Requirements for the Packaging of Geminivirus Circular Single-Stranded DNA: Effect of DNA Length and Coat Protein Sequence
Abstract
1. Introduction
2. Materials and Methods
2.1. Plasmids for the Expression of Geminivirus Coat Proteins
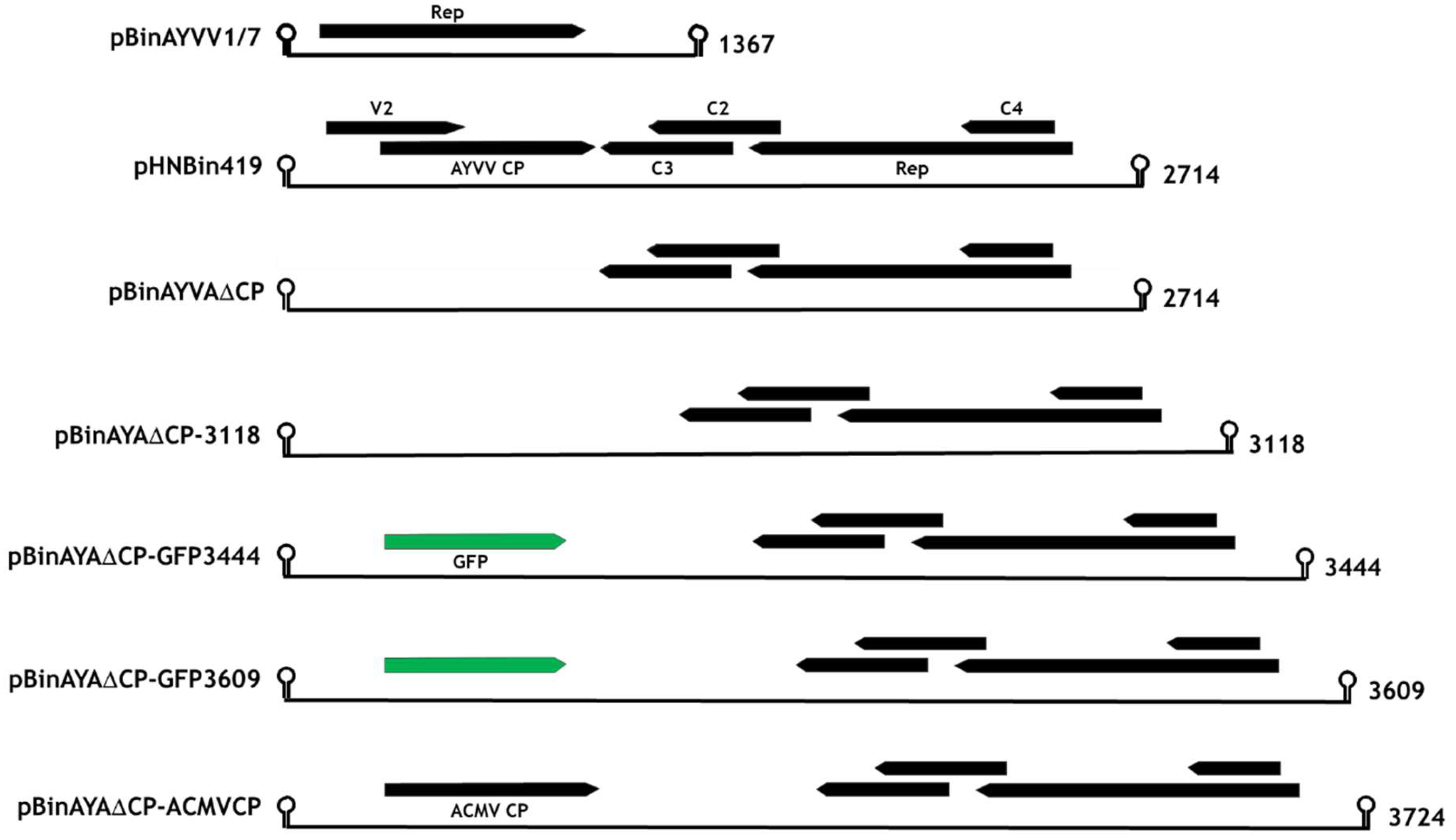
2.2. Replicating Gene Constructs
2.3. Isolation of Virus-Like Particles
2.4. Negative Stain Transmission Electron Microscopy
3. Results
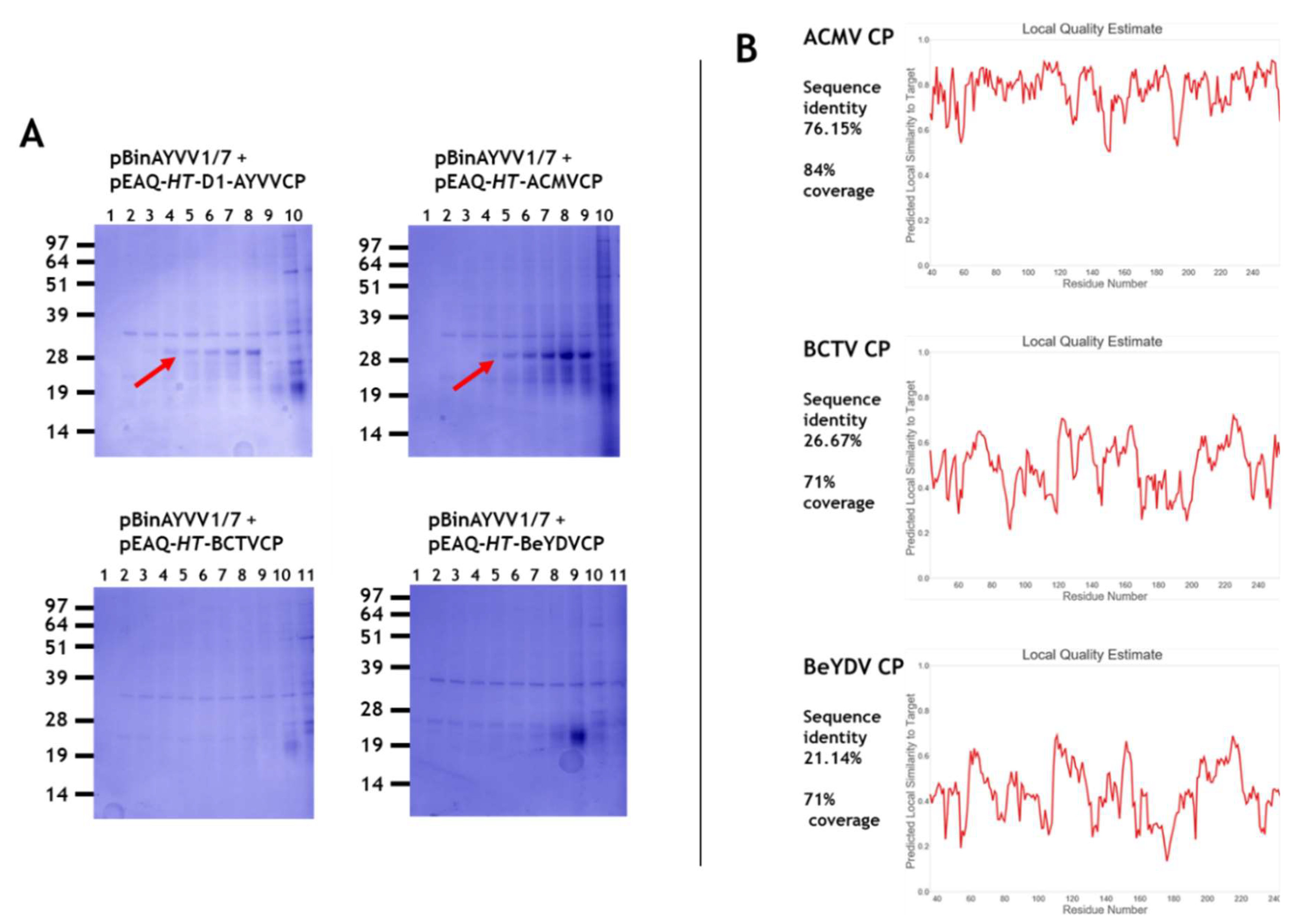
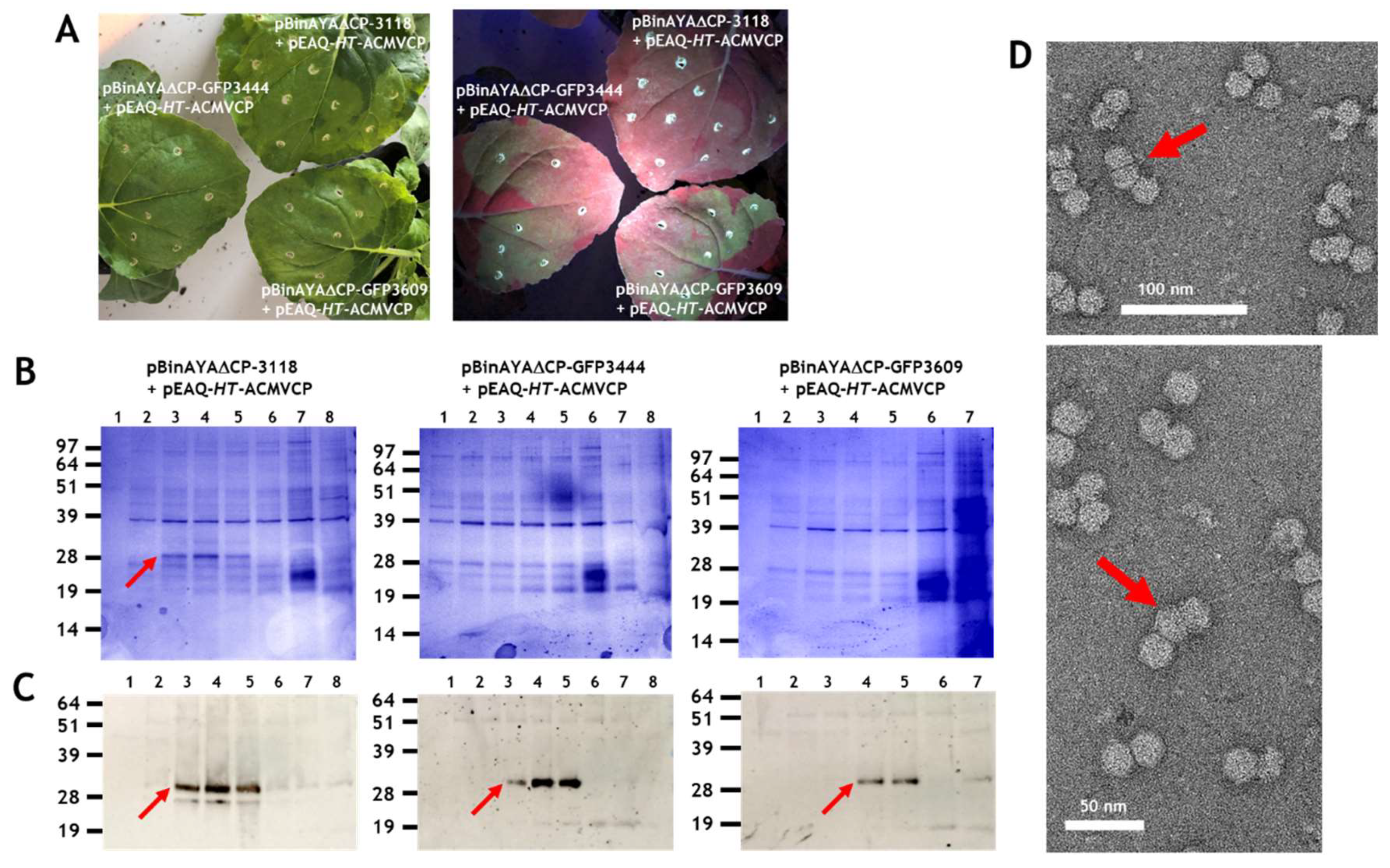
3.1. Transencapsidation of Alphasatellite DNA by Geminivirus Coat Proteins
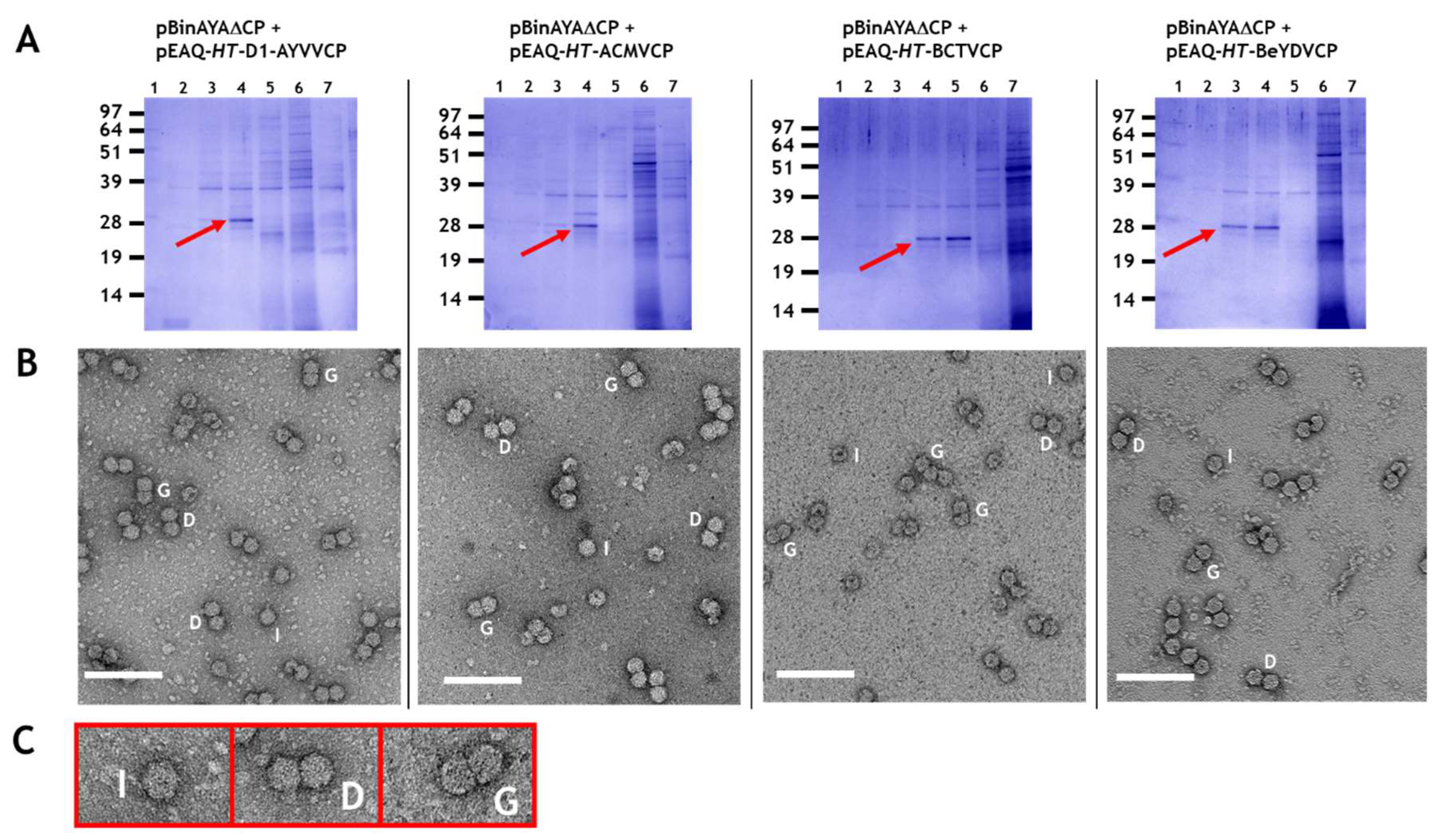
3.2. Transencapsidation of AYVV Genome-Length DNA
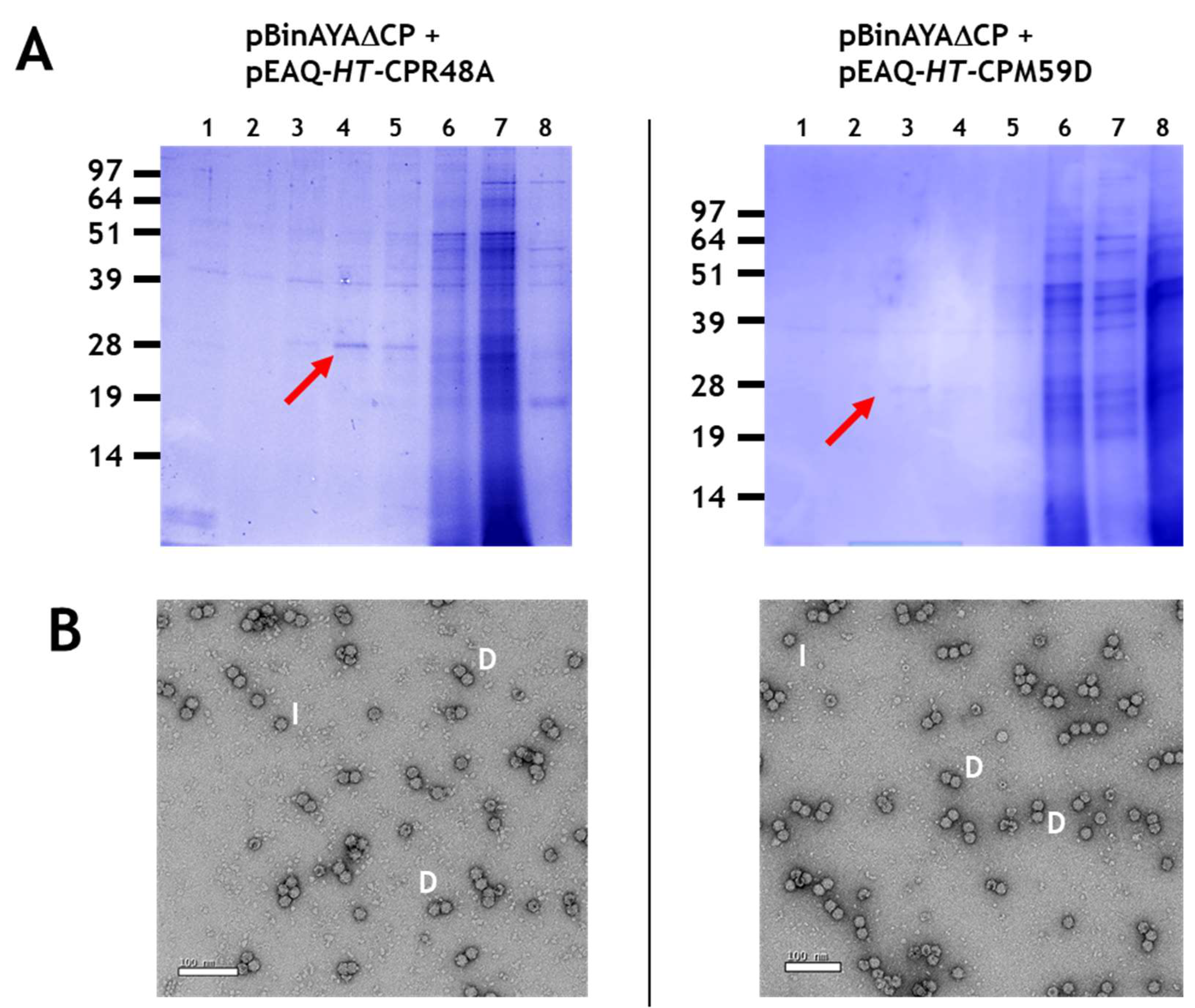
3.3. Effect on Particle Morphology of Increasing DNA Length
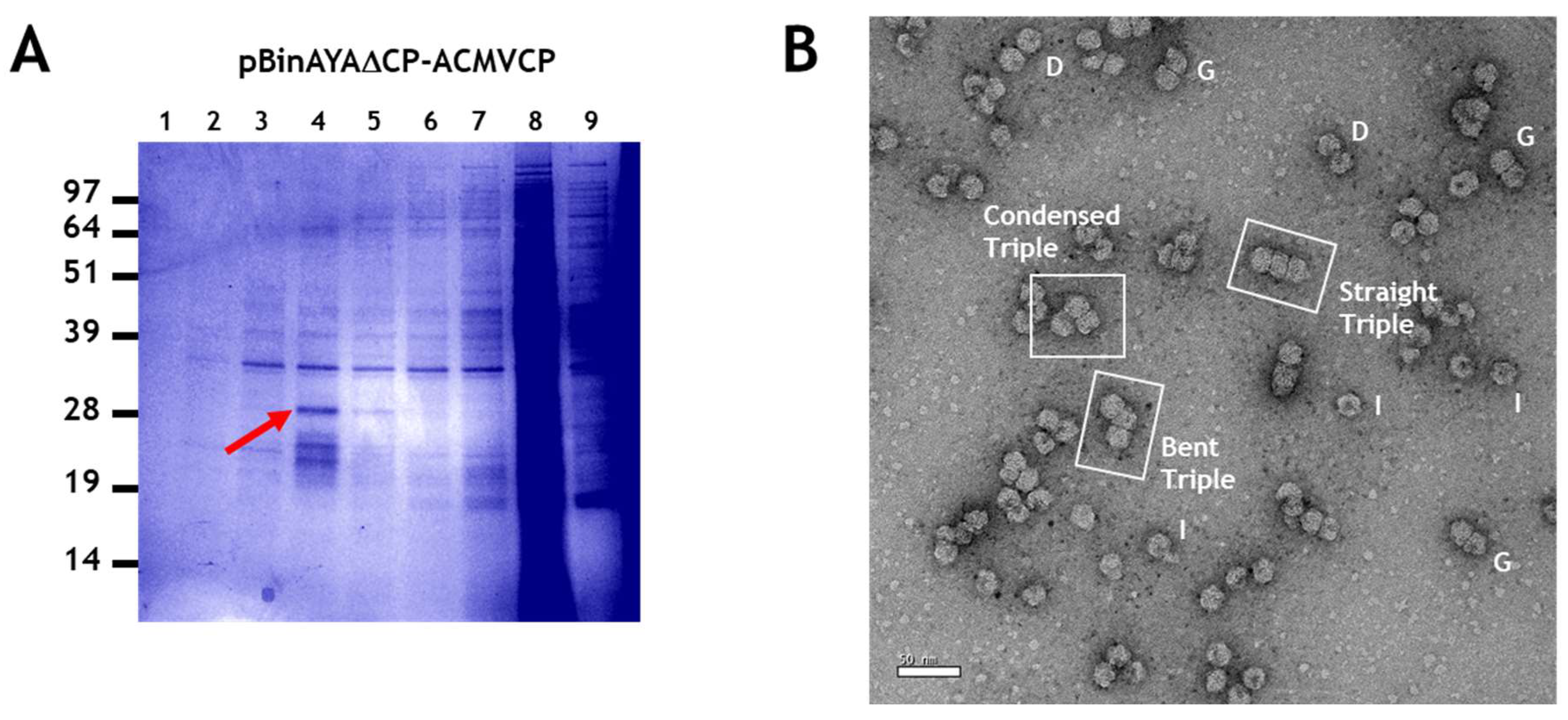
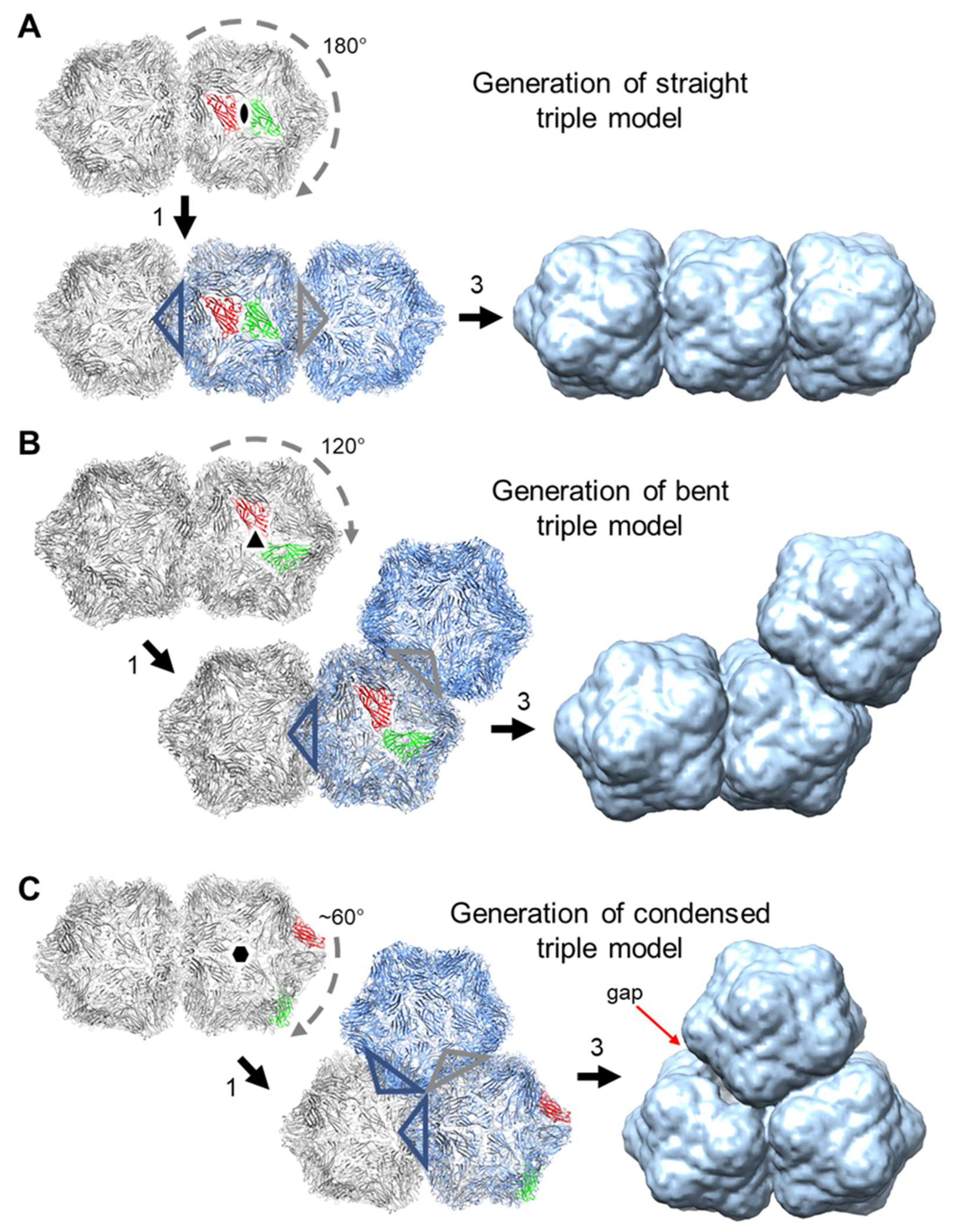
3.4. Generation of Triple Geminate Structures Using a Replicating Vector
3.5. Single Particle Analysis of Triple Particles
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Zerbini, F.M.; Briddon, R.W.; Idris, A.; Martin, D.P.; Moriones, E.; Navas-Castillo, J.; Rivera-Bustamante, R.; Roumagnac, P.; Varsani, A.; ICTV Report Consortium. ICTV virus taxonomy profile: Geminiviridae. J. Gen. Virol. 2017, 98, 131–133. [Google Scholar] [CrossRef]
- Varsani, A.; Roumagnac, P.; Fuchs, M.; Navas-Castillo, J.; Moriones, E.; Idris, A.; Briddon, R.W.; Rivera-Bustamante, R.; Murilo Zerbini, F.; Martin, D.P. Capulavirus and grablovirus: Two new genera in the family geminiviridae. Arch. Virol. 2017, 162, 1819–1831. [Google Scholar] [CrossRef] [PubMed]
- Rojas, M.R.; Macedo, M.A.; Maliano, M.R.; Soto-Aguilar, M.; Souza, J.O.; Briddon, R.W.; Kenyon, L.; Rivera-Bustamante, R.F.; Zerbini, F.M.; Adkins, S.; et al. World management of geminiviruses. Annu. Rev. Phytopathol. 2018, 56, 637–677. [Google Scholar] [CrossRef] [PubMed]
