A Diverse Virome of Leafroll-Infected Grapevine Unveiled by dsRNA Sequencing
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Material
2.2. dsRNA Extraction, Libraries Construction and Sequencing
2.3. Bioinformatic and Statistical Analysis
2.3.1. Raw Data Treatment
2.3.2. Analysis of the Diversity of the Virome and Association between Viruses and Symptom Development
3. Results
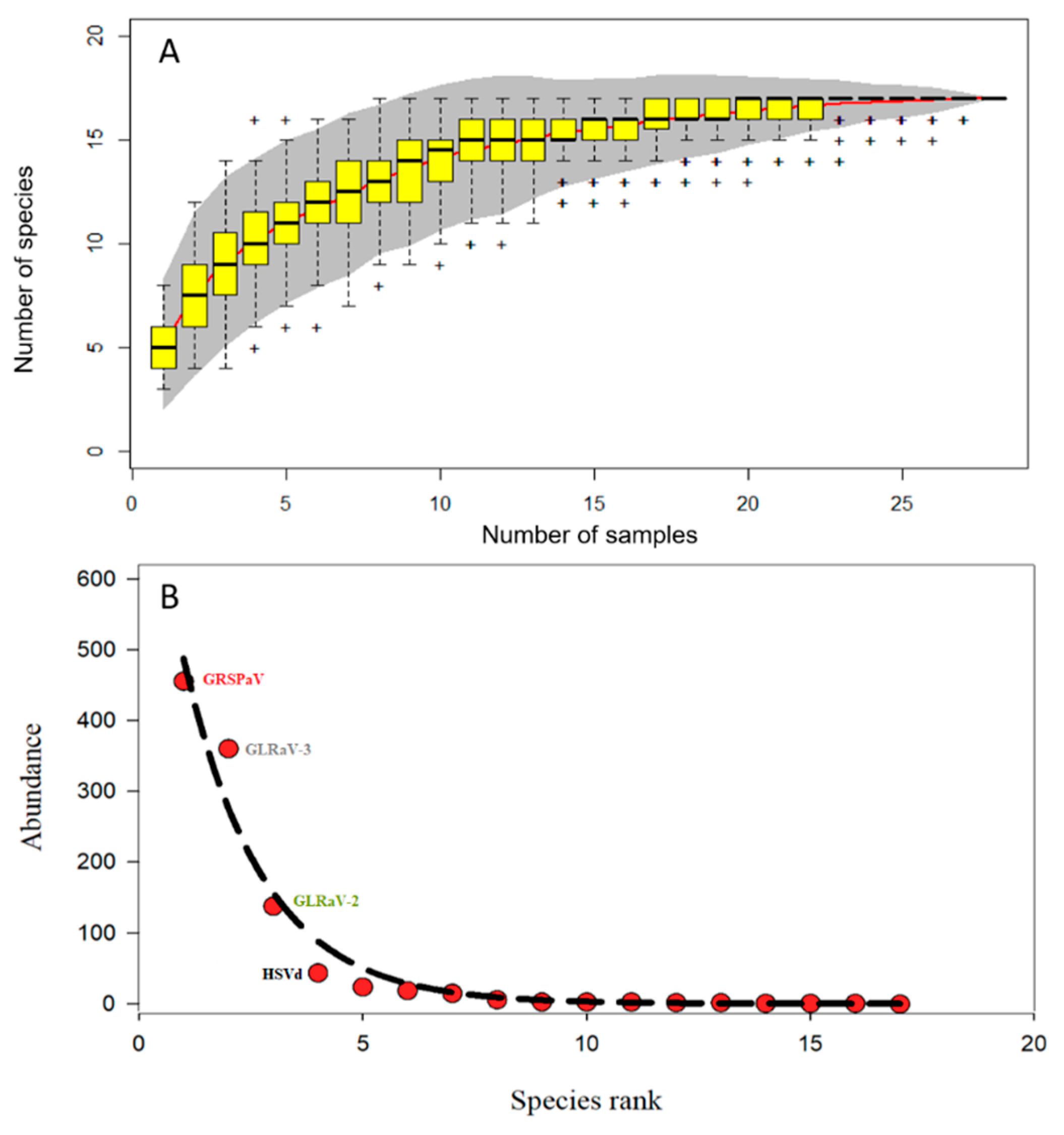
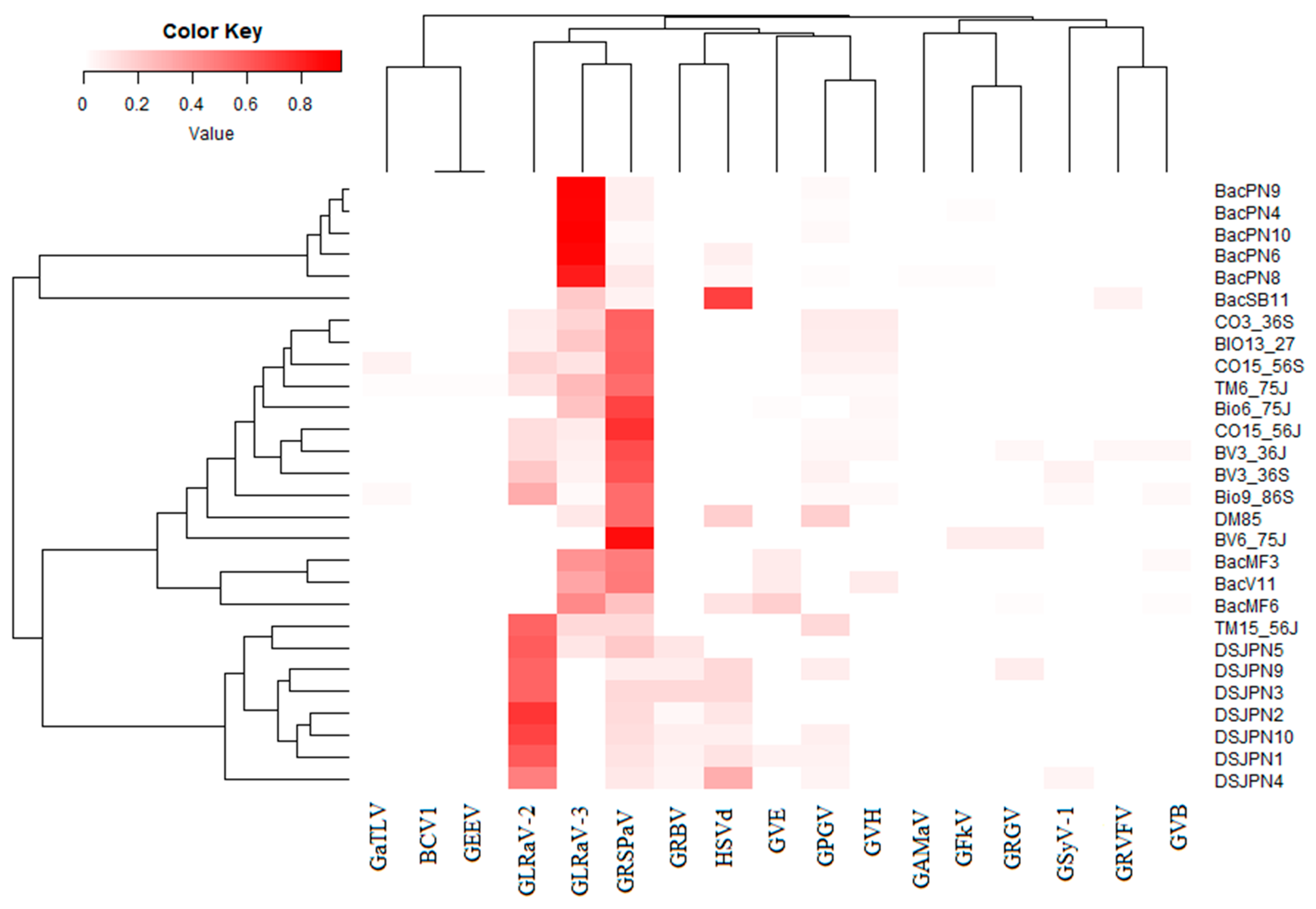
3.1. Diversity of Virome in Leafroll-Infected Leaves of Different Grapevine Cultivars
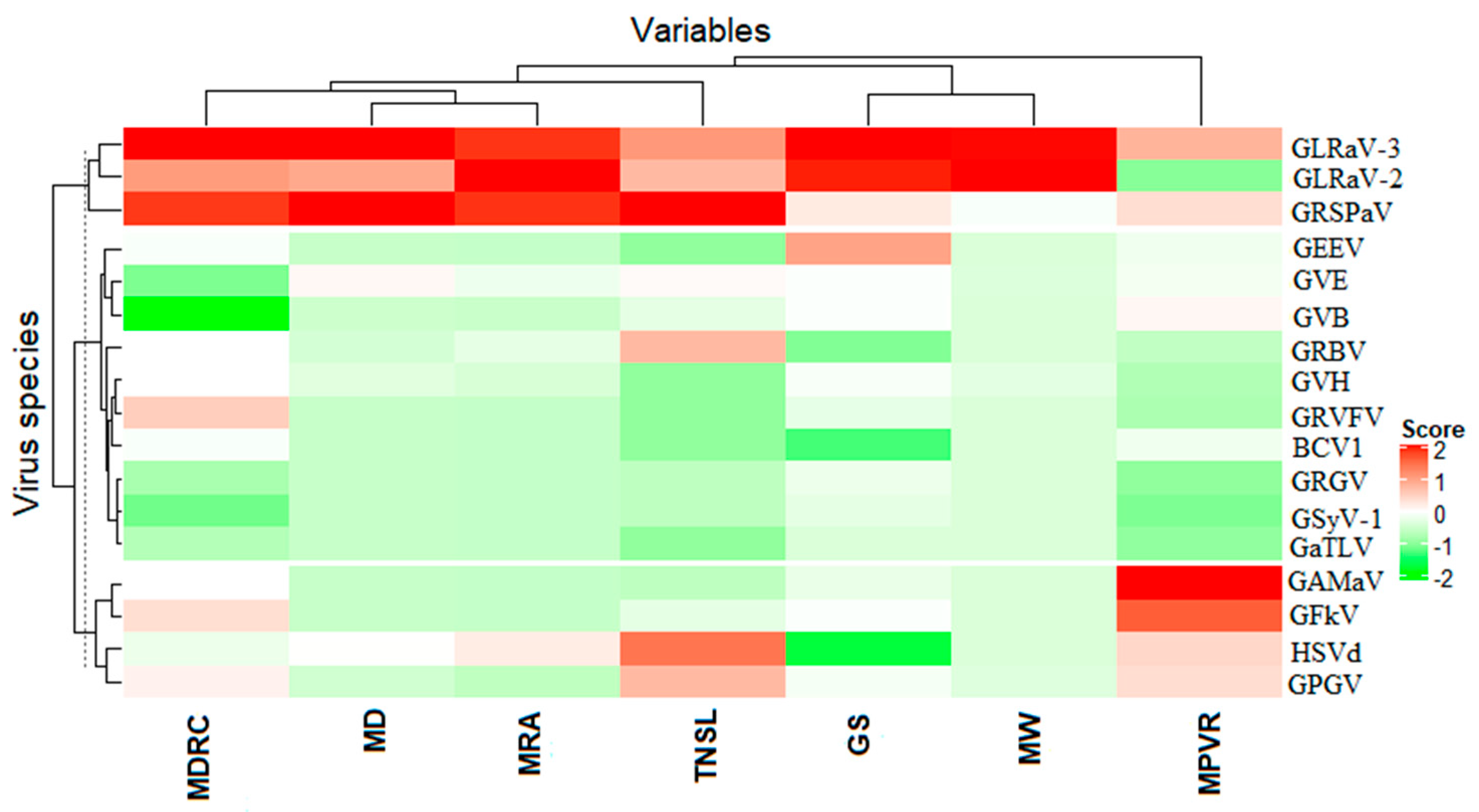
3.2. Association between Viruses and Virus-Like Symptom Development
3.3. The Genetic Diversity of Viruses and Viroids Detected for the First Time in Quebec
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Statistic Canada. Table 32-10-0364-01 Area, Production and Farm Gate Value of Marketed Fruits. Available online: https://doi.org/10.25318/3210036401-eng (accessed on 17 June 2020).
