A Mutation Network Method for Transmission Analysis of Human Influenza H3N2
Abstract
1. Introduction
2. Materials and Methods
2.1. Simulations
2.2. Bedford Data
2.3. Nucleotide Sequence Data
2.4. Mutation Network Construction
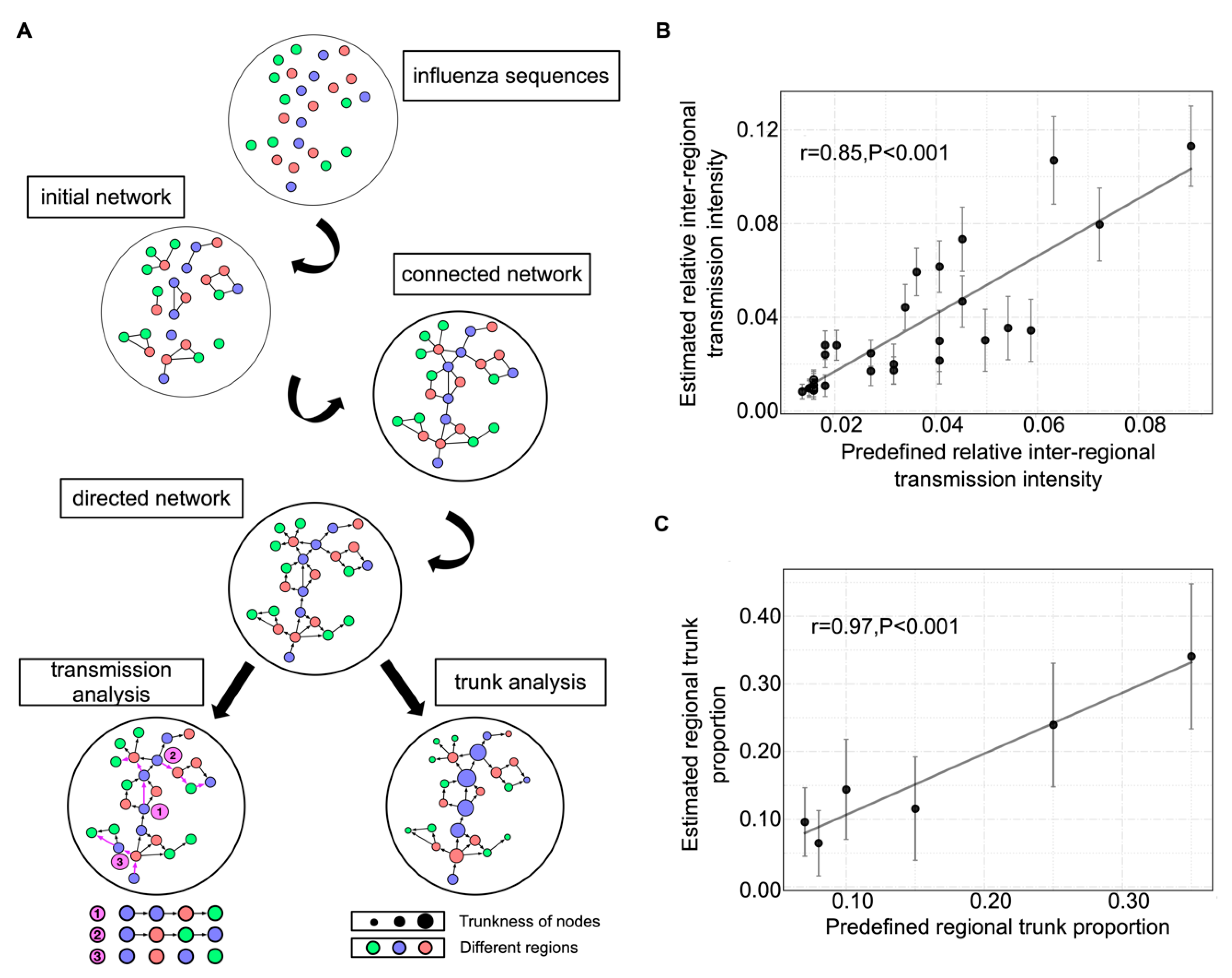
2.4.1. Initial Network
2.4.2. Fully Connected Network