- Navas-Castillo, J.; Fiallo-Olivé, E.; Sánchez-Campos, S. Emerging virus diseases transmitted by whiteflies. Annu. Rev. Phytopathol. 2011, 49, 219–248. [Google Scholar] [CrossRef] [PubMed]
- Gilbertson, R.L.; Batuman, O.; Webster, C.G.; Adkins, S. Role of the insect supervectors Bemisia tabaci and Frankliniella occidentalis in the emergence and global spread of plant viruses. Annu. Rev. Virol. 2015, 2, 67–93. [Google Scholar] [CrossRef] [PubMed]
- Harrison, B.D.; Barker, H.; Bock, K.R.; Guthrie, E.J.; Meredith, G.; Atkinson, M. Plant-viruses with circular single-stranded-DNA. Nature 1977, 270, 760–762. [Google Scholar] [CrossRef]
- Jeske, H. Geminiviruses. Curr. Top. Microbiol. Immunol. 2009, 331, 185–226. [Google Scholar]
- Saunders, K.; Bedford, I.D.; Briddon, R.W.; Markham, P.G.; Wong, S.M.; Stanley, J. A unique virus complex causes Ageratum yellow vein disease. Proc. Natl. Acad. Sci. USA 2000, 97, 6890–6895. [Google Scholar] [CrossRef]
- Briddon, R.W.; Mansoor, S.; Bedford, I.D.; Pinner, M.S.; Saunders, K.; Stanley, J.; Zafar, Y.; Malik, K.A.; Markham, P.G. Identification of DNA components required for induction of cotton leaf curl disease. Virology 2001, 285, 234–243. [Google Scholar] [CrossRef]
- Mansoor, S.; Khan, S.H.; Bashir, A.; Saeed, M.; Zafar, Y.; Malik, K.A.; Briddon, R.; Stanley, J.; Markham, P.G. Identification of a novel circular single-stranded DNA associated with cotton leaf curl disease in Pakistan. Virology 1999, 259, 190–199. [Google Scholar] [CrossRef] [PubMed]
- Saunders, K.; Stanley, J. A nanovirus-like DNA component associated with yellow vein disease of Ageratum conyzoides: Evidence for interfamilial recombination between plant DNA viruses. Virology 1999, 264, 142–152. [Google Scholar] [PubMed]