- Martelli, G.P. Directory of virus and virus-like diseases of the grapevine and their agents. J. Plant Pathol. 2014, 96, 1–136. [Google Scholar]
- Fuchs, M. Grapevine viruses: A multitude of diverse species with simple but overall poorly adopted management solutions in the vineyard. J. Plant Pathol. 2020, 102, 643–653. [Google Scholar] [CrossRef]
- Alabi, O.J.; Casassa, L.F.; Gutha, L.R.; Larsen, R.C.; Henick-Kling, T.; Harbertson, J.F.; Naidu, R.A. Impacts of Grapevine Leafroll Disease on Fruit Yield and Grape and Wine Chemistry in a Wine Grape (Vitis vinifera L.) Cultivar. PLoS ONE 2016, 11, e0149666. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Atallah, S.S.; Gomez, M.I.; Fuchs, M.F.; Martinson, T.E. Economic Impact of Grapevine Leafroll Disease on Vitis vinifera cv. Cabernet franc in Finger Lakes Vineyards of New York. Am. J. Enol. Vitic. 2012, 63, 73–79. [Google Scholar] [CrossRef] [Green Version]
- Martelli, G.P.; Boudon-Padieu, E. Directory of infectious diseases of grapevines. International centre for advanced mediterranean agronomic studies. Options Méditerr. Ser. B Stud. Res. 2006, 55, 59–75. [Google Scholar]
- Poojari, S.; Alabi, O.J.; Fofanov, V.Y.; Naidu, R.A. A leafhopper-transmissible DNA virus with novel evolutionary lineage in the family geminiviridae implicated in grapevine redleaf disease by next-generation sequencing. PLoS ONE 2013, 8, e64194. [Google Scholar] [CrossRef]
- Almeida, R.P.; Daane, K.M.; Bell, V.A.; Blaisdell, G.K.; Cooper, M.L.; Herrbach, E.; Pietersen, G. Ecology and management of grapevine leafroll disease. Front. Microbiol. 2013, 4, 94. [Google Scholar] [CrossRef] [Green Version]
- Beuve, M.; Hily, J.M.; Alliaume, A.; Reinbold, C.; Le Maguet, J.; Candresse, T.; Herrbach, E.; Lemaire, O. A complex virome unveiled by deep sequencing analysis of RNAs from a French Pinot noir grapevine exhibiting strong leafroll symptoms. Arch. Virol. 2018, 163, 2937–2946. [Google Scholar] [CrossRef] [Green Version]
- Fuchs, M.; Marsella-Herrick, P.; Loeb, G.M.; Martinson, T.E.; Hoch, H.C. Diversity of ampeloviruses in mealybug and soft scale vectors and in grapevine hosts from leafroll-affected vineyards. Phytopathology 2009, 99, 1177–1184. [Google Scholar] [CrossRef] [Green Version]
- Golino, D.A.; Weber, E.; Sim, S.; Rowhani, A. Leafroll disease is spreading rapidly in a Napa Valley vineyard. Calif. Agric. 2008, 62, 156–160. [Google Scholar] [CrossRef] [Green Version]
- Le Maguet, J.; Beuve, M.; Herrbach, E.; Lemaire, O. Transmission of six ampeloviruses and two vitiviruses to grapevine by Phenacoccus aceris. Phytopathology 2012, 102, 717–723. [Google Scholar] [CrossRef] [Green Version]
- Poojari, S.; Moreau, D.L.; Kahl, D.; Ritchie, M.; Ali, S.; Úrbez-Torres, J.R. Disease incidence and genetic variability of economically important grapevine viruses in Nova Scotia. Can. J. Plant Pathol. 2020, 1–11. [Google Scholar] [CrossRef]