2.4.3. Directed Network
2.5. Transmission Analysis
2.6. Trunk Analysis
2.7. Statistical Analysis
3. Result
3.1. Mutation Network Framework and Simulation Validation
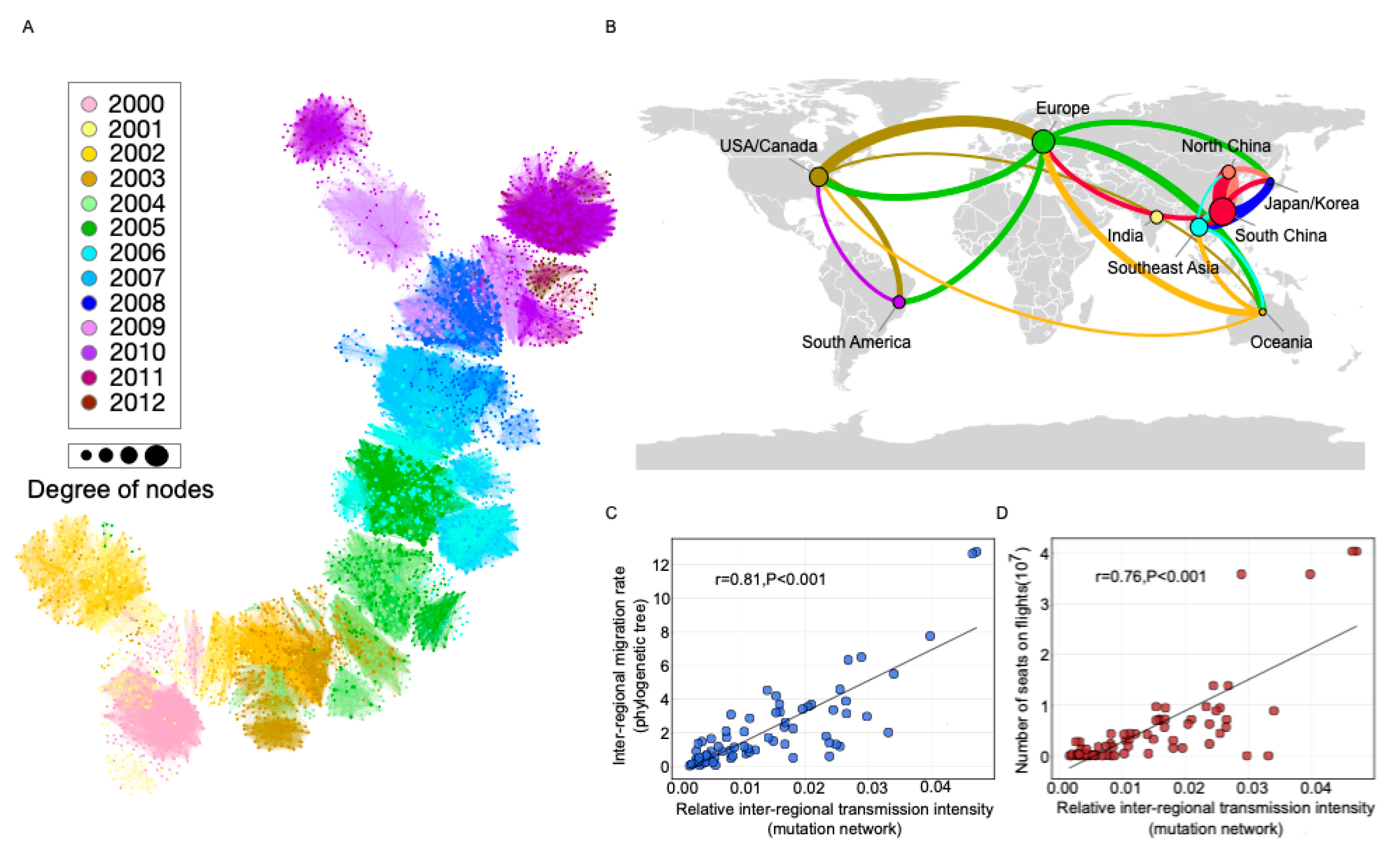
3.2. Transmission Analysis for Bedford Data
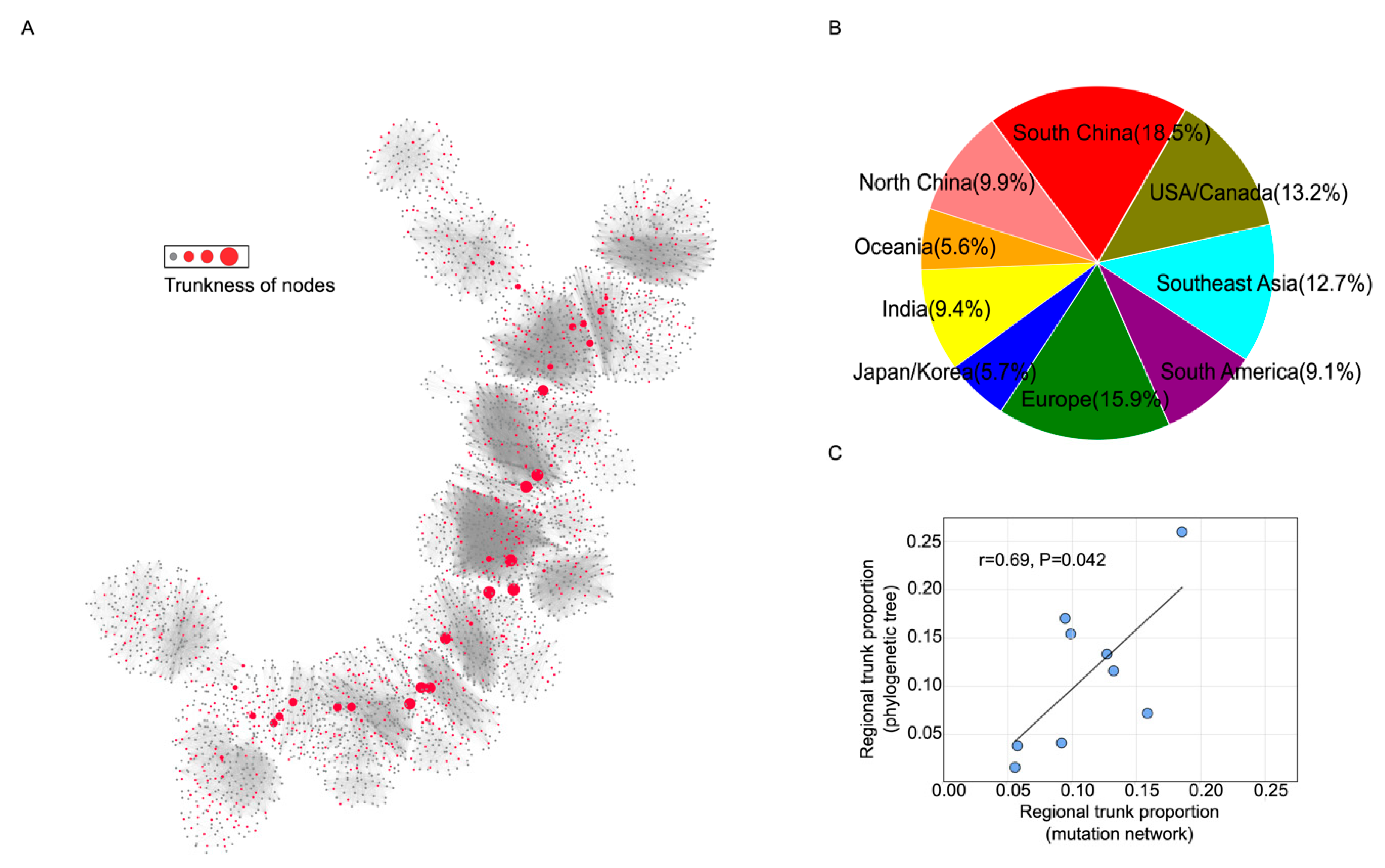
