Characterization of a DCL2-Insensitive Tomato Bushy Stunt Virus Isolate Infecting Arabidopsis thaliana
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material
2.2. Virus Infection and Transgene Transient Expression
2.3. Virus Isolation and HTS Sequence Analyses
2.4. Peroxisome Isolation
2.5. Immunoprecipitation
2.6. Protein Extraction and Analysis
2.7. RNA Extraction and Analysis
2.8. Mass Spectrometry Protein Analysis
2.9. Fluorescence Microscopy
2.10. Electron Microscopy
3. Results
3.1. Characterization of a Tomato Bushy Stunt Virus (TBSV) Isolate Infecting Arabidopsis thaliana
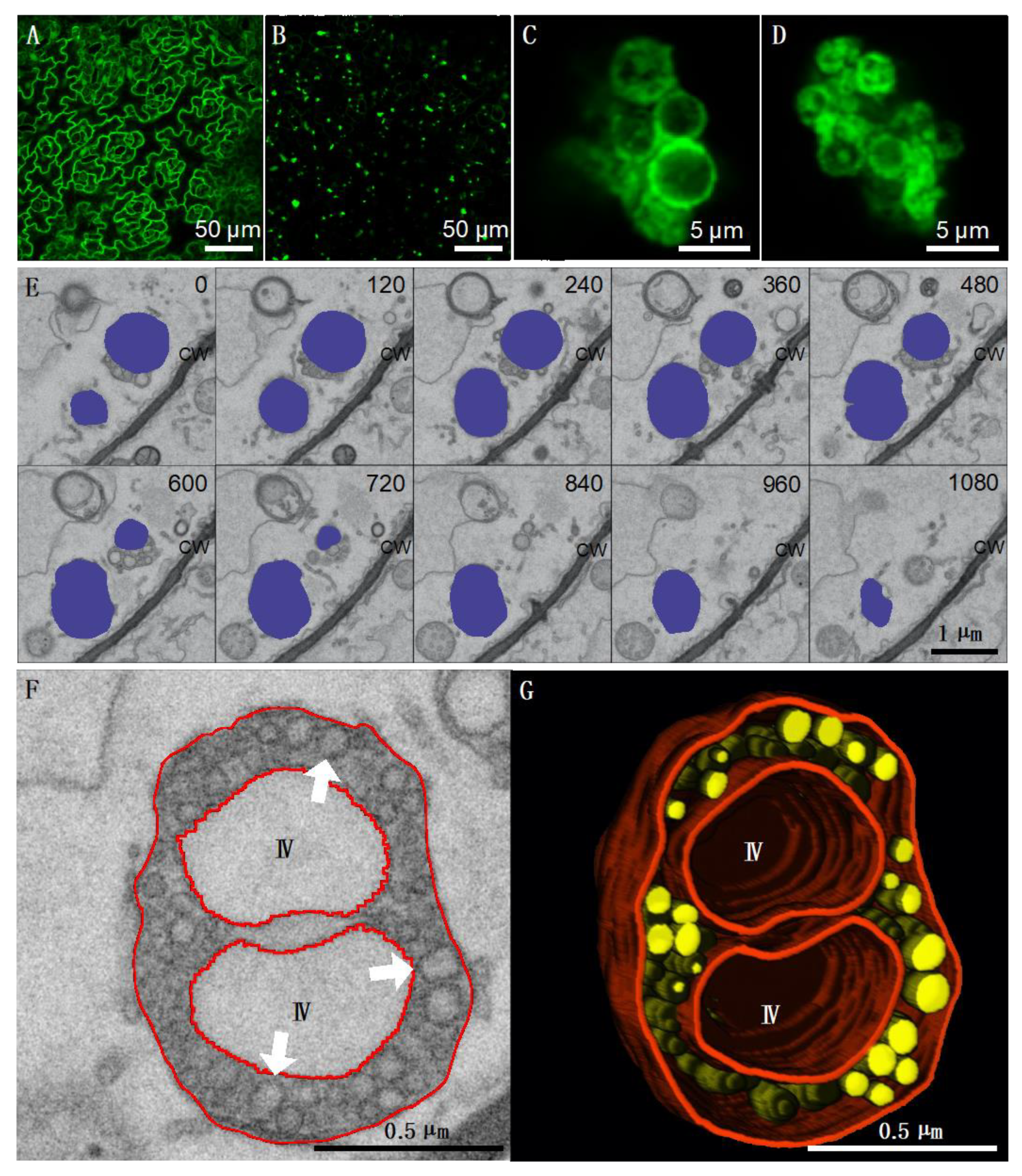
3.2. 3D electron Microscopy of TBSV Replication Vesicles on Peroxisomes from Infected Plants
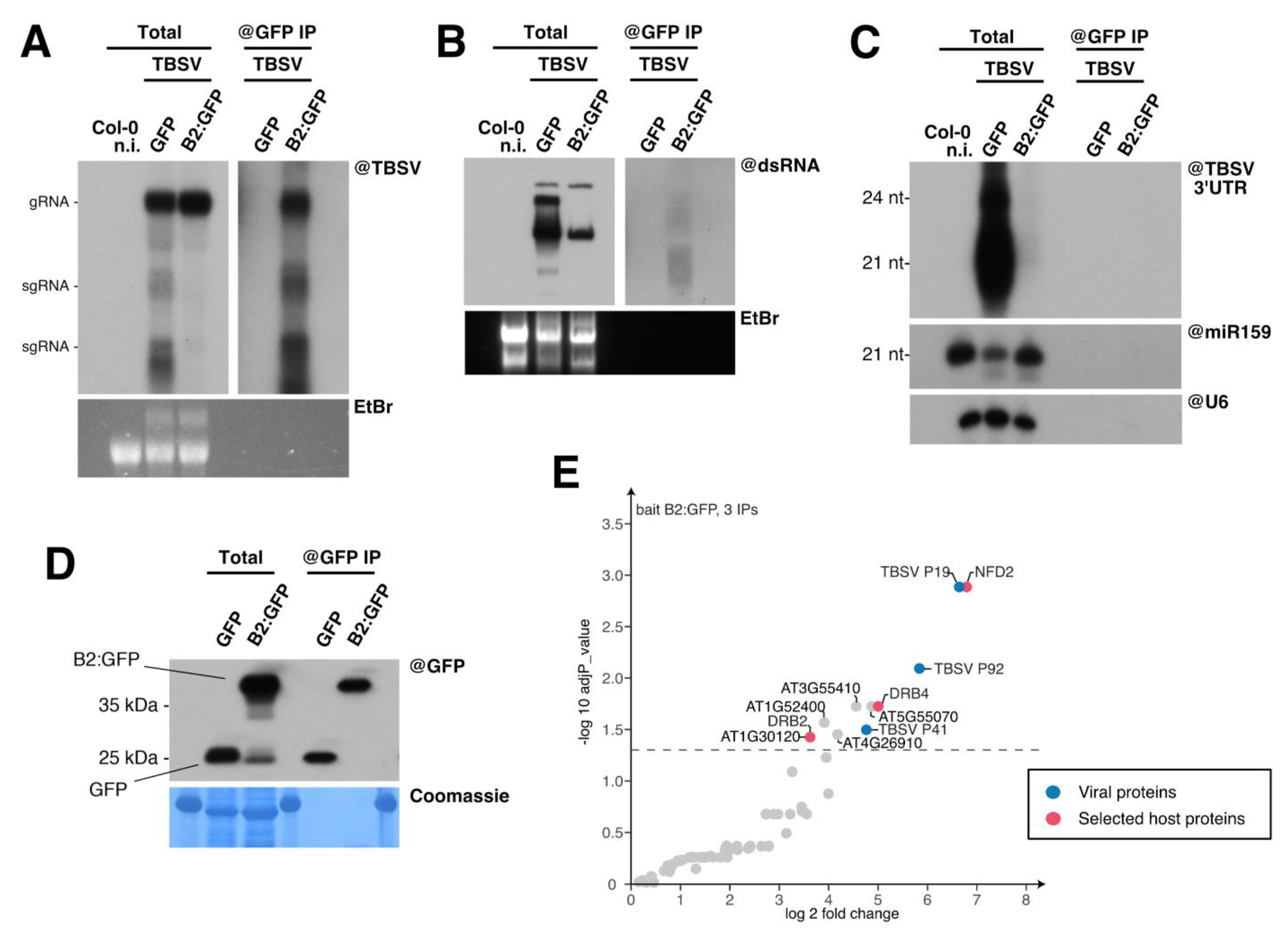
3.3. Immunoprecipitation of TBSV Replication Complexes
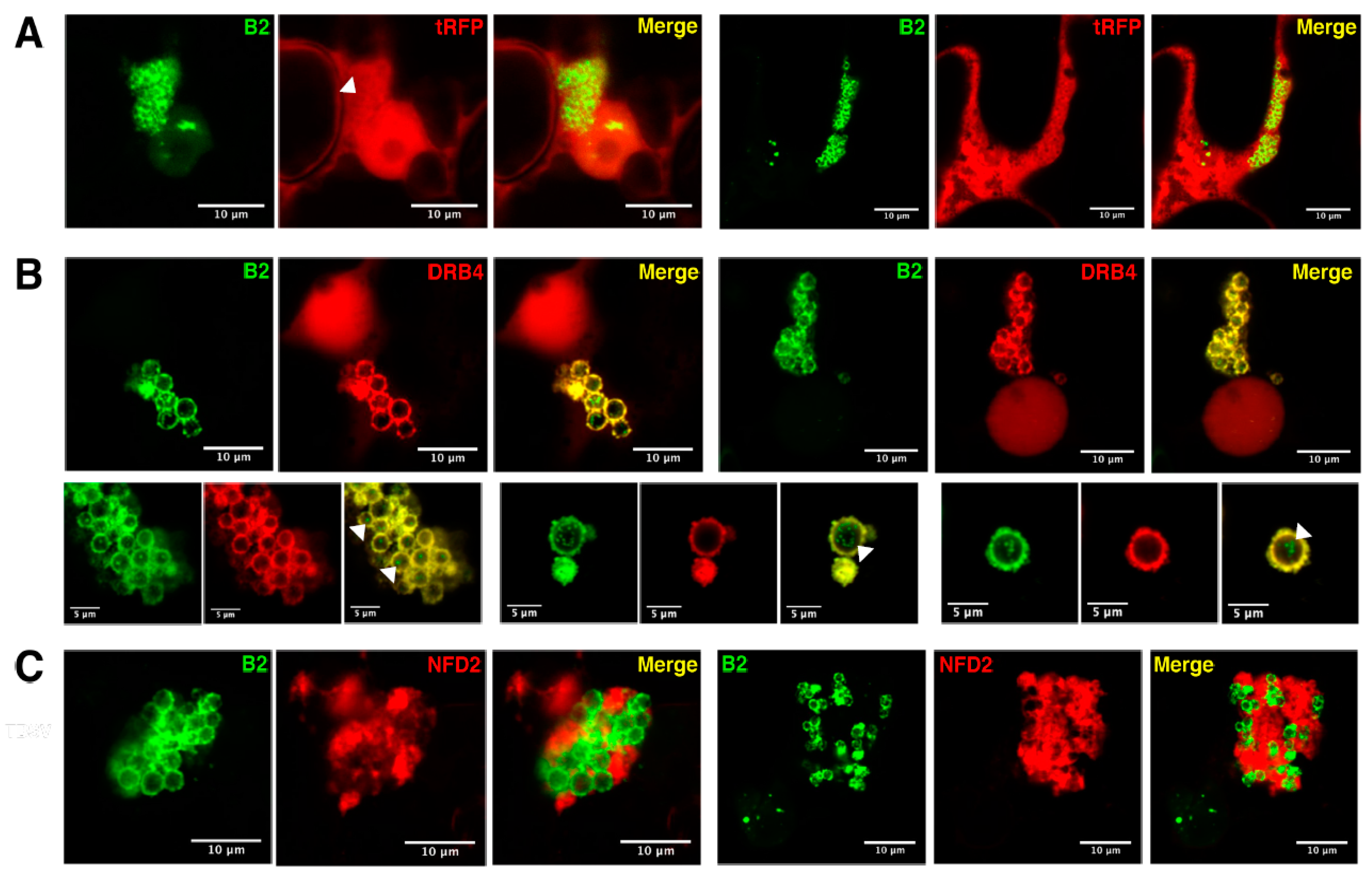
3.4. Host Double-Stranded RNA-Binding Proteins Associate to TBSV Replication Complexes
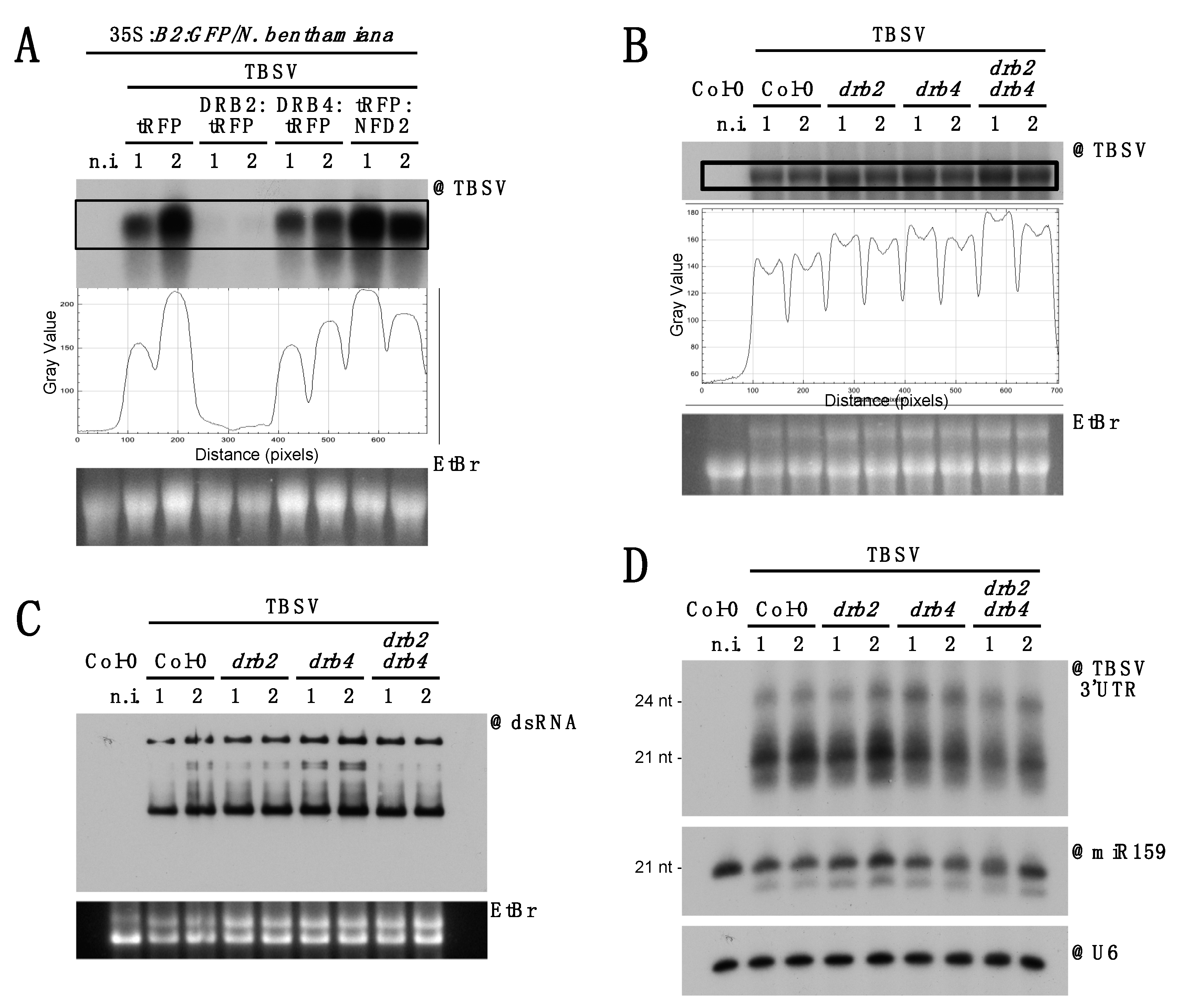
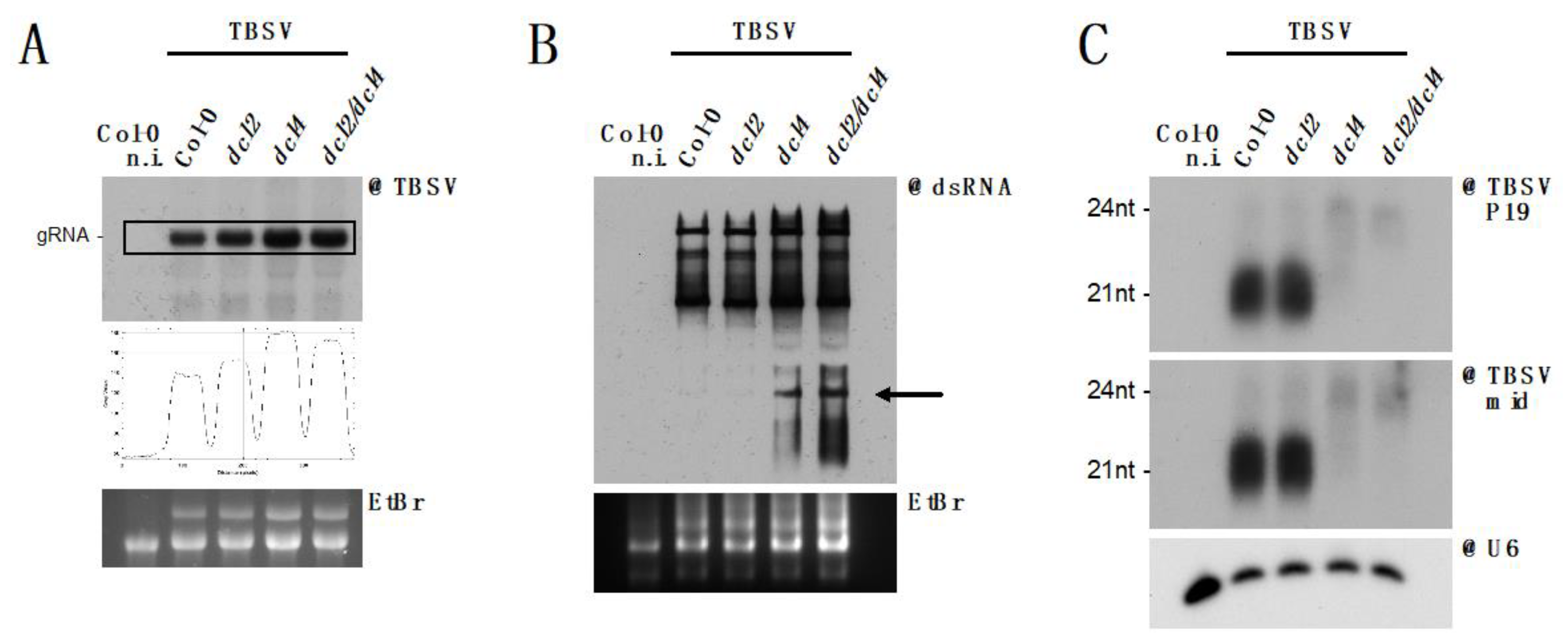
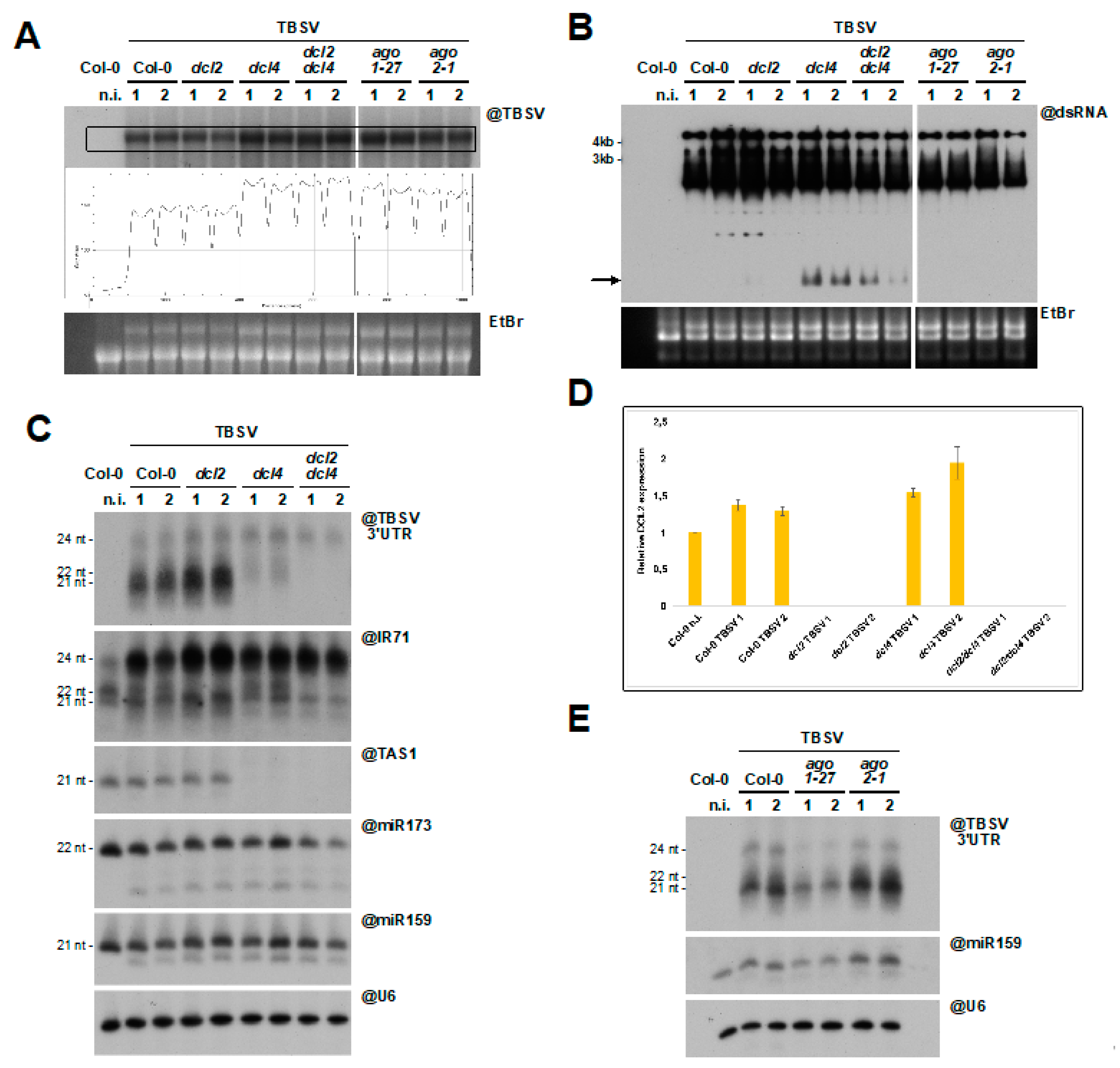
3.5. DCL2 Does not Effectively Process TBSV dsRNA
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Smith, K.M.; Bald, J. A description of a necrotic virus disease affecting tobacco and other plants. Parasitology 1935, 27, 231–245. [Google Scholar] [CrossRef]
- Martelli, G.; Gallitelli, D.; Russo, M. Tombusviruses. In The Plant Viruses; Springer: New York, NY, USA, 1988; pp. 13–72. [Google Scholar]
- Bally, J.; Jung, H.; Mortimer, C.; Naim, F.; Philips, J.G.; Hellens, R.; Bombarely, A.; Goodin, M.M.; Waterhouse, P.M. The Rise and Rise of Nicotiana benthamiana: A Plant for All Reasons. Annu. Rev. Phytopathol. 2018, 56, 405–426. [Google Scholar] [CrossRef] [PubMed]
- Bally, J.; Nakasugi, K.; Jia, F.; Jung, H.; Ho, S.Y.; Wong, M.; Paul, C.M.; Naim, F.; Wood, C.C.; Crowhurst, R.N.; et al. The extremophile Nicotiana benthamiana has traded viral defence for early vigour. Nat. Plants 2015, 1, 15165. [Google Scholar] [CrossRef] [PubMed]
- Provart, N.J.; Alonso, J.; Assmann, S.M.; Bergmann, D.; Brady, S.M.; Brkljacic, J.; Browse, J.; Chapple, C.; Colot, V.; Cutler, S.; et al. 50 years of Arabidopsis research: Highlights and future directions. New Phytol. 2016, 209, 921–944. [Google Scholar] [CrossRef]
- Russo, M.; Burgyan, J.; Martelli, G.P. Molecular biology of Tombusviridae. In Advances in Virus Research; Elsevier: Amsterdam, The Netherlands, 1994; Volume 44, pp. 381–428. [Google Scholar]
