The Antiviral Small-Interfering RNA Pathway Induces Zika Virus Resistance in Transgenic Aedes aegypti
Abstract
1. Introduction
2. Materials and Methods
2.1. Mosquito Rearing and Maintenance
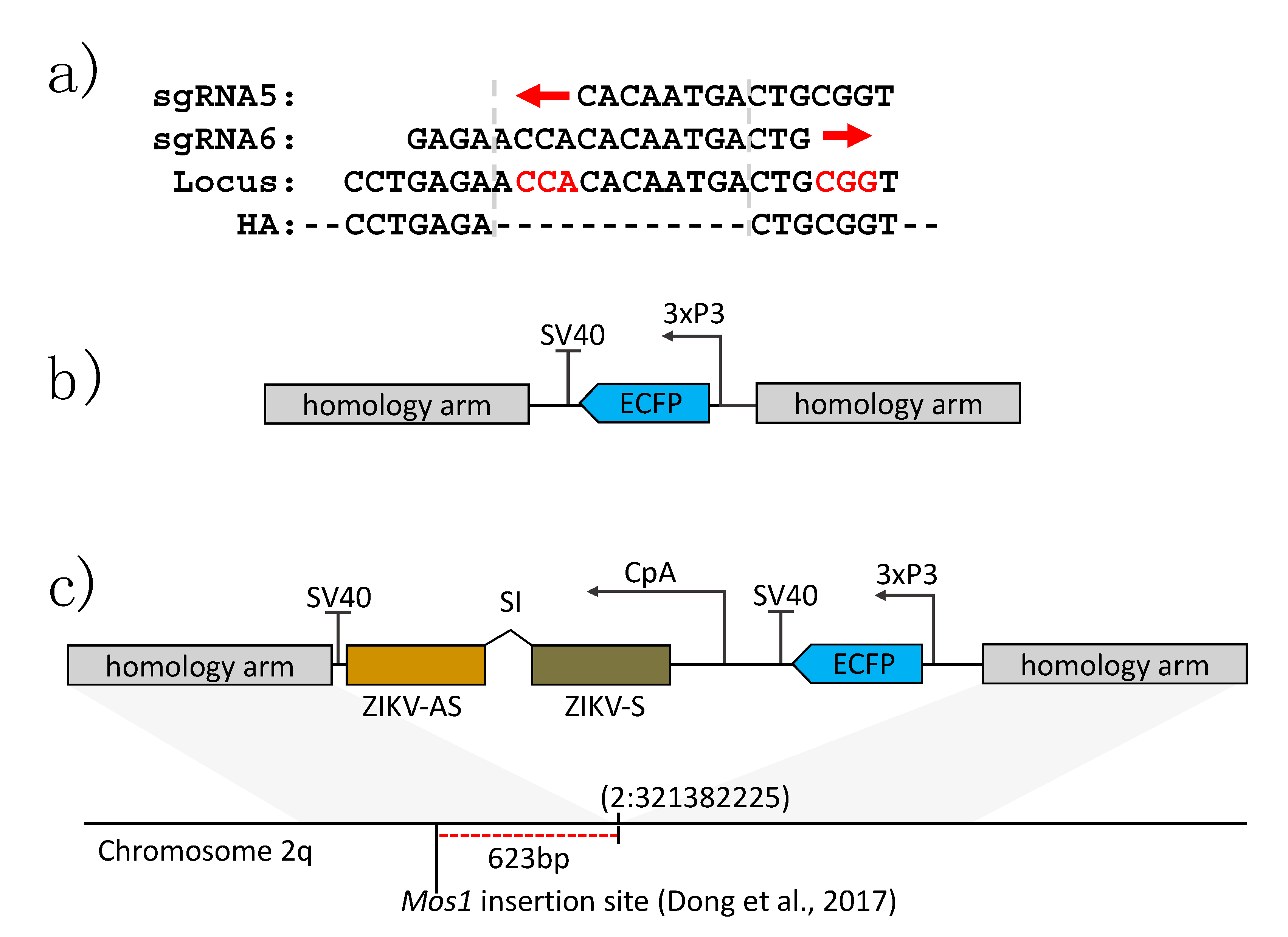
2.2. Identification of Active sgRNA Target Sites
2.3. Construction of Donor Plasmid DNAs
2.4. Establishment of a Transgenic Line of Ae. aegypti Containing an Anti-ZIKV IR Effector
2.5. Small RNA Sequencing
2.6. Virus Challenge Experiments
2.7. Mosquito Tissue Plaque Assays for ZIKV Detection
2.8. Intrathoracic Inoculation of ZIKV
2.9. ZIKV Transmission Assays
2.10. Immunofluorescence Assays to Detect ZIKV Antigen
2.11. Statistical Analyses
3. Results
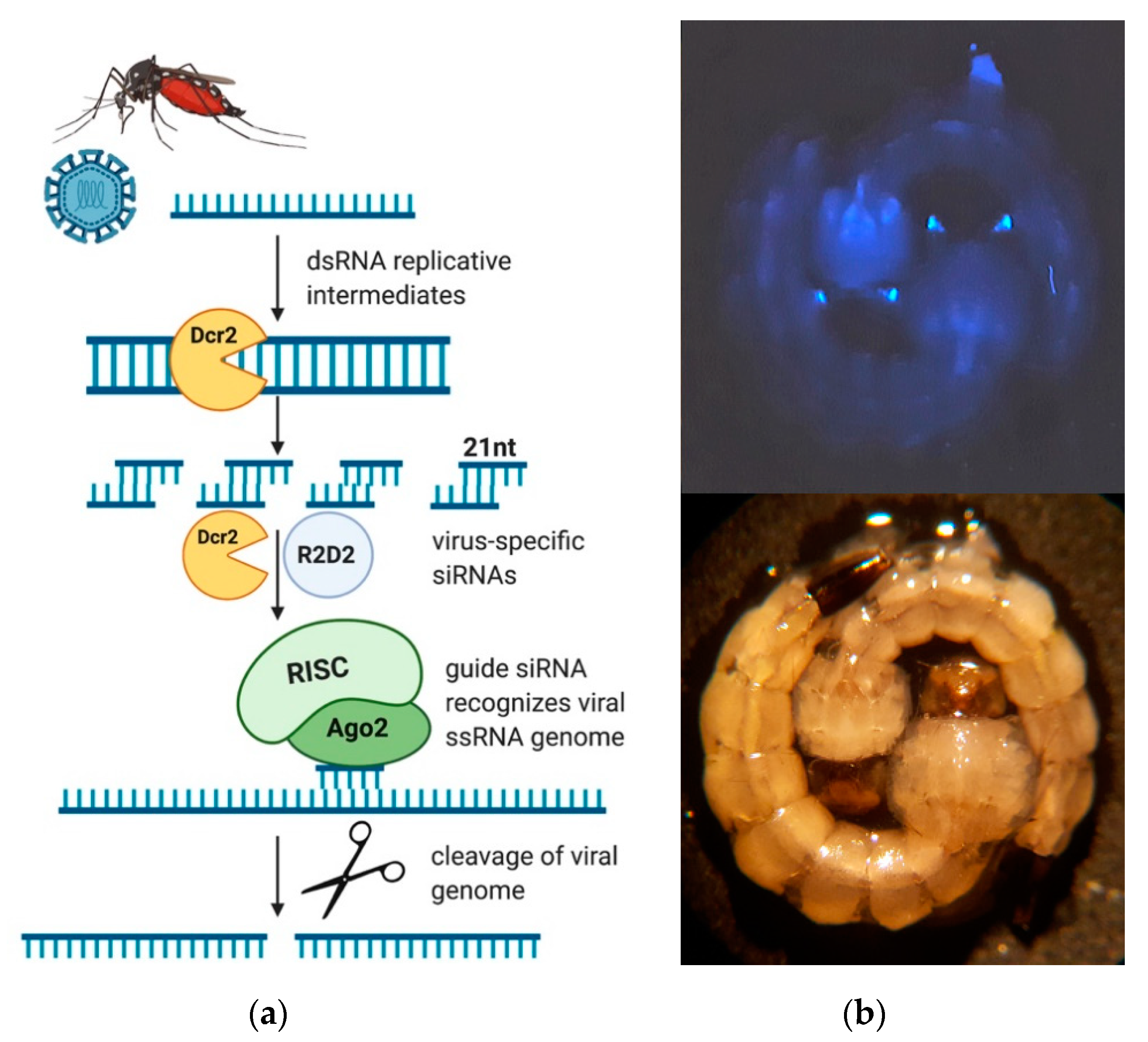
3.1. Generation of Transgenic Ae. aegypti Expressing a ZIKV-Specific IR Effector
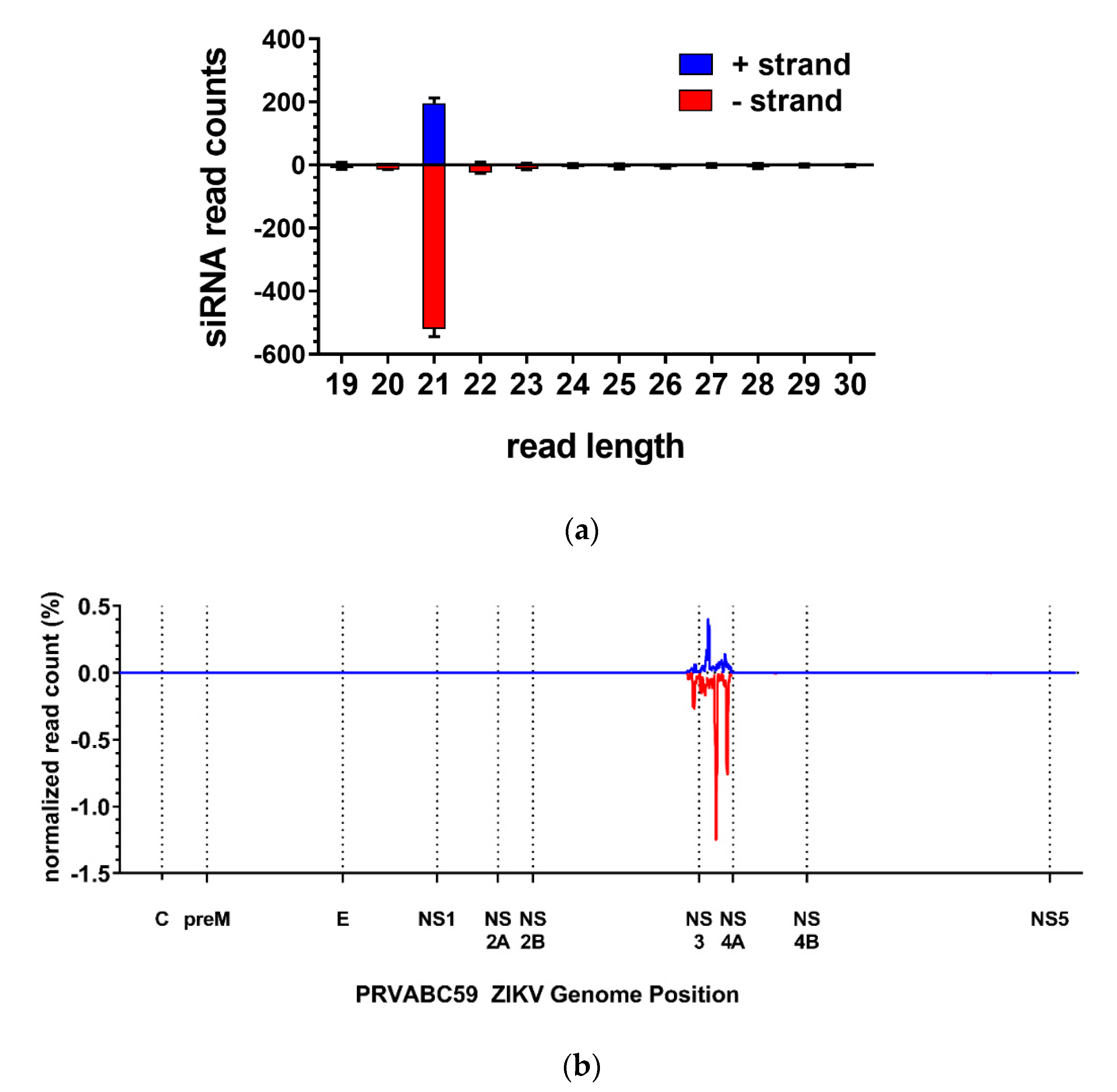