- Shahid, M.S.; Ikegami, M.; Waheed, A.; Briddon, R.W.; Natsuake, K.T. Association of an alphasatellite with tomato yellow leaf curl virus and ageratum yellow vein virus in Japan is suggestive of a recent introduction. Viruses 2014, 6, 189–200. [Google Scholar]
- Gelbart, D.; Chen, L.; Alon, T.; Dobrinin, S.; Levin, I.; Lapidot, M. The recent association of a DNA betasatellite with tomato yellow leaf curl virus in Israel—A new threat to tomato production. Crop. Prot. 2019, 128, 104995. [Google Scholar]
- Conlon, D.; Granier, M.; Tiendrebeogo, F.; Gentit, P.; Peterschmitt, M.; Urbino, C. Accumulation and transmission of alphasatellite, betasatellite and tomato yellow leaf curl virus in susceptible and Ty-1-rsistant tomato plants. Virus Res. 2018, 253, 124–134. [Google Scholar]
- Ito, T.; Kimbara, J.; Sharma, P.; Ikegami, M. Interaction of tomato yellow leaf curl virus with diverse betasatellites enhances symptom severity. Arch. Virol. 2009, 154, 1233–1239. [Google Scholar]
- Claverie, S.; Varsani, A.; Hoareau, M.; Filloux, D.; Roumagnac, P.; Martin, D.P.; Lefeuvre, P.; Lett, J.-M. Sorghum mastrevirus-associated alphasatellites: New geminialphasatellites associated with an African streak mastrevirus infecting wild Poaceae plants on Reunion Island. Arch. Virol. 2020. [Google Scholar] [CrossRef]
- Patil, B.L.; Dutt, N.; Briddon, R.W.; Bull, S.E.; Rothenstein, D.; Borah, B.K.; Dasgupta, I.; Stanley, J.; Jeske, H. Deletion and recombination events between the DNA-A and DNA-B components of Indian cassava-infecting geminiviruses generate defective molecules in Nicotiana benthamiana. Virus Res. 2007, 124, 59–67. [Google Scholar]
- Stanley, J.; Saunders, K.; Pinner, M.S.; Wong, S.M. Novel defective interfering DNAs associated with ageratum yellow vein geminivirus infection of Ageratum conyzoides. Virology 1997, 239, 87–96. [Google Scholar]
- Saunders, K.; Bedford, I.D.; Stanley, J. Pathogenicity of a natural recombinant associated with ageratum yellow vein disease: Implications for geminivirus evolution and disease aetiology. Virology 2001, 282, 38–47. [Google Scholar]