- Harrison, S.; Olson, A.; Schutt, C.; Winkler, F.; Bricogne, G. Tomato bushy stunt virus at 2.9 Å resolution. Nature 1978, 276, 368–373. [Google Scholar] [CrossRef]
- Olson, A.J.; Bricogne, G.; Harrison, S.C. Structure of tomato bushy stunt virus IV. J. Mol. Biol. 1983, 171, 61–93. [Google Scholar] [CrossRef]
- Hayes, R.J.; Brunt, A.A.; Buck, K.W. Gene-Mapping and Expression of Tomato Bushy Stunt Virus. J. Gen. Virol. 1988, 69, 3047–3057. [Google Scholar] [CrossRef]
- Scholthof, K.-B.G.; Scholthof, H.B.; Jackson, A. The tomato bushy stunt virus replicase proteins are coordinately expressed and membrane associated. Virology 1995, 208, 365–369. [Google Scholar] [CrossRef][Green Version]
- Nagy, P.D. Tombusvirus-Host Interactions: Co-Opted Evolutionarily Conserved Host Factors Take Center Court. Annu. Rev. Virol. 2016, 3, 491–515. [Google Scholar] [CrossRef]
- Zhang, Z.; He, G.; Filipowicz, N.A.; Randall, G.; Belov, G.A.; Kopek, B.G.; Wang, X. Host Lipids in Positive-Strand RNA Virus Genome Replication. Front. Microbiol. 2019, 10, 286. [Google Scholar] [CrossRef]
- Jiang, J.; Laliberté, J.-F. Membrane Association for Plant Virus Replication and Movement. In Current Research Topics in Plant Virology; Wang, A., Zhou, X., Eds.; Springer International Publishing: Cham, Switzerland, 2016; pp. 67–85. [Google Scholar]
- Fernandez de Castro, I.; Fernandez, J.J.; Barajas, D.; Nagy, P.D.; Risco, C. Three-dimensional imaging of the intracellular assembly of a functional viral RNA replicase complex. J. Cell Sci. 2017, 130, 260–268. [Google Scholar] [CrossRef]
- Nagy, P.D. Viral sensing of the subcellular environment regulates the assembly of new viral replicase complexes during the course of infection. J. Virol. 2015, 89, 5196–5199. [Google Scholar] [CrossRef]
- Qu, F.; Morris, T.J. Efficient infection of Nicotiana benthamiana by Tomato bushy stunt virus is facilitated by the coat protein and maintained by p19 through suppression of gene silencing. Mol. Plant Microbe Interact. 2002, 15, 193–202. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Scholthof, H.B. The Tombusvirus-encoded P19: From irrelevance to elegance. Nat. Rev. Microbiol. 2006, 4, 405–411. [Google Scholar] [CrossRef] [PubMed]
- Vargason, J.M.; Szittya, G.; Burgyan, J.; Hall, T.M. Size selective recognition of siRNA by an RNA silencing suppressor. Cell 2003, 115, 799–811. [Google Scholar] [CrossRef]