3.2. The ZIKV-Specific IR Effector Is Processed by the Endogenous siRNA Machinery of the Mosquito
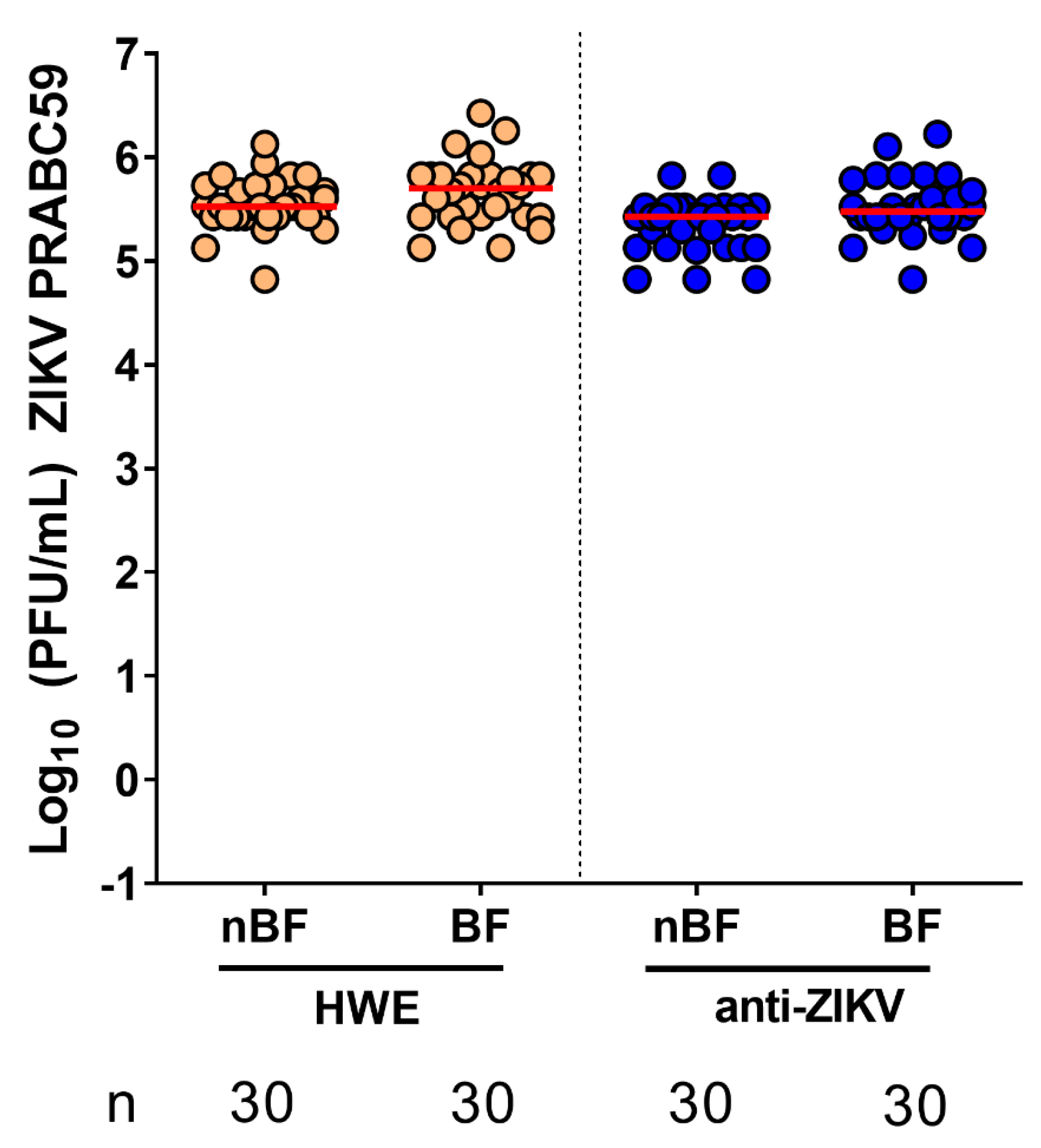
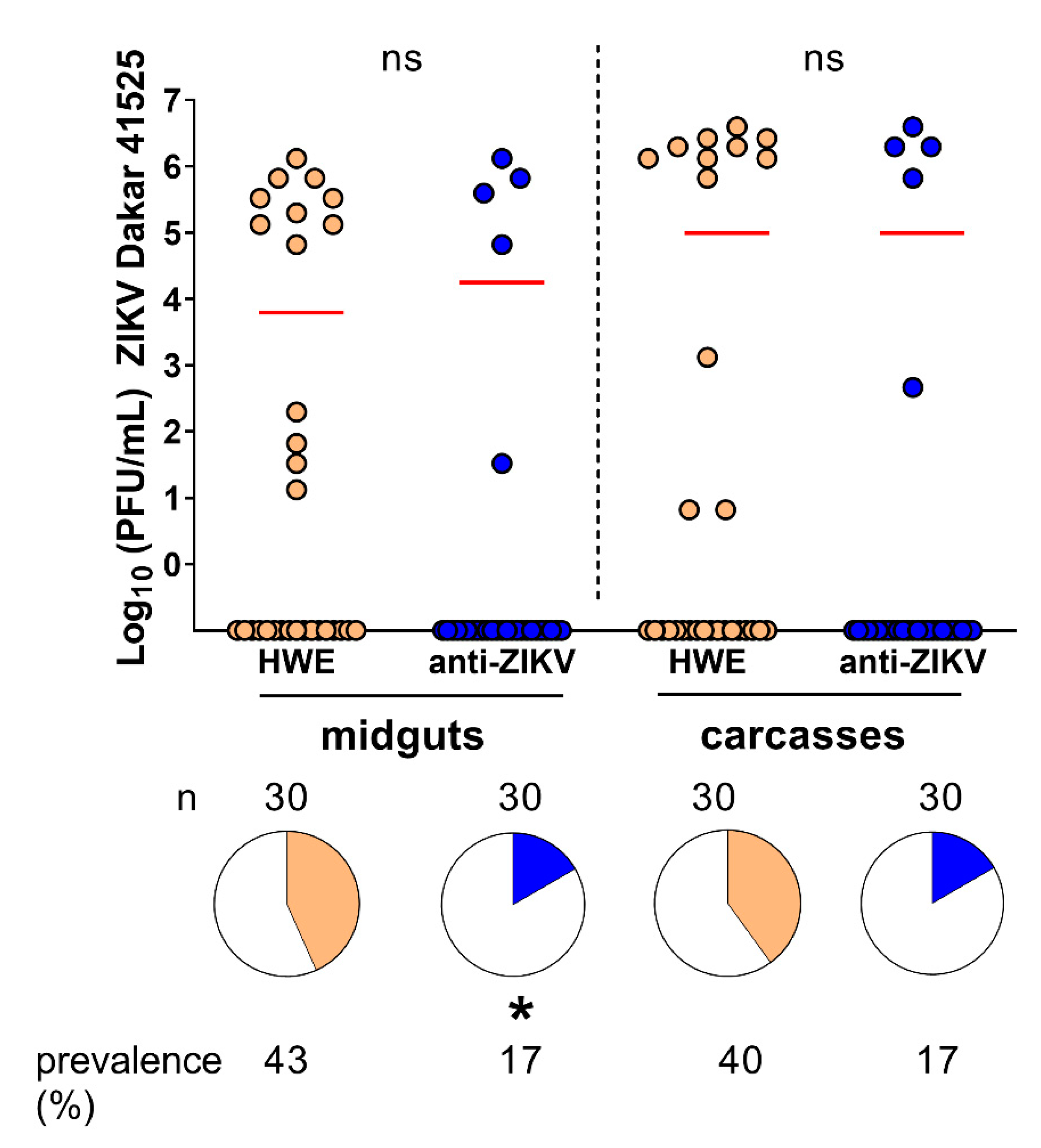
3.3. Ae. aegypti Expressing the Anti-ZIKV IR Effector Are Resistant to ZIKV
3.4. Ae. aegypti Expressing the Anti-ZIKV IR Effector Lose Their Resistance to the Virus When Their Midgut Infection Barriers Are Bypassed
3.5. Resistance of Anti-ZIKV-NS3/4A Transgenics to ZIKV Shows a Tendency to Be Virus Strain-Specific
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Girard, M.; Nelson, C.B.; Picot, V.; Gubler, D.J. Arboviruses: A global public health threat. Vaccine 2020, 38, 3989–3994. [Google Scholar] [CrossRef]
- Wilder-Smith, A.; Gubler, D.J.; Weaver, S.C.; Monath, T.P.; Heymann, D.L.; Scott, T.W. Epidemic arboviral diseases: Priorities for research and public health. Lancet Infect. Dis. 2017, 17, 101–106. [Google Scholar] [CrossRef]