- Briddon, R.W.; Watts, J.; Markham, P.G.; Stanley, J. The coat protein of beet curly top virus is essential for infectivity. Virology 1989, 172, 628–633. [Google Scholar]
- He, Y.-Z.; Wang, Y.-M.; Yin, T.-Y.; Fiallo-Olive, E.; Liu, Y.-Q.; Hanley-Bowdoin, L.; Wang, X.-W. A plant DNA virus replicates in the salivary glands of its insect vector via recruitment of host DNA synthesis machinery. Proc. Natl. Acad. Sci. USA 2020, 117, 16928–16937. [Google Scholar]
- Zhang, W.; Olson, N.H.; Baker, T.S.; Faulkner, L.; Agbandje-McKenna, M.; Boulton, M.I.; Davies, J.W.; McKenna, R. Structure of the maize streak virus geminate particle. Virology 2001, 279, 471–477. [Google Scholar] [CrossRef] [PubMed]
- Bottcher, B.; Unseld, S.; Ceulemans, H.; Russell, R.B.; Jeske, H. Geminate structures of African cassava mosaic virus. J. Virol. 2004, 78, 6758–6765. [Google Scholar] [CrossRef] [PubMed]
- Hipp, K.; Grimm, C.; Jeske, H.; Bottcher, B. Near-atomic resolution structure of a plant geminivirus determined by electron cryomicroscopy. Structure 2017, 25, 1303–1309. [Google Scholar] [CrossRef]
- Casado, C.G.; Javier Ortiz, G.; Padron, E.; Bean, S.J.; McKenna, R.; Agbandje-McKenna, M.; Boulton, M.I. Isolation and characterization of subgenomic DNAs encapsidated in “single” T = 1 isometric particles of maize streak virus. Virology 2004, 323, 164–171. [Google Scholar] [CrossRef] [PubMed]
- Stanley, J.; Townsend, R. Characterisation of DNA forms associated with cassava latent virus infection. Nucleic Acids Res. 1985, 13, 2189–2206. [Google Scholar] [CrossRef]
- Gronenborn, B. Nanoviruses: Genome organisation and protein function. Vet. Microbiol. 2004, 98, 103–109. [Google Scholar] [CrossRef]