3.3. Trunk Analysis for Bedford Data
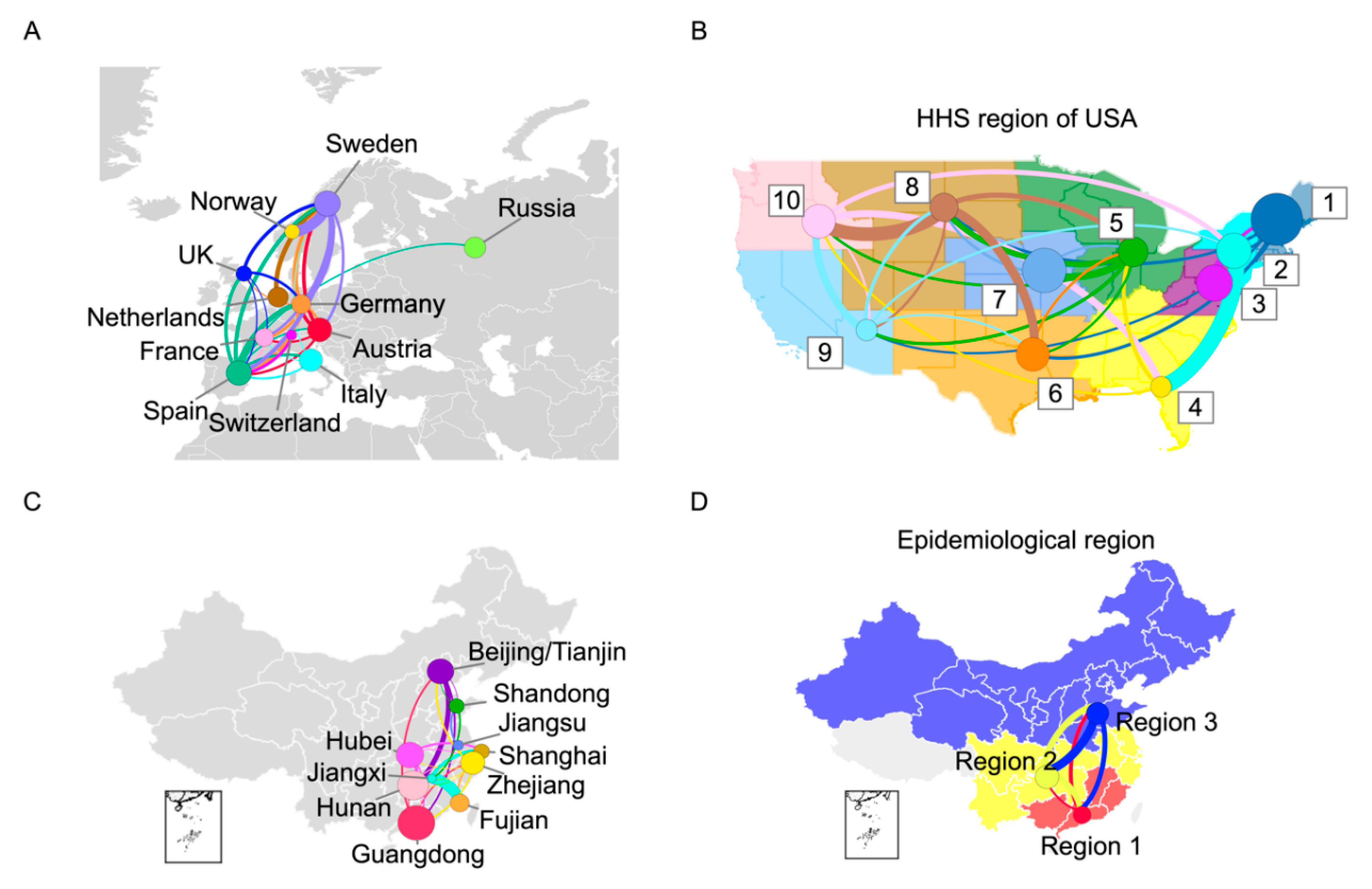
3.4. Transmission Analysis for Europe, the United States, and China
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| WHO | World Health Organization |
| HA | hemagglutinin |
| GISAID | Global Avian Influenza Data Sharing Initiative |
| HHS | US Department of Health and Human Services |
| BC | betweenness centrality |
| SARS-CoV-2 | the 2019 novel coronavirus |
References
- World Health Organization (2018) Fact sheet Number 211. Available online: http://www.who.int/mediacentre/factsheets/fs211/en/ (accessed on 6 November 2018).