- Moyes, C.L.; Vontas, J.; Martins, A.J.; Ng, L.C.; Koou, S.Y.; Dusfour, I.; Raghavendra, K.; Pinto, J.; Corbel, V.; David, J.-P.; et al. Contemporary status of insecticide resistance in the major Aedes vectors of arboviruses infecting humans. PLoS Negl. Trop. Dis. 2017, 11, e0005625. [Google Scholar] [CrossRef] [PubMed]
- Luckhart, S.; Lindsay, S.W.; James, A.A.; Scott, T.W. Reframing critical needs in vector biology and management of vector-borne disease. PLoS Negl. Trop. Dis. 2010, 4, e566. [Google Scholar] [CrossRef]
- Curtis, C.F. Possible use of translocations to fix desirable genes in insect pest populations. Nature 1968, 218, 368–369. [Google Scholar] [CrossRef] [PubMed]
- Roiz, D.; Wilson, A.L.; Scott, T.W.; Fonseca, D.M.; Jourdain, F.; Müller, P.; Corbel, R.V. Integrated Aedes management for the control of Aedes-borne diseases. PLoS Negl. Trop. Dis. 2018, 12, e0006845. [Google Scholar] [CrossRef]
- Williams, A.E.; Franz, A.W.E.; Reid, W.R.; Olson, K.E. Antiviral Effectors and Gene Drive Strategies for Mosquito Population Suppression or Replacement to Mitigate Arbovirus Transmission by Aedes aegypti. Insects 2020, 11, 52. [Google Scholar] [CrossRef] [PubMed]
- Bargielowski, I.; Nimmo, D.; Alphey, L.; Koella, J.C. Comparison of life history characteristics of the genetically modified OX513A line and a wild type strain of Aedes aegypti. PLoS ONE 2011, 6, e20699. [Google Scholar] [CrossRef] [PubMed]
- Harris, A.F.; Nimmo, D.; McKemey, A.R.; Kelly, N.; Scaife, S.; A Donnelly, C.; Beech, C.; Petrie, W.D.; Alphey, L. Field performance of engineered male mosquitoes. Nat. Biotechnol. 2011, 29, 1034–1037. [Google Scholar] [CrossRef]