- Larsen, R.C.; Duffus, J.E. A simplified procedure for the purification of curly top virus and the isolation of its monomer and dimer particles. Phytopathology 1983, 73, 114–118. [Google Scholar] [CrossRef]
- Frischmuth, T.; Ringel, M.; Kocher, C. The size of encapsidated single-stranded DNA determines the multiplicity of African cassava mosaic virus particles. J. Gen. Virol. 2001, 82, 673–676. [Google Scholar] [CrossRef]
- Jovel, J.; Preiß, W.; Jeske, H. Characterization of DNA intermediates of an arising geminivirus. Virus Res. 2007, 130, 63–70. [Google Scholar] [CrossRef]
- Hooker, W.J.; Salazar, L.F. A new plant virus from the high jungle of the Eastern Andes; Solanum apical leaf curling virus (SALCV). Ann. Appl. Biol. 1983, 103, 449–454. [Google Scholar] [CrossRef]
- Hooker, W.J.; Salazar, L.F.; Brown, C.R. Field infection of potato by the Solanum apical leaf curling virus (SALCV). Am. Potato J. 1985, 62, 263–272. [Google Scholar] [CrossRef]
- Harrison, B.D.; Duncan, G.H.; Roberts, I.M.; Robinson, D.J. Genome and Serological Relationships among Geminiviruses; Fourth Annual Report 1984; Scottish Crop Research Institute: Dundee, UK, 1985; pp. 179–180. ISSN 0263-7200. [Google Scholar]
- Loconsole, G.; Saldarelli, P.; Doddapaneni, H.; Savino, V.; Martelli, G.P.; Saponari, M. Identification of a single-stranded DNA virus associated with citrus chlorotic dwarf disease, a new member in the family geminiviridae. Virology 2012, 432, 162–172. [Google Scholar] [CrossRef]
- Zhang, S.; Shen, P.; Li, M.; Tian, X.; Zhou, C.; Cao, M. Discovery of a novel geminivirus associated with camellia chlorotic dwarf disease. Arch. Virol. 2018, 163, 1709–1712. [Google Scholar]
- Sudarshana, M.R.; Perry, K.L.; Fuchs, M.F. Grapevine red blotch-associated virus, an emerging threat to the grapevine industry. Phytopathology 2015, 105, 1026–1032. [Google Scholar] [CrossRef] [PubMed]