- Kovacs, L.G.; Hanami, H.; Fortenberry, M.; Kaps, M.L. Latent infection by leafroll agent GLRaV-3 is linked to lower fruit quality in french-american hybrid grapevines Vidal blanc and St Vincent. Am J Enol Vitic. 2001, 52, 254–259. [Google Scholar]
- MacKenzie, D.J.; Johnson, R.C.; Warner, C. Incidence of four important viral pathogens in Canadian vineyards. Plant Dis. 1996, 80, 955–958. [Google Scholar] [CrossRef]
- Poojari, S.; Boule, J.; DeLury, N.; Lowery, D.T.; Rott, M.; Schmidt, A.M.; Urbez-Torres, J.R. Epidemiology and Genetic Diversity of Grapevine Leafroll-Associated Viruses in British Columbia. Plant Dis. 2017, 101, 2088–2097. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Poojari, S.; Lowery, D.T.; Rott, M.; Schmidt, A.M.; Úrbez-Torres, J.R. Incidence, distribution and genetic diversity of Grapevine red blotch virus in British Columbia. Can. J. Plant Pathol. 2017, 39, 201–211. [Google Scholar] [CrossRef]
- Xiao, H.; Shabanian, M.; Moore, C.; Li, C.; Meng, B. Survey for major viruses in commercial Vitis vinifera wine grapes in Ontario. Virol. J. 2018, 15, 127. [Google Scholar] [CrossRef]
- du Québec, G. Profil Sectoriel de L’industrie Bioalimentaire au Québec; Institut de la statistique du Québec: Québec, QC, Canada, 2020. [Google Scholar]
- Gouvernement du Québec. Profil Sectoriel de L’industrie Bioalimentaire au Québec, 2008th ed.; Institut de la statistique du Québec: Québec, QC, Canada, 2009; pp. 80–81. [Google Scholar]
- Xiao, H.; Li, C.; Al Rwahnih, M.; Dolja, V.; Meng, B. Metagenomic Analysis of Riesling Grapevine Reveals a Complex Virome Including Two New and Divergent Variants of Grapevine leafroll-associated virus 3. Plant Dis. 2019, 103, 1275–1285. [Google Scholar] [CrossRef]
- Ben Moussa, I.E.; Lemoyne, P.; Fall, M.L. Virus and grapevine, unbreakable relationship: Biovigilance is more than require. Phytoprotection 2019, 99, 15–20. [Google Scholar] [CrossRef]
- Carisse, O.; Fall, M.L.; Vincent, C. Using a biovigilance approach for pest and disease management in Quebec vineyards. Can. J. Plant Pathol. 2017, 39, 393–404. [Google Scholar] [CrossRef]
- Alkowni, R.; Zhang, Y.P.; Rowhani, A.; Uyemoto, J.K.; Minafra, A. Biological, molecular, and serological studies of a novel strain of grapevine leafroll-associated virus 2. Virus Genes 2011, 43, 102–110. [Google Scholar] [CrossRef] [PubMed]
- Vionnet, L.; De Vrieze, M.; Agnès, D.; Gfeller, A.; Lüthi, A.; L’Haridon, F.; Weisskopf, L. Microbial life in the grapevine: What can we expect from the leaf microbiome? OENO One 2018, 52, 219–224. [Google Scholar] [CrossRef]
- Xiao, H.; Kim, W.S.; Meng, B. A highly effective and versatile technology for the isolation of RNAs from grapevines and other woody perennials for use in virus diagnostics. Virol. J. 2015, 12, 171. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Poojari, S.; Alabi, O.J.; Okubara, P.A.; Naidu, R.A. SYBR((R)) Green-based real-time quantitative reverse-transcription PCR for detection and discrimination of grapevine viruses. J. Virol. Methods 2016, 235, 112–118. [Google Scholar] [CrossRef] [Green Version]