- De Andrade, P.P.; Aragão, F.J.L.; Colli, W.; Dellagostin, O.A.; Finardi-Filho, F.; Hirata, M.H.; Lira-Neto, A.D.C.; De Melo, M.A.; Nepomuceno, A.L.; Da Nóbrega, F.G.; et al. Use of transgenic Aedes aegypti in Brazil: Risk perception and assessment. Bull. World Health Organ. 2016, 94, 766–771. [Google Scholar] [CrossRef]
- Facchinelli, L.; Valerio, L.; Ramsey, J.M.; Gould, F.; Walsh, R.K.; Bond, G.; Robert, M.A.; Lloyd, A.L.; James, A.A.; Alphey, L.; et al. Field Cage Studies and Progressive Evaluation of Genetically Engineered Mosquitoes. PLoS Negl. Trop. Dis. 2013, 7, e2001. [Google Scholar] [CrossRef] [PubMed]
- Bargielowski, I.; Kaufmann, C.; Alphey, L.; Reiter, P.; Koella, J. Flight performance and teneral energy reserves of two genetically modified and one wild-type strain of the yellow fever mosquito Aedes aegypti. Vector Borne Zoonotic Dis. 2012, 12, 1053–1058. [Google Scholar] [CrossRef] [PubMed]
- Oxitec. Available online: oxitech.com/florida (accessed on 24 August 2020).