- Fontenele, R.S.; Abreu, R.A.; Lamas, N.S.; Alves-Freitas, D.M.T.; Vida, A.H.; Poppiel, R.R.; Melo, F.L.; Lacorte, C.; Martin, D.P.; Campos, M.A.; et al. Passion Fruit Chlorotic Mottle Virus: Molecular Characterization of a New Divergent Geminivirus in Brazil. Viruses 2018, 10, 169. [Google Scholar] [CrossRef]
- Hesketh, E.L.; Saunders, K.; Fisher, C.; Potze, J.; Stanley, J.; Lomonossoff, G.P.; Ranson, N.A. The 3.3 A structure of a plant geminivirus using cryo-EM. Nat. Commun. 2018, 9, 2369. [Google Scholar]
- Sainsbury, F.; Thuenemann, E.C.; Lomonossoff, G.P. pEAQ: Versatile expression vectors for easy and quick transient expression of heterologous proteins in plants. Plant. Biotechnol. J. 2009, 7, 682–693. [Google Scholar] [CrossRef] [PubMed]
- Klinkenberg, F.A.; Ellwood, S.; Stanley, J. Fate of African cassava mosaic virus coat protein deletion mutants after agroinoculation. J. Gen. Virol. 1995, 70, 1837–1844. [Google Scholar] [CrossRef]
- Liu, L.; van Tonder, T.; Pieterersen, G.; Davies, J.W.; Stanley, J. Molecular characterization of a subgroup I geminivirus from a legume in South Africa. J. Gen. Virol. 1997, 78, 2113–2117. [Google Scholar] [CrossRef] [PubMed]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; de Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef] [PubMed]
- Tan, P.H.N.; Wong, S.M.; Wu, M.; Bedford, I.D.; Saunders, K.; Stanley, J. Genome organization of Ageratum yellow vein virus, a monopartite whitefly transmitted geminivirus isolated from a common weed. J. Gen. Virol. 1995, 76, 2915–2922. [Google Scholar] [CrossRef]
- van Engelen, F.A.; Molthoff, J.W.; Conner, A.J.; Nap, J.-P.; Pereira, A.; Stiekema, W.J. pBINPLUS: An improved plant transformation vector based on pBIN19. Transgenic Res. 1995, 4, 288–290. [Google Scholar] [CrossRef] [PubMed]
- Punjani, A.; Rubinstein, J.L.; Fleet, D.J.; Brubaker, M.A. CryoSPARC: Algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 2017, 14, 290–296. [Google Scholar] [CrossRef]