- Kontra, L.; Csorba, T.; Tavazza, M.; Lucioli, A.; Tavazza, R.; Moxon, S.; Tisza, V.; Medzihradszky, A.; Turina, M.; Burgyan, J. Distinct Effects of p19 RNA Silencing Suppressor on Small RNA Mediated Pathways in Plants. PLoS Pathog. 2016, 12, e1005935. [Google Scholar] [CrossRef]
- Scholthof, H.B. Plant virus transport: Motions of functional equivalence. Trends Plant Sci. 2005, 10, 376–382. [Google Scholar] [CrossRef] [PubMed]
- Zhao, R.Y. Yeast for virus research. Microb. Cell 2017, 4, 311. [Google Scholar] [CrossRef]
- Panavas, T.; Nagy, P.D. Yeast as a model host to study replication and recombination of defective interfering RNA of Tomato bushy stunt virus. Virology 2003, 314, 315–325. [Google Scholar] [CrossRef]
- Sasvari, Z.; Lin, W.; Inaba, J.I.; Xu, K.; Kovalev, N.; Nagy, P.D. Co-opted Cellular Sac1 Lipid Phosphatase and PI(4)P Phosphoinositide Are Key Host Factors during the Biogenesis of the Tombusvirus Replication Compartment. J. Virol. 2020, 94, JVI.01979-19. [Google Scholar] [CrossRef]
- Nagy, P.D.; Lin, W. Taking over Cellular Energy-Metabolism for TBSV Replication: The High ATP Requirement of an RNA Virus within the Viral Replication Organelle. Viruses 2020, 12, 56. [Google Scholar] [CrossRef] [PubMed]
- Nagy, P.D. Exploitation of a surrogate host, Saccharomyces cerevisiae, to identify cellular targets and develop novel antiviral approaches. Curr. Opin. Virol. 2017, 26, 132–140. [Google Scholar] [CrossRef]
- Wang, A. Dissecting the molecular network of virus-plant interactions: The complex roles of host factors. Annu. Rev. Phytopathol. 2015, 53, 45–66. [Google Scholar] [CrossRef] [PubMed]
- Incarbone, M.; Dunoyer, P. RNA silencing and its suppression: Novel insights from in planta analyses. Trends Plant Sci. 2013, 18, 382–392. [Google Scholar] [CrossRef] [PubMed]
- Csorba, T.; Kontra, L.; Burgyán, J. Viral silencing suppressors: Tools forged to fine-tune host-pathogen coexistence. Virology 2015, 479, 85–103. [Google Scholar] [CrossRef] [PubMed]
- Guo, Z.; Li, Y.; Ding, S.W. Small RNA-based antimicrobial immunity. Nat. Rev. Immunol. 2019, 19, 31–44. [Google Scholar] [CrossRef] [PubMed]
- Kovalev, N.; Inaba, J.I.; Li, Z.; Nagy, P.D. The role of co-opted ESCRT proteins and lipid factors in protection of tombusviral double-stranded RNA replication intermediate against reconstituted RNAi in yeast. PLoS Pathog. 2017, 13, e1006520. [Google Scholar] [CrossRef] [PubMed]