- Kesanakurti, P.; Belton, M.; Saeed, H.; Rast, H.; Boyes, I.; Rott, M. Screening for plant viruses by next generation sequencing using a modified double strand RNA extraction protocol with an internal amplification control. J. Virol. Methods 2016, 236, 35–40. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [Green Version]
- Rott, M.; Xiang, Y.; Boyes, I.; Belton, M.; Saeed, H.; Kesanakurti, P.; Hayes, S.; Lawrence, T.; Birch, C.; Bhagwat, B.; et al. Application of Next Generation Sequencing for Diagnostic Testing of Tree Fruit Viruses and Viroids. Plant Dis. 2017, 101, 1489–1499. [Google Scholar] [CrossRef] [Green Version]
- Ho, T.; Tzanetakis, I.E. Development of a virus detection and discovery pipeline using next generation sequencing. Virology 2014, 471–473, 54–60. [Google Scholar] [CrossRef] [Green Version]
- Kembel, S.W.; Cahill, J.F., Jr. Independent evolution of leaf and root traits within and among temperate grassland plant communities. PLoS ONE 2011, 6, e19992. [Google Scholar] [CrossRef]
- Kaufman, L.; Rousseeuw, P.J. Partitioning Around Medoids (Program PAM). In An Introduction to Cluster Analysis; John and Wily and Sons: Hoboken, NJ, USA, 1990; pp. 68–125. [Google Scholar]
- Veech, J.A.; Peres-Neto, P. A probabilistic model for analysing species co-occurrence. Glob. Ecol. Biogeogr. 2013, 22, 252–260. [Google Scholar] [CrossRef]
- Griffith, D.M.; Veech, J.A.; Marsh, C.J. Cooccur: Probabilistic Species Co-Occurrence Analysis inR. J. Stat. Softw. 2016, 69, 1–17. [Google Scholar] [CrossRef]
- Al Rwahnih, M.; Daubert, S.; Golino, D.; Islas, C.; Rowhani, A. Comparison of Next-Generation Sequencing Versus Biological Indexing for the Optimal Detection of Viral Pathogens in Grapevine. Phytopathology 2015, 105, 758–763. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eichmeier, A.; Kominkova, M.; Kominek, P.; Baranek, M. Comprehensive Virus Detection Using Next Generation Sequencing in Grapevine Vascular Tissues of Plants Obtained from the Wine Regions of Bohemia and Moravia (Czech Republic). PLoS ONE 2016, 11, e0167966. [Google Scholar] [CrossRef] [PubMed]
- Fajardo, T.V.M.; Silva, F.N.; Eiras, M.; Nickel, O. High-throughput sequencing applied for the identification of viruses infecting grapevines in Brazil and genetic variability analysis. Trop. Plant Pathol. 2017, 42, 250–260. [Google Scholar] [CrossRef] [Green Version]
- Giampetruzzi, A.; Roumi, V.; Roberto, R.; Malossini, U.; Yoshikawa, N.; La Notte, P.; Terlizzi, F.; Credi, R.; Saldarelli, P. A new grapevine virus discovered by deep sequencing of virus- and viroid-derived small RNAs in Cv Pinot gris. Virus Res. 2012, 163, 262–268. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v4: Recent updates and new developments. Nucleic Acids Res. 2019, 47, W256–W259. [Google Scholar] [CrossRef] [Green Version]