- Nelson, M.; Holmes, E. The evolution of epidemic influenza. Nat. Rev. Genet. 2007, 8, 196–205. [Google Scholar] [CrossRef] [PubMed]
- Russell, C.A.; Jones, T.C.; Barr, I.G.; Cox, N.J.; Garten, R.J.; Gregory, V.; Gust, I.D.; Hampson, A.W.; Hay, A.J.; Hurt, A.C.; et al. The global circulation of seasonal influenza A (H3N2) viruses. Science 2008, 320, 340–346. [Google Scholar] [CrossRef] [PubMed]
- Cox, N.J.; Brammer, T.L.; Regnery, H.L. Influenza: Global surveillance for epidemic and pandemic variants. Eur. J. Epidemiol. 1994, 10, 467–470. [Google Scholar] [CrossRef] [PubMed]
- Chan, J.; Holmes, A.; Rabadan, R. Network Analysis of Global Influenza Spread. PLoS Comput. Biol. 2010, 6. [Google Scholar] [CrossRef] [PubMed]
- Lemey, P.; Rambaut, A.; Bedford, T.; Faria, N.; Bielejec, F.; Baele, G.; Russell, C.A.; Smith, D.J.; Pybus, O.G.; Brockmann, D.; et al. Unifying viral genetics and human transportation data to predict the global transmission dynamics of human influenza H3N2. PLoS Pathog 2014, 10, e1003932. [Google Scholar] [CrossRef]
- Bedford, T.; Riley, S.; Barr, I.G.; Broor, S.; Chadha, M.; Cox, N.J.; Daniels, R.S.; Gunasekaran, C.P.; Hurt, A.C.; Kelso, A.; et al. Global circulation patterns of seasonal influenza viruses vary with antigenic drift. Nature 2015, 523, 217–220. [Google Scholar] [CrossRef]
- Venton, D. Phylogeography. Proc. Natl. Acad. Sci. USA 2013, 110, 4158. [Google Scholar] [CrossRef]
- Bielejec, F.; Rambaut, A.; Suchard, M.A.; Lemey, P. SPREAD: Spatial phylogenetic reconstruction of evolutionary dynamics. Bioinformatics 2011, 27, 2910–2912. [Google Scholar] [CrossRef]
- Riddle, B.; Hafner, D. 7 Phylogeography in Historical Biogeography: Investigating the Biogeographic Histories of Populations, Species, and Young Biotas. Biogeogr. Chang. World 2007, 1, 161–176. [Google Scholar]
- Kunin, V.; Goldovsky, L.; Darzentas, N.; Ouzounis, C.A. The net of life: Reconstructing the microbial phylogenetic network. Genome Res. 2005, 15, 954–959. [Google Scholar] [CrossRef]
- Derrible, S.; Kennedy, C. Applications of Graph Theory and Network Science to Transit Network Design. Transport. Rev. 2011, 31, 495–519. [Google Scholar] [CrossRef]
- Wedgwood, K.C.A.; Satin, L.S. Six degrees of depolarization: Comment on "Network science of biological systems at different scales: A review" by Marko Gosak et al. Phys. Life Rev. 2018, 24, 136–139. [Google Scholar] [CrossRef] [PubMed]
- Bandelt, H.J.; Forster, P.; Rohl, A. Median-joining networks for inferring intraspecific phylogenies. Mol. Biol. Evol. 1999, 16, 37–48. [Google Scholar] [CrossRef] [PubMed]