- Deleris, A.; Gallego-Bartolome, J.; Bao, J.; Kasschau, K.D.; Carrington, J.C.; Voinnet, O. Hierarchical action and inhibition of plant Dicer-like proteins in antiviral defense. Science 2006, 313, 68–71. [Google Scholar] [CrossRef]
- Cao, M.; Du, P.; Wang, X.; Yu, Y.Q.; Qiu, Y.H.; Li, W.; Gal-On, A.; Zhou, C.; Li, Y.; Ding, S.W. Virus infection triggers widespread silencing of host genes by a distinct class of endogenous siRNAs in Arabidopsis. Proc. Natl. Acad. Sci. USA 2014, 111, 14613–14618. [Google Scholar] [CrossRef]
- Garcia-Ruiz, H.; Takeda, A.; Chapman, E.J.; Sullivan, C.M.; Fahlgren, N.; Brempelis, K.J.; Carrington, J.C. Arabidopsis RNA-dependent RNA polymerases and dicer-like proteins in antiviral defense and small interfering RNA biogenesis during Turnip Mosaic Virus infection. Plant Cell 2010, 22, 481–496. [Google Scholar] [CrossRef]
- Incarbone, M.; Zimmermann, A.; Hammann, P.; Erhardt, M.; Michel, F.; Dunoyer, P. Neutralization of mobile antiviral small RNA through peroxisomal import. Nat. Plants 2017, 3, 17094. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.B.; Jovel, J.; Udomporn, P.; Wang, Y.; Wu, Q.; Li, W.X.; Gasciolli, V.; Vaucheret, H.; Ding, S.W. The 21-nucleotide, but not 22-nucleotide, viral secondary small interfering RNAs direct potent antiviral defense by two cooperative argonautes in Arabidopsis thaliana. Plant Cell 2011, 23, 1625–1638. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Ruiz, H.; Carbonell, A.; Hoyer, J.S.; Fahlgren, N.; Gilbert, K.B.; Takeda, A.; Giampetruzzi, A.; Garcia Ruiz, M.T.; McGinn, M.G.; Lowery, N.; et al. Roles and programming of Arabidopsis ARGONAUTE proteins during Turnip mosaic virus infection. PLoS Pathog. 2015, 11, e1004755. [Google Scholar] [CrossRef] [PubMed]
- Brosseau, C.; Bolaji, A.; Roussin-Léveillée, C.; Zhao, Z.; Biga, S.; Moffett, P. Natural variation in the Arabidopsis AGO2 gene is associated with susceptibility to potato virus X. New Phytol. 2020, 226, 866–878. [Google Scholar] [CrossRef]
- Lakatos, L.; Csorba, T.; Pantaleo, V.; Chapman, E.J.; Carrington, J.C.; Liu, Y.P.; Dolja, V.V.; Calvino, L.F.; Lopez-Moya, J.J.; Burgyan, J. Small RNA binding is a common strategy to suppress RNA silencing by several viral suppressors. EMBO J. 2006, 25, 2768–2780. [Google Scholar] [CrossRef]
- Incarbone, M.; Ritzenthaler, C.; Dunoyer, P. Peroxisomal Targeting as a Sensitive Tool to Detect Protein-Small RNA Interactions through in Vivo Piggybacking. Front. Plant Sci. 2018, 9, 135. [Google Scholar] [CrossRef]