- Maree, H.J.; Almeida, R.P.; Bester, R.; Chooi, K.M.; Cohen, D.; Dolja, V.V.; Fuchs, M.F.; Golino, D.A.; Jooste, A.E.; Martelli, G.P.; et al. Grapevine leafroll-associated virus 3. Front. Microbiol. 2013, 4, 82. [Google Scholar] [CrossRef] [Green Version]
- Saldarelli, P.; Giampetruzzi, A.; Maree, H.J.; Al Rwahnih, M. High-Throughput Sequencing: Advantages Beyond Virus Identification. In Grapevine Viruses: Molecular Biology, Diagnostics and Management; Meng, B., Martelli, G.P., Golino, D.A., Fuchs, M., Eds.; Springer: Berlin/Heidelberg, Germany, 2017. [Google Scholar]
- Blanchet, F.G.; Cazelles, K.; Gravel, D. Co-occurrence is not evidence of ecological interactions. Ecol. Lett. 2020, 23, 1050–1063. [Google Scholar] [CrossRef]
- Monis, J.; Bestwick, R.K. Detection and Localization of Grapevine Leafroll Associated Closteroviruses in Greenhouse and Tissue Culture Grown Plants. Am. J. Enol. Vitic. 1996, 47, 199–205. [Google Scholar]
- Bertazzon, N.; Borgo, M.; Vanin, S.; Angelini, E. Genetic variability and pathological properties of Grapevine Leafroll-associated Virus 2 isolates. Eur. J. Plant Pathol. 2010, 127, 185–197. [Google Scholar] [CrossRef]
- Thompson, B.D.; Dahan, J.; Lee, J.; Martin, R.R.; Karasev, A.V. A Novel Genetic Variant of Grapevine leafroll-associated virus-3 (GLRaV-3) from Idaho Grapevines. Plant Dis. 2019, 103, 509–518. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hily, J.M.; Beuve, M.; Vigne, E.; Demangeat, G.; Candresse, T.; Lemaire, O. A genome-wide diversity study of grapevine rupestris stem pitting-associated virus. Arch. Virol. 2018, 163, 3105–3111. [Google Scholar] [CrossRef] [PubMed]
- Reynolds, A.G.; Lanterman, W.S.; Wardle, D.A. Yield and Berry Composition of Five Vitis Cultivars as Affected by Rupestris Stem Pitting Virus. Am. J. Enol. Vitic. 1997, 48, 449–458. [Google Scholar]
- Hily, J.M.; Candresse, T.; Garcia, S.; Vigne, E.; Tanniere, M.; Komar, V.; Barnabe, G.; Alliaume, A.; Gilg, S.; Hommay, G.; et al. High-Throughput Sequencing and the Viromic Study of Grapevine Leaves: From the Detection of Grapevine-Infecting Viruses to the Description of a New Environmental Tymovirales Member. Front. Microbiol. 2018, 9, 1782. [Google Scholar] [CrossRef]
- Cabaleiro, C.; Segura, A. Temporal Analysis of Grapevine leafroll associated virus 3 Epidemics. Eur. J. Plant Pathol. 2006, 114, 441–446. [Google Scholar] [CrossRef]
- Habili, N.; Nutter, F.W., Jr. Temporal and Spatial Analysis of Grapevine Leafroll-Associated Virus 3 in Pinot noir Grapevines in Australia. Plant Dis. 1997, 81, 625–628. [Google Scholar] [CrossRef] [Green Version]





| Virus Name | Species (ICTV) a | Abbreviation | NGS b | PCR c | Sampling Sites | ||
|---|---|---|---|---|---|---|---|
| Fr | Hem | SJM | |||||
| Beet cryptic virus 1 | Beet cryptic virus 1 | BCV1 | + | NT | + | - | - |
| Grapevine endophyte endornavirus | Grapevine endophyte alphaendornavirus | GEEV | + | NT | + | - | - |
| Grapevine associated tymo like virus | Not been approved yet | GaTLV | + | NT | + | - | - |