- Campbell, E.M.; Jia, H.W.; Shankar, A.; Hanson, D.; Luo, W.; Masciotra, S.; Owen, S.M.; Oster, A.M.; Galang, R.R.; Spiller, M.W.; et al. Detailed Transmission Network Analysis of a Large Opiate-Driven Outbreak of HIV Infection In the United States. J. Infect. Dis. 2017, 216, 1053–1062. [Google Scholar] [CrossRef]
- de Bernardi Schneider, A.; Ford, C.T.; Hostager, R.; Williams, J.; Cioce, M.; Çatalyürek, Ü.V.; Wertheim, J.O.; Janies, D. StrainHub: A phylogenetic tool to construct pathogen transmission networks. Bioinformatics 2020, 36, 945–947. [Google Scholar] [CrossRef]
- Legendre, P.; Makarenkov, V. Reconstruction of biogeographic and evolutionary networks using reticulograms. Syst. Biol. 2002, 51, 199–216. [Google Scholar] [CrossRef]
- Huson, D.H.; Scornavacca, C. A Survey of Combinatorial Methods for Phylogenetic Networks. Genome Biol. Evol. 2010, 3, 23–35. [Google Scholar] [CrossRef]
- Yang, Z. Estimating the pattern of nucleotide substitution. J. Mol. Evol. 1994, 39, 105–111. [Google Scholar] [CrossRef]
- Lofgren, E.; Fefferman, N.; Naumov, Y.; Gorski, J.; Naumova, E. Influenza Seasonality: Underlying Causes and Modeling Theories. J. Virol. 2007, 81, 5429–5436. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree 2--approximately maximum-likelihood trees for large alignments. PLoS One 2010, 5, e9490. [Google Scholar] [CrossRef] [PubMed]
- Brin, S.; Page, L. The anatomy of a large-scale hypertextual Web search engine. Comput. Netw. ISDN Syst. 1998, 30, 107–117. [Google Scholar] [CrossRef]
- Bastian, M.; Heymann, S.; Jacomy, M. Gephi: An Open Source Software for Exploring and Manipulating Networks. ICWSM 2009, 8, 361–362. [Google Scholar]
- Barabasi, A.-L. Scale-Free Networks: A Decade and Beyond. Science 2009, 325, 412–413. [Google Scholar] [CrossRef]
- Thiemann, C.; Theis, F.; Grady, D.; Brune, R.; Brockmann, D. The structure of borders in a small world. PLoS One 2010, 5, e15422. [Google Scholar] [CrossRef]
- Yu, H.J.; Alonso, W.J.; Feng, L.Z.; Tan, Y.; Shu, Y.L.; Yang, W.Z.; Viboud, C. Characterization of Regional Influenza Seasonality Patterns in China and Implications for Vaccination Strategies: Spatio-Temporal Modeling of Surveillance Data. PLoS Med. 2013, 10. [Google Scholar] [CrossRef]
- Charu, V.; Zeger, S.; Gog, J.; Bjørnstad, O.N.; Kissler, S.; Simonsen, L.; Grenfell, B.T.; Viboud, C. Human mobility and the spatial transmission of influenza in the United States. PLoS Comput. Biol. 2017, 13, e1005382. [Google Scholar] [CrossRef]
- Zinder, D.; Bedford, T.; Baskerville, E.B.; Woods, R.J.; Roy, M.; Pascual, M. Seasonality in the migration and establishment of H3N2 Influenza lineages with epidemic growth and decline. Bmc Evol. Biol. 2014, 14. [Google Scholar] [CrossRef][Green Version]
- Du, X.J.; Dong, L.B.; Lan, Y.; Peng, Y.S.; Wu, A.P.; Zhang, Y.; Huang, W.J.; Wang, D.Y.; Wang, M.; Guo, Y.J.; et al. Mapping of H3N2 influenza antigenic evolution in China reveals a strategy for vaccine strain recommendation. Nat. Commun. 2012, 3. [Google Scholar] [CrossRef]