- Chen, C.H.; Huang, H.; Ward, C.M.; Su, J.T.; Schaeffer, L.V.; Guo, M.; Hay, B.A. A Synthetic Maternal-Effect Selfish Genetic Element Drives Population Replacement in Drosophila. Science 2007, 316, 597–600. [Google Scholar] [CrossRef]
- Gantz, V.M.; Jasinskiene, N.; Tatarenkova, O.; Fazekas, A.; Macias, V.M.; Bier, E.; James, A.A. Highly efficient Cas9-mediated gene drive for population modification of the malaria vector mosquito Anopheles stephensi. Proc. Natl. Acad. Sci. USA 2015, 112, 6736–6743. [Google Scholar] [CrossRef] [PubMed]
- Hammond, A.; Galizi, R.; Kyrou, K.; Simoni, A.; Siniscalchi, C.; Katsanos, D.; Gribble, M.; Baker, D.; Marois, E.; Russell, S.; et al. A CRISPR-Cas9 gene drive system targeting female reproduction in the malaria mosquito vector Anopheles gambiae. Nat. Biotechnol. 2016, 34, 78–83. [Google Scholar] [CrossRef]
- Li, M.; Yang, T.; Kandul, N.P.; Bui, M.; Gamez, S.; Raban, R.; Bennett, J.; Sanchez, C.H.M.; Lanzaro, G.C.; Schmidt, H.; et al. Development of a confinable gene drive system in the human disease vector Aedes aegypti. Elife 2020, 9, e51701. [Google Scholar] [CrossRef] [PubMed]
- Sinkins, S.P.; Gould, F. Gene drive systems for insect disease vectors. Nat. Rev. Genet. 2006, 7, 427–435. [Google Scholar] [CrossRef]
- Franz, A.W.E.; Sanchez-Vargas, I.; Adelman, Z.N.; Blair, C.D.; Beaty, B.J.; James, A.A.; Olson, K.E. Engineering RNA interference-based resistance to dengue virus type 2 in genetically modified Aedes aegypti. Proc. Natl. Acad. Sci. USA 2006, 103, 4198–4203. [Google Scholar] [CrossRef]
- Franz, A.W.E.; Sanchez-Vargas, I.; Raban, R.R.; Iv, W.C.B.; James, A.A.; Olson, K.E. Fitness impact and stability of a transgene conferring resistance to dengue-2 virus following introgression into a genetically diverse Aedes aegypti strain. PLoS Negl. Trop. Dis. 2014, 8, e2833. [Google Scholar] [CrossRef]
- Mathur, G.; Sanchez-Vargas, I.; Alvarez, D.; Olson, K.E.; Marinotti, O.; James, A.A. Transgene-mediated suppression of dengue viruses in the salivary glands of the yellow fever mosquito, Aedes aegypti. Insect Mol. Biol. 2010, 19, 753–763. [Google Scholar] [CrossRef]
- Pinkerton, A.C.; Michel, K.; O’Brochta, D.A.; Atkinson, P.W. Green fluorescent protein as a genetic marker in transgenic Aedes aegypti. Insect Mol. Biol. 2000, 9, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Atkinson, P.W.; Pinkerton, A.C.; O’Brochta, D.A. Genetic Transformation Systems in Insects. Annu. Rev. Entomol. 2001, 46, 317–346. [Google Scholar] [CrossRef] [PubMed]
- Kokoza, V.; Ahmed, A.; Wimmer, E.A.; Raikhel, A.S. Efficient transformation of the yellow fever mosquito Aedes aegypti using the piggyBac transposable element vector pBac[3 xP3-EGFP afm]. Insect Biochem. Mol. Biol. 2001, 31, 1137–1143. [Google Scholar] [CrossRef]
- Wilson, R.; Orsetti, J.; Klocko, A.D.; Aluvihare, C.; Peckham, E.; Atkinson, P.W.; Lehane, M.J.; O’Brochta, D.A. Post-integration behavior of a Mos1 mariner gene vector in Aedes aegypti. Insect Biochem. Mol. Biol. 2003, 33, 853–863. [Google Scholar] [CrossRef]
- Franz, A.W.; Sanchez-Vargas, I.; Piper, J.; Smith, M.R.; Khoo, C.C.H.; James, A.A.; Olson, K.E. Stability and loss of a virus resistance phenotype over time in transgenic mosquitoes harbouring an antiviral effector gene. Insect Mol. Biol. 2009, 18, 661–672. [Google Scholar] [CrossRef]
- Moreira, L.A.; Edwards, M.J.; Adhami, F.; Jasinskiene, N.; James, A.A.; Jacobs-Lorena, M. Robust gut-specific gene expression in transgenic Aedes aegypti mosquitoes. Proc. Natl. Acad. Sci. USA 2000, 97, 10895–10898. [Google Scholar] [CrossRef] [PubMed]