- Ye, K.; Malinina, L.; Patel, D.J. Recognition of small interfering RNA by a viral suppressor of RNA silencing. Nature 2003, 426, 874–878. [Google Scholar] [CrossRef]
- Lakatos, L.; Szittya, G.; Silhavy, D.; Burgyan, J. Molecular mechanism of RNA silencing suppression mediated by p19 protein of tombusviruses. EMBO J. 2004, 23, 876–884. [Google Scholar] [CrossRef]
- Curtin, S.J.; Watson, J.M.; Smith, N.A.; Eamens, A.L.; Blanchard, C.L.; Waterhouse, P.M. The roles of plant dsRNA-binding proteins in RNAi-like pathways. FEBS Lett. 2008, 582, 2753–2760. [Google Scholar] [CrossRef]
- Xie, Z.; Allen, E.; Wilken, A.; Carrington, J.C. DICER-LIKE 4 functions in trans-acting small interfering RNA biogenesis and vegetative phase change in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2005, 102, 12984–12989. [Google Scholar] [CrossRef]
- Xie, Z.; Johansen, L.K.; Gustafson, A.M.; Kasschau, K.D.; Lellis, A.D.; Zilberman, D.; Jacobsen, S.E.; Carrington, J.C. Genetic and functional diversification of small RNA pathways in plants. PLoS Biol. 2004, 2, e104. [Google Scholar] [CrossRef]
- Lobbes, D.; Rallapalli, G.; Schmidt, D.D.; Martin, C.; Clarke, J. SERRATE: A new player on the plant microRNA scene. EMBO Rep. 2006, 7, 1052–1058. [Google Scholar] [CrossRef]
- Incarbone, M.; Clavel, M.; Monsion, B.; Kuhn, L.; Scheer, H.; Poignavent, V.; Dunoyer, P.; Genschik, P.; Ritzenthaler, C. Immunocapture of dsRNA-bound proteins provides insight into tobacco rattle virus replication complexes and reveals Arabidopsis DRB2 to be a wide-spectrum antiviral effector. bioRxiv 2020, 2020, 842666. [Google Scholar]
- Monsion, B.; Incarbone, M.; Hleibieh, K.; Poignavent, V.; Ghannam, A.; Dunoyer, P.; Daeffler, L.; Tilsner, J.; Ritzenthaler, C. Efficient Detection of Long dsRNA in Vitro and in Vivo Using the dsRNA Binding Domain from FHV B2 Protein. Front. Plant Sci. 2018, 9, 70. [Google Scholar] [CrossRef] [PubMed]
- Galetzka, D.; Russo, M.; Rubino, L.; Krczal, G. Molecular characterization of a Tombusvirus associated with a disease of statice [Goniolimon tataricum (L.) boiss.]. J. Plant Pathol. 2000, 82, 151–155. [Google Scholar]
- Burgyán, J.; Russo, M. Tombusvirus isolation and RNA extraction. In Plant Virology Protocols; Springer: New York, NY, USA, 1998; pp. 225–230. [Google Scholar]