- Caini, S.; Alonso, W.J.; Séblain, C.E.-G.; Schellevis, F.; Paget, J. The spatiotemporal characteristics of influenza A and B in the WHO European Region: Can one define influenza transmission zones in Europe? Eurosurveillance 2017, 22. [Google Scholar] [CrossRef]
- Paget, J.; Marquet, R.; Meijer, A.; van der Velden, K. Influenza activity in Europe during eight seasons (1999-2007): An evaluation of the indicators used to measure activity and an assessment of the timing, length and course of peak activity (spread) across Europe. BMC Infect. Dis 2007, 7, 141. [Google Scholar] [CrossRef] [PubMed]
- Forster, P.; Forster, L.; Renfrew, C.; Forster, M. Phylogenetic network analysis of SARS-CoV-2 genomes. Proc. Natl. Acad. Sci. USA 2020, 117, 9241–9243. [Google Scholar] [CrossRef] [PubMed]
- Kong, S.; Sánchez-Pacheco, S.J.; Murphy, R.W. On the use of median-joining networks in evolutionary biology. Cladistics 2016, 32, 691–699. [Google Scholar] [CrossRef]
- Wagner, A. A genotype network reveals homoplastic cycles of convergent evolution in influenza A (H3N2) haemagglutinin. Proc. R. Soc. B Biol. Sci. 2014, 281. [Google Scholar] [CrossRef]
- Wang, C.; Lyu, N.; Deng, L.; Wang, J.; Gu, W.; Ding, H.; Wu, Y.; Luo, J.; Wang, L.; Lyv, X.; et al. Evolutionary Pattern and Large-Scale Architecture of Mutation Networks of 2009 A (H1N1) Influenza A Virus. Front. Genet. 2018, 9, 204. [Google Scholar] [CrossRef]
- Dolev, S.; Elovici, Y.; Puzis, R. Routing Betweenness Centrality. J. ACM 2010, 57. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, C.; Wang, Y.; Chen, C.; Long, H.; Bai, J.; Zeng, J.; Cao, Z.; Zhang, B.; Shen, W.; Tang, F.; et al. A Mutation Network Method for Transmission Analysis of Human Influenza H3N2. Viruses 2020, 12, 1125. https://doi.org/10.3390/v12101125
Zhang C, Wang Y, Chen C, Long H, Bai J, Zeng J, Cao Z, Zhang B, Shen W, Tang F, et al. A Mutation Network Method for Transmission Analysis of Human Influenza H3N2. Viruses. 2020; 12(10):1125. https://doi.org/10.3390/v12101125
Chicago/Turabian StyleZhang, Chi, Yinghan Wang, Cai Chen, Haoyu Long, Junbo Bai, Jinfeng Zeng, Zicheng Cao, Bing Zhang, Wei Shen, Feng Tang, and et al. 2020. "A Mutation Network Method for Transmission Analysis of Human Influenza H3N2" Viruses 12, no. 10: 1125. https://doi.org/10.3390/v12101125
APA StyleZhang, C., Wang, Y., Chen, C., Long, H., Bai, J., Zeng, J., Cao, Z., Zhang, B., Shen, W., Tang, F., Liang, S., Sun, C., Shu, Y., & Du, X. (2020). A Mutation Network Method for Transmission Analysis of Human Influenza H3N2. Viruses, 12(10), 1125. https://doi.org/10.3390/v12101125