- Dong, S.; Balaraman, V.; Kantor, A.M.; Lin, J.; Grant, D.G.; Held, N.L.; Franz, A.W.E. Chikungunya virus dissemination from the midgut of Aedes aegypti is associated with temporal basal lamina degradation during bloodmeal digestion. PLoS Negl. Trop. Dis. 2017, 11, e0005976. [Google Scholar] [CrossRef]
- Magalhaes, T.; Bergren, N.A.; Bennett, S.L.; Borland, E.M.; Hartman, D.A.; Lymperopoulos, K.; Sayre, R.; Borlee, B.R.; Campbell, C.L.; Foy, B.D.; et al. Induction of RNA interference to block Zika virus replication and transmission in the mosquito Aedes aegypti. Insect Biochem. Mol. Biol. 2019, 111, 103169. [Google Scholar] [CrossRef]
- Labun, K.; Montague, T.G.; Gagnon, J.A.; Thyme, S.B.; Valen, E. CHOPCHOP v2: A web tool for the next generation of CRISPR genome engineering. Nucleic Acids Res. 2016, 44, 272–276. [Google Scholar] [CrossRef]
- Labun, K.; Montague, T.G.; Krause, M.; Cleuren, Y.N.T.; Tjeldnes, H.; Valen, E. CHOPCHOP v3: Expanding the CRISPR web toolbox beyond genome editing. Nucleic Acids Res. 2019, 47, 171–174. [Google Scholar] [CrossRef]
- Wendell, M.D.; Wilson, T.G.; Higgs, S.; Black, W.C. Chemical and gamma-ray mutagenesis of the white gene in Aedes aegypti. Insect Mol. Biol. 2000, 9, 119–125. [Google Scholar] [CrossRef]
- Horn, C.; Jaunich, B.; Wimmer, E.A. Highly sensitive, fluorescent transformation marker for Drosophila transgenesis. Dev. Genes Evol. 2000, 210, 623–629. [Google Scholar] [CrossRef] [PubMed]
- Franz, A.W.; Jasinskiene, N.; Sanchez-Vargas, I.; Isaacs, A.; Smith, M.R.; Khoo, C.C.; Heersink, M.S.; James, A.A.; Olson, K.E. Comparison of transgene expression in Aedes aegypti generated by mariner Mos1 transposition and ΦC31 site-directed recombination. Insect Mol. Biol. 2011, 20, 587–598. [Google Scholar] [CrossRef]
- Beerntsen, B.T.; Champagne, D.E.; Coleman, J.L.; Campos, Y.A.; James, A.A. Characterization of the Sialokinin I gene encoding the salivary vasodilator of the yellow fever mosquito, Aedes aegypti. Insect Mol. Biol. 1999, 8, 459–467. [Google Scholar] [CrossRef]
- Edwards, M.J.; Moskalyk, L.A.; Donelly-Doman, M.; Vlaskova, M.; Noriega, F.G.; Walker, V.K.; Jacobs-Lorena, M. Characterization of a carboxypeptidase A gene from the mosquito, Aedes aegypti. Insect Mol. Biol. 2000, 9, 33–38. [Google Scholar] [CrossRef]
- Rückert, C.; Prasad, A.N.; Garcia-Luna, S.M.; Robison, A.; Grubaugh, N.D.; Weger-Lucarelli, J.; Ebel, G.D. Small RNA responses of Culex mosquitoes and cell lines during acute and persistent virus infection. Insect Biochem. Mol. Biol. 2019, 109, 13–23. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.; Domrachev, M.; Lash, A.E. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002, 30, 207–210. [Google Scholar] [CrossRef] [PubMed]
- Rosen, L.; Gubler, D. The use of mosquitoes to detect and propagate dengue viruses. Am. J. Trop. Med. Hyg. 1974, 23, 1153–1160. [Google Scholar] [CrossRef]
- Dubrulle, M.; Mousson, L.; Moutailler, S.; Vazeille, M.; Failloux, A.-B. Chikungunya virus and Aedes mosquitoes: Saliva is infectious as soon as two days after oral infection. PLoS ONE 2009, 4, e5895. [Google Scholar] [CrossRef]
- Sanchez-Vargas, I.; Harrington, L.C.; Black, W.C.; Olson, K.E. Analysis of Salivary Glands and Saliva from Aedes albopictus and Aedes aegypti Infected with Chikungunya Viruses. Insects 2019, 10, 39. [Google Scholar] [CrossRef]
