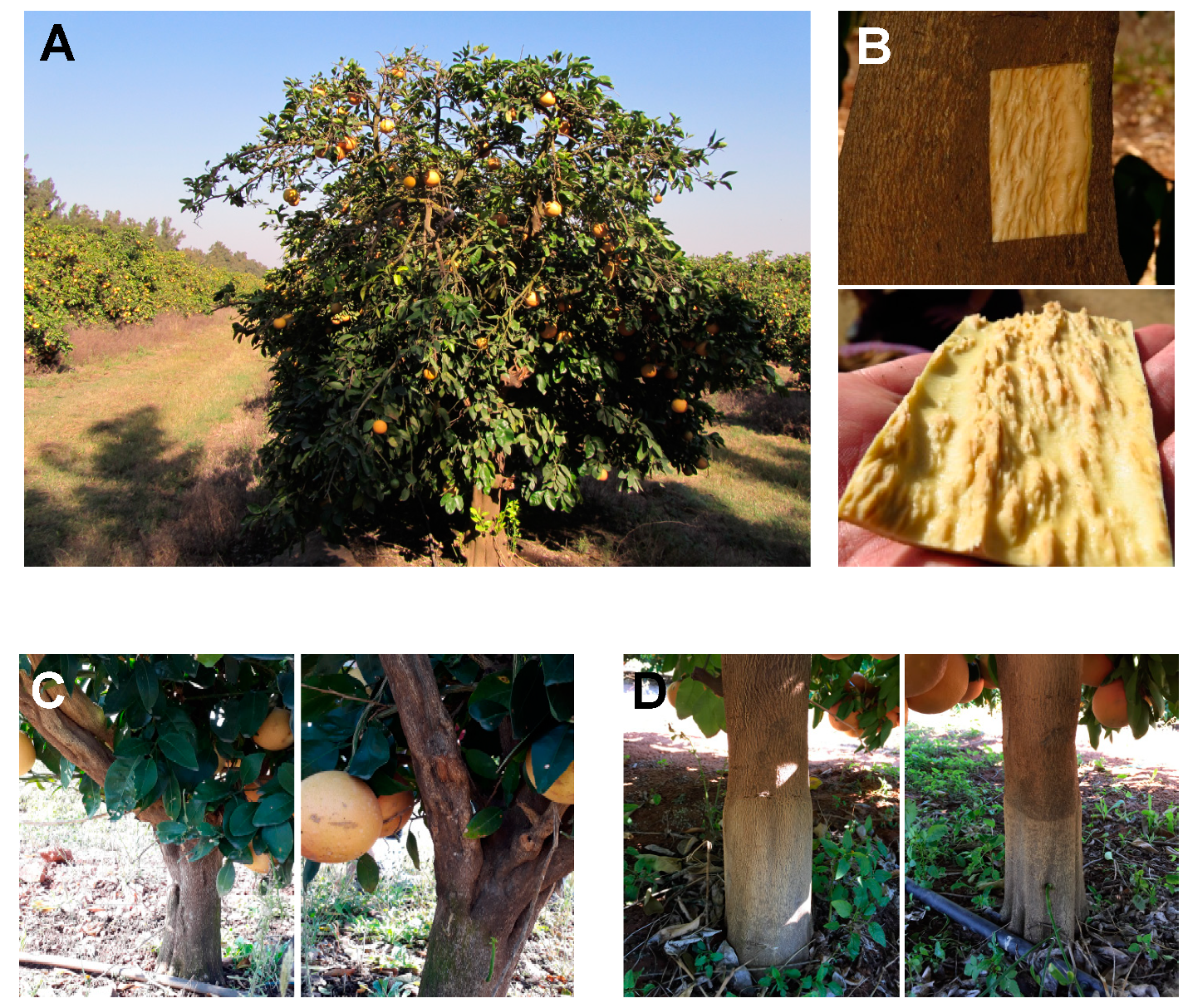
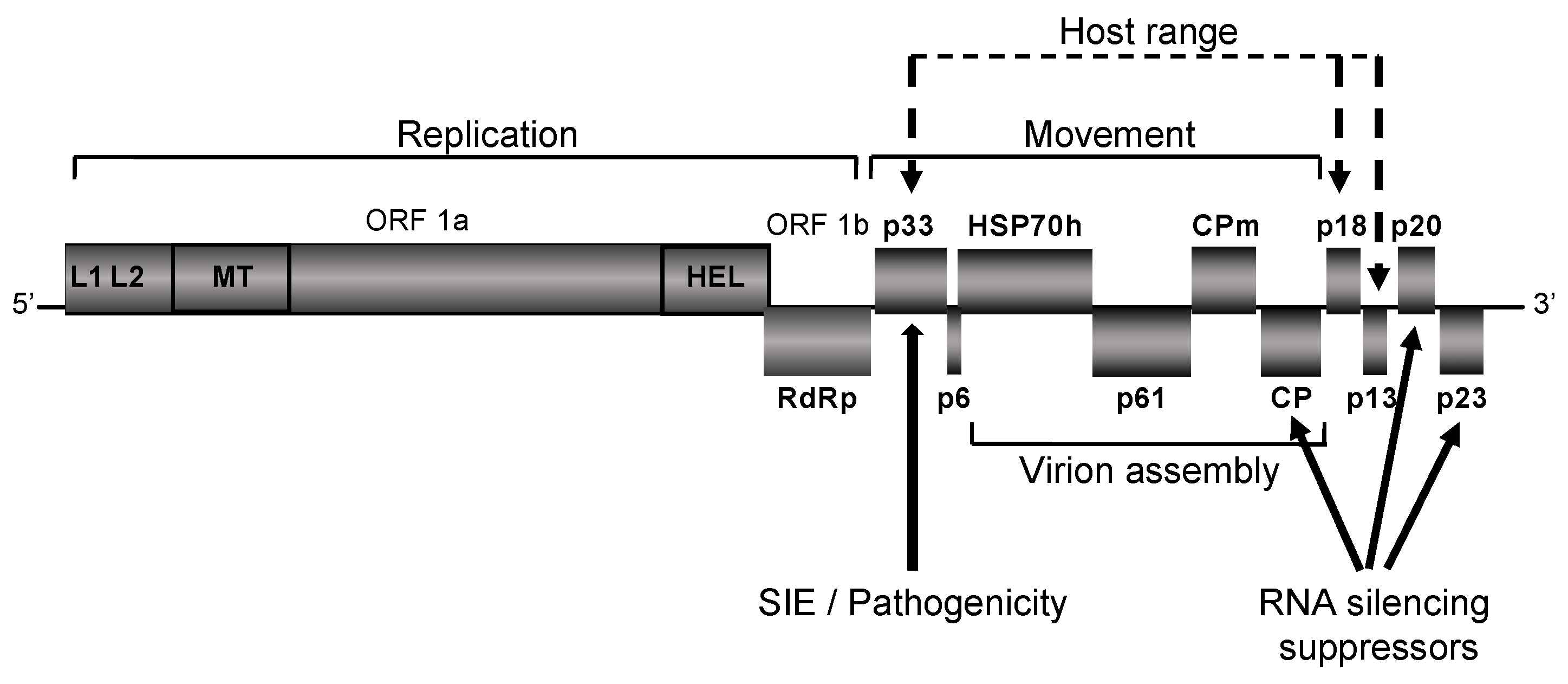
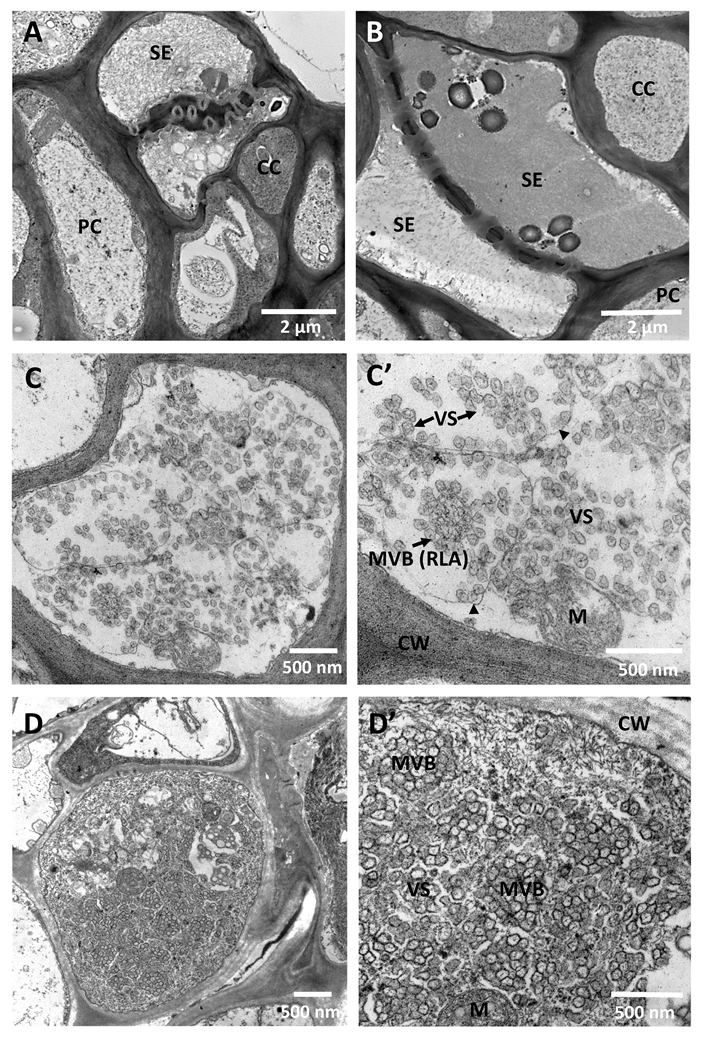
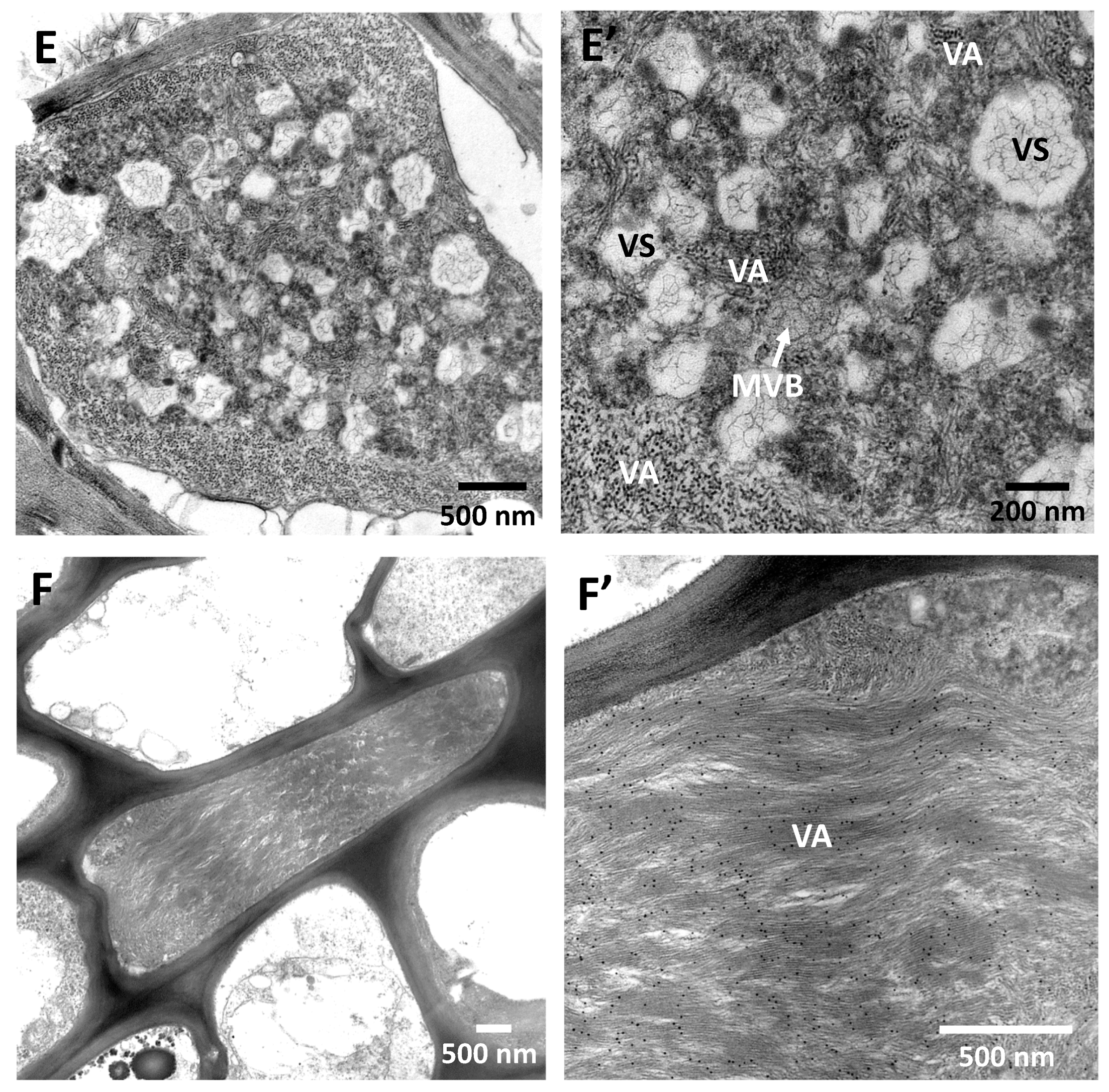
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Folimonova, S.Y.; Achor, D.; Bar-Joseph, M. Walking Together: Cross-Protection, Genome Conservation, and the Replication Machinery of Citrus tristeza virus. Viruses 2020, 12, 1353. https://doi.org/10.3390/v12121353
Folimonova SY, Achor D, Bar-Joseph M. Walking Together: Cross-Protection, Genome Conservation, and the Replication Machinery of Citrus tristeza virus. Viruses. 2020; 12(12):1353. https://doi.org/10.3390/v12121353
Chicago/Turabian StyleFolimonova, Svetlana Y., Diann Achor, and Moshe Bar-Joseph. 2020. "Walking Together: Cross-Protection, Genome Conservation, and the Replication Machinery of Citrus tristeza virus" Viruses 12, no. 12: 1353. https://doi.org/10.3390/v12121353
APA StyleFolimonova, S. Y., Achor, D., & Bar-Joseph, M. (2020). Walking Together: Cross-Protection, Genome Conservation, and the Replication Machinery of Citrus tristeza virus. Viruses, 12(12), 1353. https://doi.org/10.3390/v12121353