- Hily, J.M.; Demaneche, S.; Poulicard, N.; Tannieres, M.; Djennane, S.; Beuve, M.; Vigne, E.; Demangeat, G.; Komar, V.; Gertz, C.; et al. Metagenomic-based impact study of transgenic grapevine rootstock on its associated virome and soil bacteriome. Plant Biotechnol. J. 2018, 16, 208–220. [Google Scholar] [CrossRef] [PubMed]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Reumann, S.; Singhal, R. Isolation of leaf peroxisomes from Arabidopsis for organelle proteome analyses. In Plant Proteomics; Springer: New York, NY, USA, 2014; pp. 541–552. [Google Scholar]
- Incarbone, M.; Ritzenthaler, C. Double-stranded RNA pull-down to characterize viral replication complexes in plants. In RNA Tagging; Heinlein, M., Ed.; Springer: New York, NY, USA, 2020. [Google Scholar]
- Chicher, J.; Simonetti, A.; Kuhn, L.; Schaeffer, L.; Hammann, P.; Eriani, G.; Martin, F. Purification of mRNA-programmed translation initiation complexes suitable for mass spectrometry analysis. Proteomics 2015, 15, 2417–2425. [Google Scholar] [CrossRef]
- Lange, H.; Ndecky, S.Y.A.; Gomez-Diaz, C.; Pflieger, D.; Butel, N.; Zumsteg, J.; Kuhn, L.; Piermaria, C.; Chicher, J.; Christie, M.; et al. RST1 and RIPR connect the cytosolic RNA exosome to the Ski complex in Arabidopsis. Nat. Commun. 2019, 10, 3871. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [PubMed]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef]
- Cardona, A.; Saalfeld, S.; Schindelin, J.; Arganda-Carreras, I.; Preibisch, S.; Longair, M.; Tomancak, P.; Hartenstein, V.; Douglas, R.J. TrakEM2 software for neural circuit reconstruction. PLoS ONE 2012, 7, e38011. [Google Scholar] [CrossRef] [PubMed]
- Ohki, T.; Uematsu, S.; Lesemann, D.-E.; Honda, Y.; Tsuda, S.; Fujisawa, I. Characterization of Tomato bushy stunt virus newly isolated from nipplefruit (Solanum mammosum) in Japan. J. Gen. Plant Pathol. 2005, 71, 74–79. [Google Scholar] [CrossRef]
- Nagy, P.D.; Strating, J.R.; van Kuppeveld, F.J. Building Viral Replication Organelles: Close Encounters of the Membrane Types. PLoS Pathog. 2016, 12, e1005912. [Google Scholar] [CrossRef] [PubMed]
- Romero-Brey, I.; Bartenschlager, R. Membranous replication factories induced by plus-strand RNA viruses. Viruses 2014, 6, 2826–2857. [Google Scholar] [CrossRef]
- den Boon, J.A.; Diaz, A.; Ahlquist, P. Cytoplasmic viral replication complexes. Cell Host Microbe 2010, 8, 77–85. [Google Scholar] [CrossRef] [PubMed]
- Russo, M.; Martelli, G.P. Ultrastructural observations on tomato bushy stunt virus in plant cells. Virology 1972, 49, 122–129. [Google Scholar] [CrossRef]