- Basu, S.; Aryan, A.; Overcash, J.M.; Samuel, G.H.; Anderson, M.A.E.; Dahlem, T.J.; Myles, K.M.; Adelman, Z.N. Silencing of end-joining repair for efficient site-specific gene insertion after TALEN/CRISPR mutagenesis in Aedes aegypti. Proc. Natl. Acad. Sci. USA 2015, 112, 4038–4043. [Google Scholar] [CrossRef]
- Weger-Lucarelli, J.; Rückert, C.; Chotiwan, N.; Nguyen, C.; Luna, S.M.G.; Fauver, J.R.; Foy, B.D.; Perera, R.; Black, W.C.; Kading, R.C.; et al. Vector Competence of American Mosquitoes for Three Strains of Zika Virus. PLoS Negl. Trop. Dis. 2016, 10, e0005101. [Google Scholar] [CrossRef]
- Roundy, C.M.; Azar, S.R.; Rossi, S.L.; Huang, J.H.; Al, C.M.R.E.; Yun, R.; Fernandez-Salas, I.; Vitek, C.J.; Paploski, I.A.; Kitron, U.; et al. Variation in Aedes aegypti Mosquito Competence for Zika Virus Transmission. Emerg. Infect. Dis. 2017, 23, 625–632. [Google Scholar] [CrossRef] [PubMed]
- Buchman, A.; Gamez, S.; Li, M.; Antoshechkin, I.; Li, H.-H.; Wang, H.-W.; Chen, C.-H.; Klein, M.J.; Duchemin, J.-B.; Paradkar, P.N.; et al. Engineered resistance to Zika virus in transgenic Aedes aegypti; expressing a polycistronic cluster of synthetic small RNAs. Proc. Natl. Acad. Sci. USA 2019, 116, 3656–3661. [Google Scholar] [CrossRef] [PubMed]
- Göertz, G.P.; Van Bree, J.W.M.; Hiralal, A.; Fernhout, B.M.; Steffens, C.; Boeren, S.; Visser, T.M.; Vogels, C.B.F.; Abbo, S.R.; Fros, J.J.; et al. Subgenomic flavivirus RNA binds the mosquito DEAD/H-box helicase ME31B and determines Zika virus transmission by Aedes aegypti. Proc. Natl. Acad. Sci. USA 2019, 116, 19136–19144. [Google Scholar] [CrossRef]
- Buchman, A.; Gamez, S.; Li, M.; Antoshechkin, I.; Li, H.H.; Wang, H.W.; Chen, C.H.; Klein, M.J.; Duchemin, J.B.; Crowe, J.E., Jr.; et al. Broad dengue neutralization in mosquitoes expressing an engineered antibody. PLoS Pathog. 2020, 16, e1008103. [Google Scholar] [CrossRef]
- Mishra, P.; Furey, C.; Balaraman, V.; Fraser, M.J. Antiviral Hammerhead Ribozymes Are Effective for Developing Transgenic Suppression of Chikungunya Virus in Aedes aegypti Mosquitoes. Viruses 2016, 8, 163. [Google Scholar] [CrossRef] [PubMed]


| Injection Mix | Embryos Injected | Male | Female | Survival (%) |
|---|---|---|---|---|
| sgRNA 5 | 735 | 19 | 22 | 5.6 |
| sgRNA 6 | 928 | 20 | 23 | 4.6 |
| sgRNAs 5 + 6 | 882 | 38 | 36 | 8.4 |
| Anti-ZIKV-IR | 1510 | 84 | 100 | 12.1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Williams, A.E.; Sanchez-Vargas, I.; Reid, W.R.; Lin, J.; Franz, A.W.E.; Olson, K.E. The Antiviral Small-Interfering RNA Pathway Induces Zika Virus Resistance in Transgenic Aedes aegypti. Viruses 2020, 12, 1231. https://doi.org/10.3390/v12111231
Williams AE, Sanchez-Vargas I, Reid WR, Lin J, Franz AWE, Olson KE. The Antiviral Small-Interfering RNA Pathway Induces Zika Virus Resistance in Transgenic Aedes aegypti. Viruses. 2020; 12(11):1231. https://doi.org/10.3390/v12111231
Chicago/Turabian StyleWilliams, Adeline E., Irma Sanchez-Vargas, William R. Reid, Jingyi Lin, Alexander W.E. Franz, and Ken E. Olson. 2020. "The Antiviral Small-Interfering RNA Pathway Induces Zika Virus Resistance in Transgenic Aedes aegypti" Viruses 12, no. 11: 1231. https://doi.org/10.3390/v12111231
APA StyleWilliams, A. E., Sanchez-Vargas, I., Reid, W. R., Lin, J., Franz, A. W. E., & Olson, K. E. (2020). The Antiviral Small-Interfering RNA Pathway Induces Zika Virus Resistance in Transgenic Aedes aegypti. Viruses, 12(11), 1231. https://doi.org/10.3390/v12111231