- Saunders, K.; Bedford, I.D.; Stanley, J. Adaptation from whitefly to leafhopper transmission of an autonomously replicating nanovirus-like DNA component associated with ageratum yellow vein disease. J. Gen. Virol. 2002, 83, 907–913. [Google Scholar] [CrossRef] [PubMed]
- Gorovits, R.; Moshe, A.; Kolot, M.; Sobol, I.; Czosnek, H. Progressive aggregation of Tomato yellow leaf curl virus coat protein in systemically infected tomato plants, susceptible and resistant to the virus. Virus Res. 2013, 171, 33–43. [Google Scholar] [CrossRef] [PubMed]
- Bennett, A.; Rodriguez, D.; Lister, S.; Boulton, M.; McKenna, R.; Agbandje-McKenna, M. Assembly and disassembly intermediates of maize streak geminivirus. Virology 2018, 525, 224–236. [Google Scholar] [CrossRef] [PubMed]
- Hipp, K.; Zikeli, K.; Kepp, G.; Schmid, L.; Shoeman, R.L.; Jurkowski, T.P.; Kleinow, T.; Jeske, H. Different forms of African cassava mosaic virus capsid protein within plants and virions. Virology 2019, 529, 81–90. [Google Scholar] [CrossRef]
- Saunders, K.; Lucy, A.; Stanley, J. DNA forms of the geminivirus African cassava mosaic virus consistent with a rolling circle mechanism of replication. Nucleic Acids Res. 1991, 19, 2325–2330. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef]
- Bennett, A.; Agbandje-McKenna, M. Geminivirus structure and assembly. Adv. Virus Res. 2020, 108, 1–32. [Google Scholar]
- Caspar, D.L.D.; Klug, A. Physical principles in the construction of regular viruses. Cold Spring Harb. Symp. Quant. Biol. 1962, 27, 1–24. [Google Scholar] [CrossRef]
- Hesketh, E.L.; Meshcheriakova, Y.; Thompson, R.F.; Lomonossoff, G.P.; Ranson, N.A. The structures of a naturally empty cowpea mosaic virus particle and its genome-containing counterpart by cryo-electron microscopy. Sci. Rep. 2017, 7, 539. [Google Scholar] [CrossRef]