| Grapevine asteroid mosaic associated virus | Not been approved yet | GAMaV | + | NT | - | + | - |
| Grapevine fleck virus | Grapevine fleck virus | GFkV | + | + | + | + | - |
| Grapevine leafroll associated virus 2 | Grapevine leafroll-associated virus 2 | GLRaV-2 | + | + | + | + | + |
| Grapevine leafroll associated virus 3 | Grapevine leafroll-associated virus 3 | GLRaV-3 | + | + | + | + | + |
| Grapevine Pinot gris virus | Grapevine Pinot gris virus | GPGV | + | + | + | + | + |
| Grapevine red blotch associated virus | Grapevine red blotch virus | GRBV | + | + | - | - | + |
| Grapevine Red Globe Virus | Grapevine red globe virus | GRGV | + | NT | + | - | + |
| Grapevine Rupestris stem pitting associated virus | Grapevine rupestris stem pitting-associated virus | GRSPaV | + | NT | + | + | + |
| Grapevine Rupestris vein feathering virus | Not been approved yet | GRVFV | + | NT | + | + | - |
| Grapevine syrah virus 1 | Not been approved yet | GSyV-1 | + | + | + | - | + |
| Grapevine virus E | Grapevine virus E | GVE | + | NT | - | + | + |
| Grapevine virus B | Grapevine virus B | GVB | + | NT | + | + | - |
| Grapevine virus H | Not been approved yet | GVH | + | NT | + | + | - |
| Hop stunt viroid | Hop stunt viroid | HSVd | + | + | - | + | + |
| Event a | Ev_Inc b | Obs_Cooccur c | Prob_Cooccur d | Exp_Cooccur e | p_lt f | p_gt g | |
|---|---|---|---|---|---|---|---|
| Symptom expression | Hybrid Vidal | 9 | 0 | 0.097 | 6.4 | 0.00000 | 1.00000 |
| V. vinifera cvs | 41 | 38 | 0.442 | 29.2 | 1.00000 | 0.00000 | |
| GaTLV | 4 | 0 | 0.043 | 2.8 | 0.00538 | 1.00000 | |
| GPGV | 26 | 15 | 0.281 | 18.5 | 0.04747 | 0.98688 | |
| GVH | 9 | 0 | 0.097 | 6.4 | 0.00000 | 1.00000 | |
| GLRaV-3 presence | GRBV | 9 | 2 | 0.079 | 5.2 | 0.02587 | 0.99662 |
| GVB | 17 | 14 | 0.148 | 9.8 | 0.99742 | 0.01517 | |
| GVH | 9 | 9 | 0.079 | 5.2 | 1.00000 | 0.00440 | |
| HSVd | 24 | 7 | 0.209 | 13.8 | 0.00050 | 0.99994 | |
| Hybrid Vidal | 9 | 8 | 0.079 | 5.2 | 0.99560 | 0.04140 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fall, M.L.; Xu, D.; Lemoyne, P.; Moussa, I.E.B.; Beaulieu, C.; Carisse, O. A Diverse Virome of Leafroll-Infected Grapevine Unveiled by dsRNA Sequencing. Viruses 2020, 12, 1142. https://doi.org/10.3390/v12101142
Fall ML, Xu D, Lemoyne P, Moussa IEB, Beaulieu C, Carisse O. A Diverse Virome of Leafroll-Infected Grapevine Unveiled by dsRNA Sequencing. Viruses. 2020; 12(10):1142. https://doi.org/10.3390/v12101142
Chicago/Turabian StyleFall, Mamadou L., Dong Xu, Pierre Lemoyne, Issam E. Ben Moussa, Carole Beaulieu, and Odile Carisse. 2020. "A Diverse Virome of Leafroll-Infected Grapevine Unveiled by dsRNA Sequencing" Viruses 12, no. 10: 1142. https://doi.org/10.3390/v12101142
APA StyleFall, M. L., Xu, D., Lemoyne, P., Moussa, I. E. B., Beaulieu, C., & Carisse, O. (2020). A Diverse Virome of Leafroll-Infected Grapevine Unveiled by dsRNA Sequencing. Viruses, 12(10), 1142. https://doi.org/10.3390/v12101142