- Rubino, L.; Russo, M.; Martelli, G. Tombusvirus-induced multivesicular bodies: Origin and role in virus–host interaction. In Plant Virus–Host Interaction; Elsevier: Amsterdam, The Netherlands, 2014; pp. 163–175. [Google Scholar]
- Jakubiec, A.; Yang, S.W.; Chua, N.H. Arabidopsis DRB4 protein in antiviral defense against Turnip yellow mosaic virus infection. Plant J. 2012, 69, 14–25. [Google Scholar] [CrossRef]
- Elvira-Matelot, E.; Hachet, M.; Shamandi, N.; Comella, P.; Saez-Vasquez, J.; Zytnicki, M.; Vaucheret, H. Arabidopsis RNASE THREE LIKE2 Modulates the Expression of Protein-Coding Genes via 24-Nucleotide Small Interfering RNA-Directed DNA Methylation. Plant Cell 2016, 28, 406–425. [Google Scholar] [CrossRef]
- Portereiko, M.F.; Sandaklie-Nikolova, L.; Lloyd, A.; Dever, C.A.; Otsuga, D.; Drews, G.N. NUCLEAR FUSION DEFECTIVE1 encodes the Arabidopsis RPL21M protein and is required for karyogamy during female gametophyte development and fertilization. Plant Physiol. 2006, 141, 957–965. [Google Scholar] [CrossRef] [PubMed]
- Fukudome, A.; Kanaya, A.; Egami, M.; Nakazawa, Y.; Hiraguri, A.; Moriyama, H.; Fukuhara, T. Specific requirement of DRB4, a dsRNA-binding protein, for the in vitro dsRNA-cleaving activity of Arabidopsis Dicer-like 4. RNA 2011, 17, 750–760. [Google Scholar] [CrossRef] [PubMed]
- Qu, F.; Ye, X.; Morris, T.J. Arabidopsis DRB4, AGO1, AGO7, and RDR6 participate in a DCL4-initiated antiviral RNA silencing pathway negatively regulated by DCL1. Proc. Natl. Acad. Sci. USA 2008, 105, 14732–14737. [Google Scholar] [CrossRef] [PubMed]
- Drinnenberg, I.A.; Weinberg, D.E.; Xie, K.T.; Mower, J.P.; Wolfe, K.H.; Fink, G.R.; Bartel, D.P. RNAi in budding yeast. Science 2009, 326, 544–550. [Google Scholar] [CrossRef] [PubMed]
- Lewsey, M.G.; Carr, J.P. Effects of DICER-like proteins 2, 3 and 4 on cucumber mosaic virus and tobacco mosaic virus infections in salicylic acid-treated plants. J. Gen. Virol. 2009, 90 Pt 12, 3010–3014. [Google Scholar] [CrossRef]
- Carbonell, A.; Carrington, J.C. Antiviral roles of plant ARGONAUTES. Curr. Opin. Plant Biol. 2015, 27, 111–117. [Google Scholar] [CrossRef]
- Kasschau, K.D.; Fahlgren, N.; Chapman, E.J.; Sullivan, C.M.; Cumbie, J.S.; Givan, S.A.; Carrington, J.C. Genome-wide profiling and analysis of Arabidopsis siRNAs. PLoS Biol. 2007, 5, e57. [Google Scholar] [CrossRef]