- Fiallo-Olivé, E.; Yamila Martínez-Zubiaur, Y.; Moriones, E.; Navas-Castillo, J. A novel class of DNA satellites associated with New World begomoviruses. Virology 2012, 426, 1–6. [Google Scholar] [CrossRef]
- Lozano, G.; Trenado, H.P.; Fiallo-Olive, E.; Chirinos, D.; Geraud-Pouey, F.; Briddon, R.W.; Navas-Castillo, J. Characterization of NONcoding DNA Satellites associated with Sweepoviruses (Genus Begomovirus, Geminiviridae)—Definition of a distinct class of Begomovirus-associated satellites. Front. Microbiol. 2016, 7, 162. [Google Scholar] [CrossRef] [PubMed]
- Dry, I.B.; Krake, L.R.; Rigden, J.E.; Rezaian, M.A. A novel subviral agent associated with a geminivirus: The first report of a DNA satellite. Proc. Natl. Acad. Sci. USA 1997, 94, 7088–7093. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Saunders, K.; Richardson, J.; Lawson, D.M.; Lomonossoff, G.P. Requirements for the Packaging of Geminivirus Circular Single-Stranded DNA: Effect of DNA Length and Coat Protein Sequence. Viruses 2020, 12, 1235. https://doi.org/10.3390/v12111235
Saunders K, Richardson J, Lawson DM, Lomonossoff GP. Requirements for the Packaging of Geminivirus Circular Single-Stranded DNA: Effect of DNA Length and Coat Protein Sequence. Viruses. 2020; 12(11):1235. https://doi.org/10.3390/v12111235
Chicago/Turabian StyleSaunders, Keith, Jake Richardson, David M. Lawson, and George P. Lomonossoff. 2020. "Requirements for the Packaging of Geminivirus Circular Single-Stranded DNA: Effect of DNA Length and Coat Protein Sequence" Viruses 12, no. 11: 1235. https://doi.org/10.3390/v12111235
APA StyleSaunders, K., Richardson, J., Lawson, D. M., & Lomonossoff, G. P. (2020). Requirements for the Packaging of Geminivirus Circular Single-Stranded DNA: Effect of DNA Length and Coat Protein Sequence. Viruses, 12(11), 1235. https://doi.org/10.3390/v12111235