- Montavon, T.; Kwon, Y.; Zimmermann, A.; Hammann, P.; Vincent, T.; Cognat, V.; Michel, F.; Dunoyer, P. A specific dsRNA-binding protein complex selectively sequesters endogenous inverted-repeat siRNA precursors and inhibits their processing. Nucleic Acids Res. 2017, 45, 1330–1344. [Google Scholar] [CrossRef]
- Pooggin, M.M. Small RNA-Omics for Plant Virus Identification, Virome Reconstruction, and Antiviral Defense Characterization. Front. Microbiol. 2018, 9, 2779. [Google Scholar] [CrossRef]
- Barton, D.A.; Roovers, E.F.; Gouil, Q.; da Fonseca, G.C.; Reis, R.S.; Jackson, C.; Overall, R.L.; Fusaro, A.F.; Waterhouse, P.M. Live Cell Imaging Reveals the Relocation of dsRNA Binding Proteins Upon Viral Infection. Mol. Plant Microbe Interact. 2017, 30, 435–443. [Google Scholar] [CrossRef]
- Fatyol, K.; Fekete, K.A.; Ludman, M. Double-Stranded-RNA-Binding Protein 2 Participates in Antiviral Defense. J. Virol. 2020, 94, JVI.00017-20. [Google Scholar] [CrossRef] [PubMed]
- Shamandi, N.; Zytnicki, M.; Charbonnel, C.; Elvira-Matelot, E.; Bochnakian, A.; Comella, P.; Mallory, A.C.; Lepere, G.; Saez-Vasquez, J.; Vaucheret, H. Plants Encode a General siRNA Suppressor That Is Induced and Suppressed by Viruses. PLoS Biol. 2015, 13, e1002326. [Google Scholar] [CrossRef] [PubMed]

© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Incarbone, M.; Scheer, H.; Hily, J.-M.; Kuhn, L.; Erhardt, M.; Dunoyer, P.; Altenbach, D.; Ritzenthaler, C. Characterization of a DCL2-Insensitive Tomato Bushy Stunt Virus Isolate Infecting Arabidopsis thaliana. Viruses 2020, 12, 1121. https://doi.org/10.3390/v12101121
Incarbone M, Scheer H, Hily J-M, Kuhn L, Erhardt M, Dunoyer P, Altenbach D, Ritzenthaler C. Characterization of a DCL2-Insensitive Tomato Bushy Stunt Virus Isolate Infecting Arabidopsis thaliana. Viruses. 2020; 12(10):1121. https://doi.org/10.3390/v12101121
Chicago/Turabian StyleIncarbone, Marco, Hélene Scheer, Jean-Michel Hily, Lauriane Kuhn, Mathieu Erhardt, Patrice Dunoyer, Denise Altenbach, and Christophe Ritzenthaler. 2020. "Characterization of a DCL2-Insensitive Tomato Bushy Stunt Virus Isolate Infecting Arabidopsis thaliana" Viruses 12, no. 10: 1121. https://doi.org/10.3390/v12101121
APA StyleIncarbone, M., Scheer, H., Hily, J.-M., Kuhn, L., Erhardt, M., Dunoyer, P., Altenbach, D., & Ritzenthaler, C. (2020). Characterization of a DCL2-Insensitive Tomato Bushy Stunt Virus Isolate Infecting Arabidopsis thaliana. Viruses, 12(10), 1121. https://doi.org/10.3390/v12101121





